Evaluating the Molecular Basis of Nanocalcium-Induced Health Regulation in Zebra Fish (Danio rerio)
Abstract
1. Introduction
2. Experimental Methods
2.1. CaO-NP Synthesis
2.2. Physiochemical Characterization of CaO-NPs
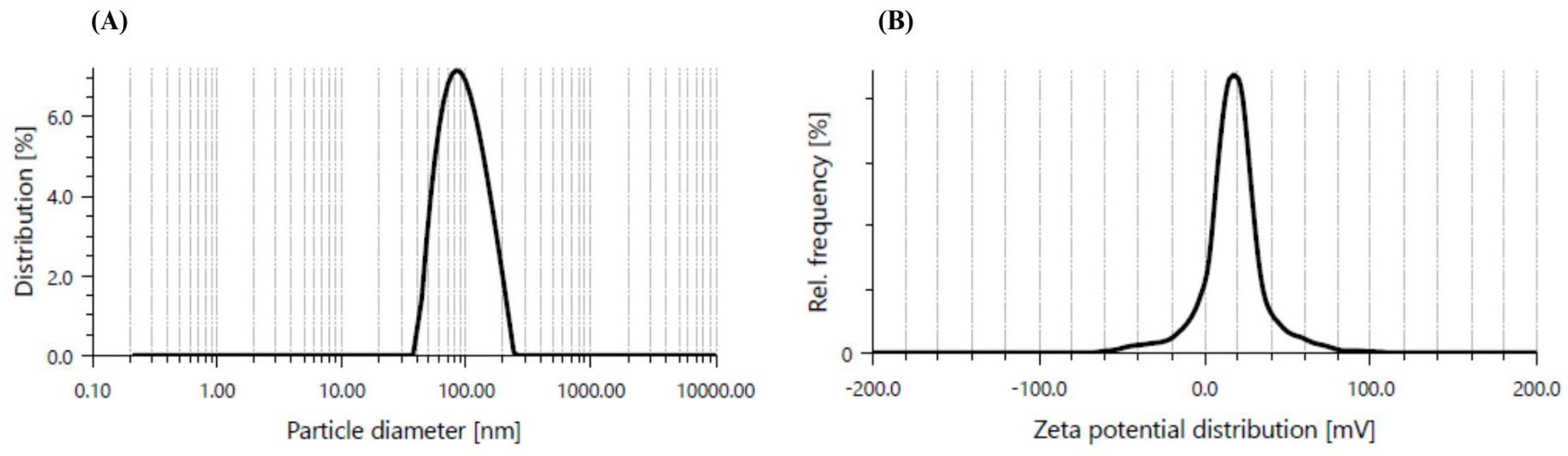
2.2.1. Dynamic Light Scattering (DLS)
2.2.2. X-Ray Diffraction (XRD) Analysis
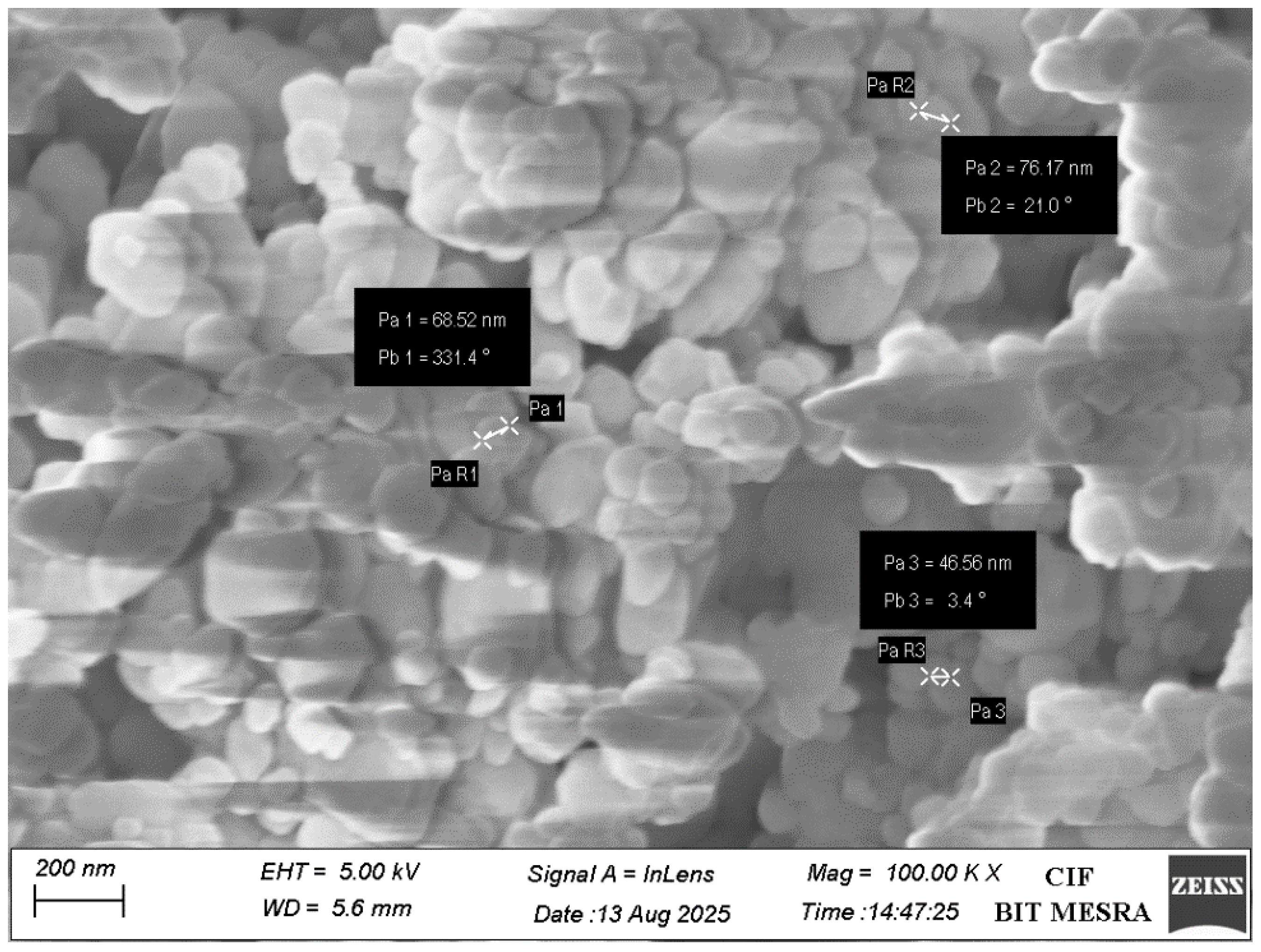
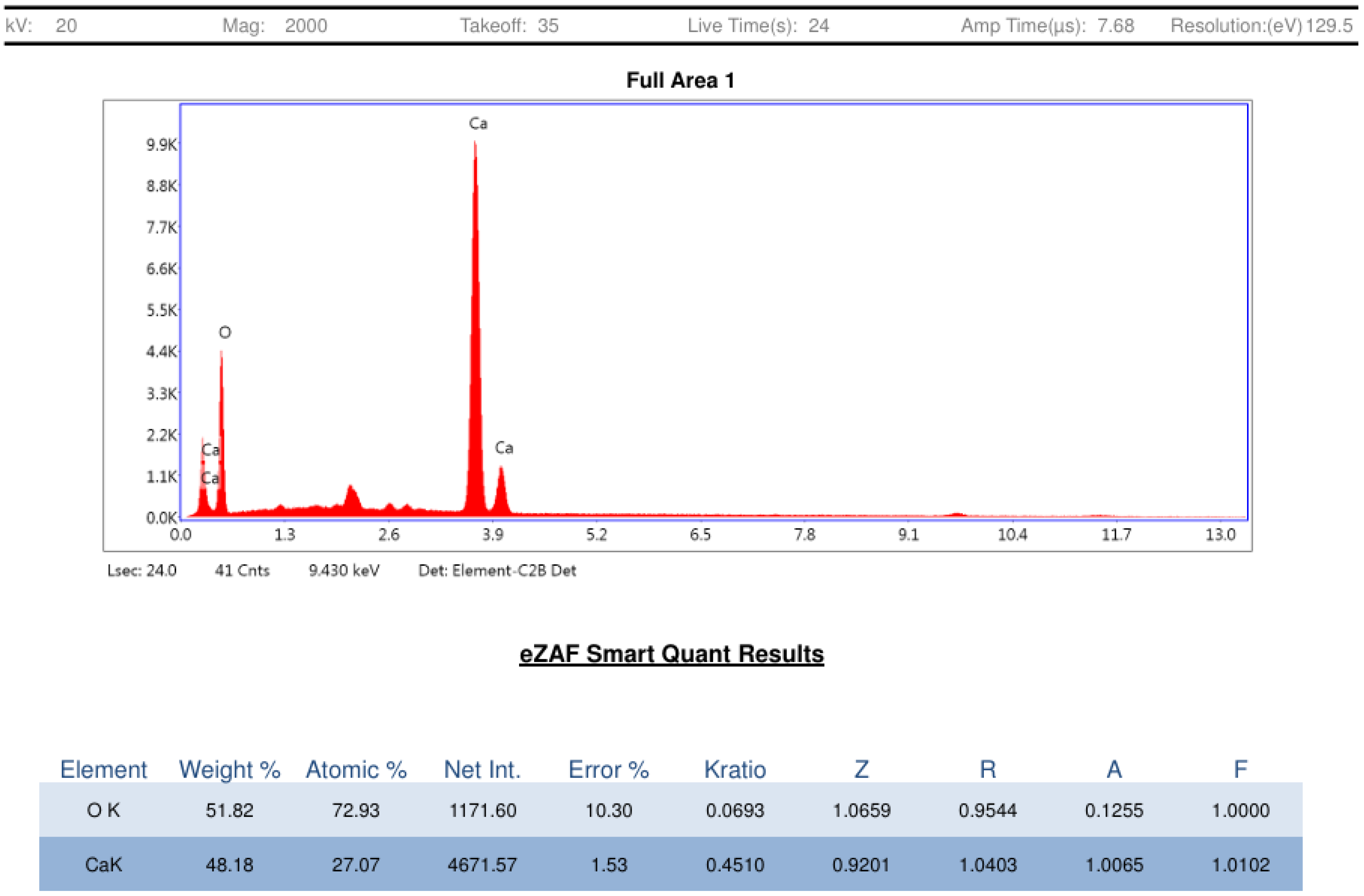
2.2.3. Field Emission Scanning Electron Microscopy (FESEM) and Energy-Dispersive Spectroscopy (EDS) Analysis
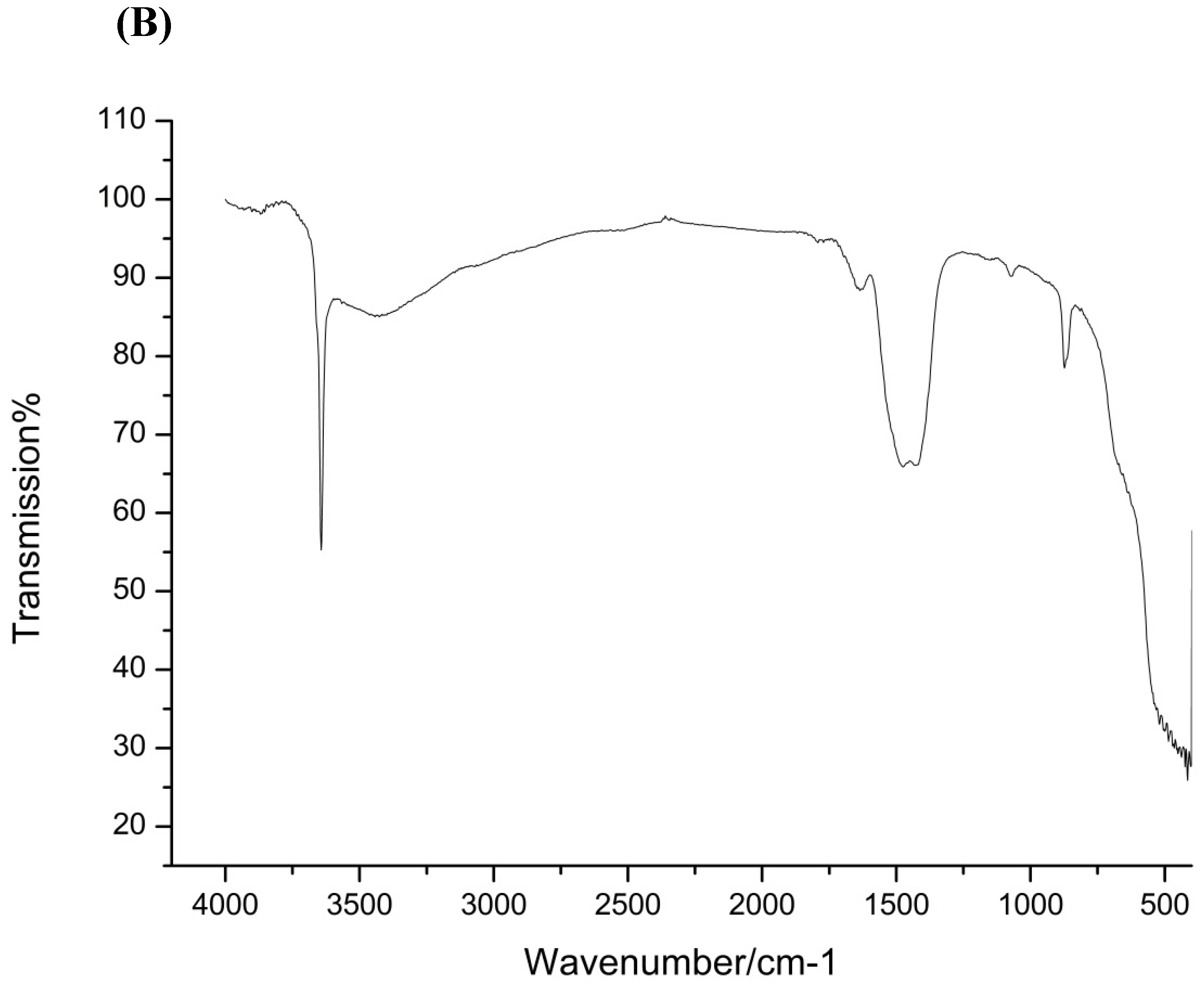
2.2.4. Fourier Transform Infrared Spectroscopy (FTIR) Analysis
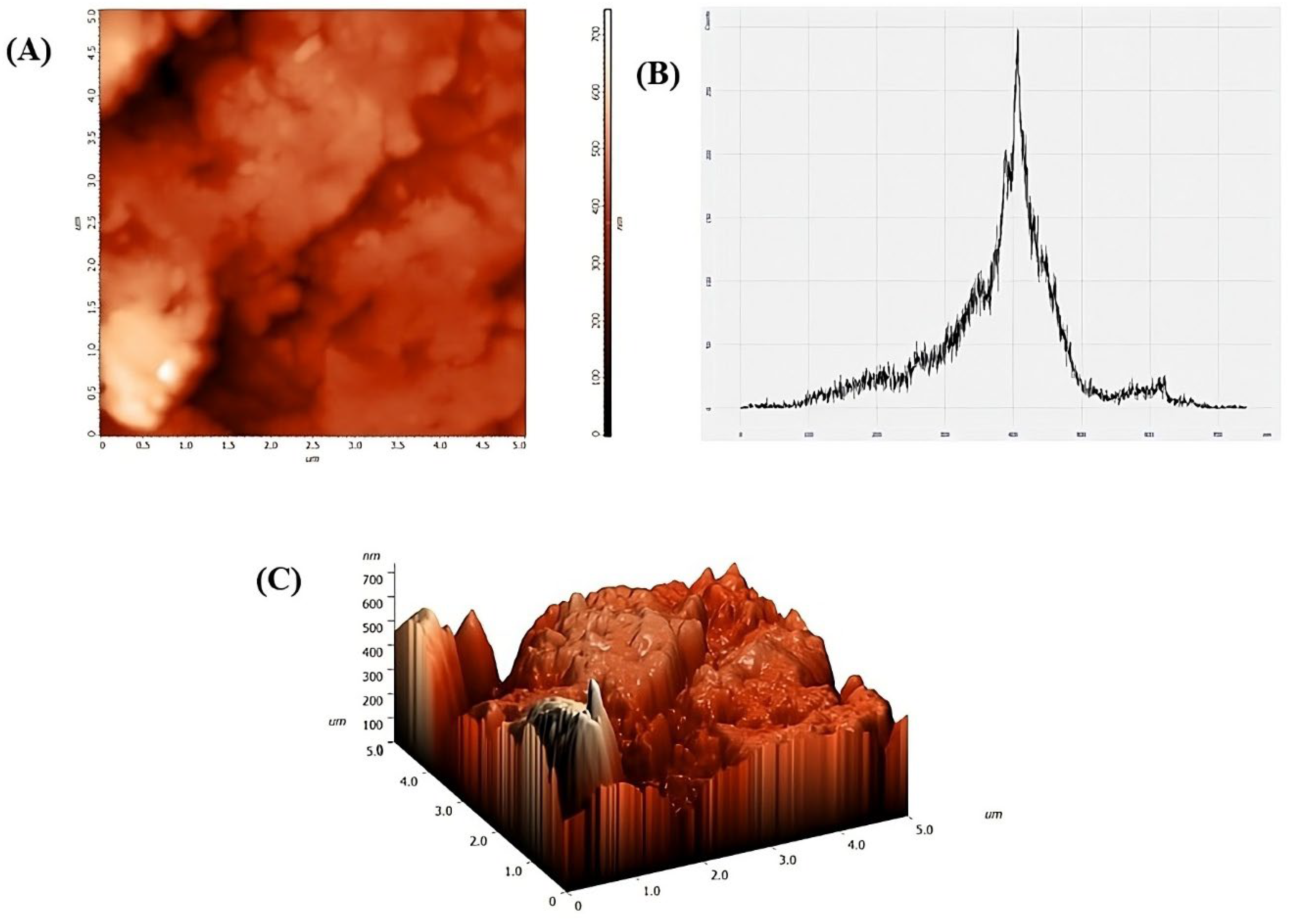
2.2.5. AFM
2.3. Physiochemical Characterization of Water Sample
2.4. Preparation of CaO-NP-Enriched Fish Diet
2.5. Acclimatization of D. rerio
2.6. Feeding Trial and Statistical Analysis on Growth Performance of D. rerio
2.7. Tissue Sample Collection
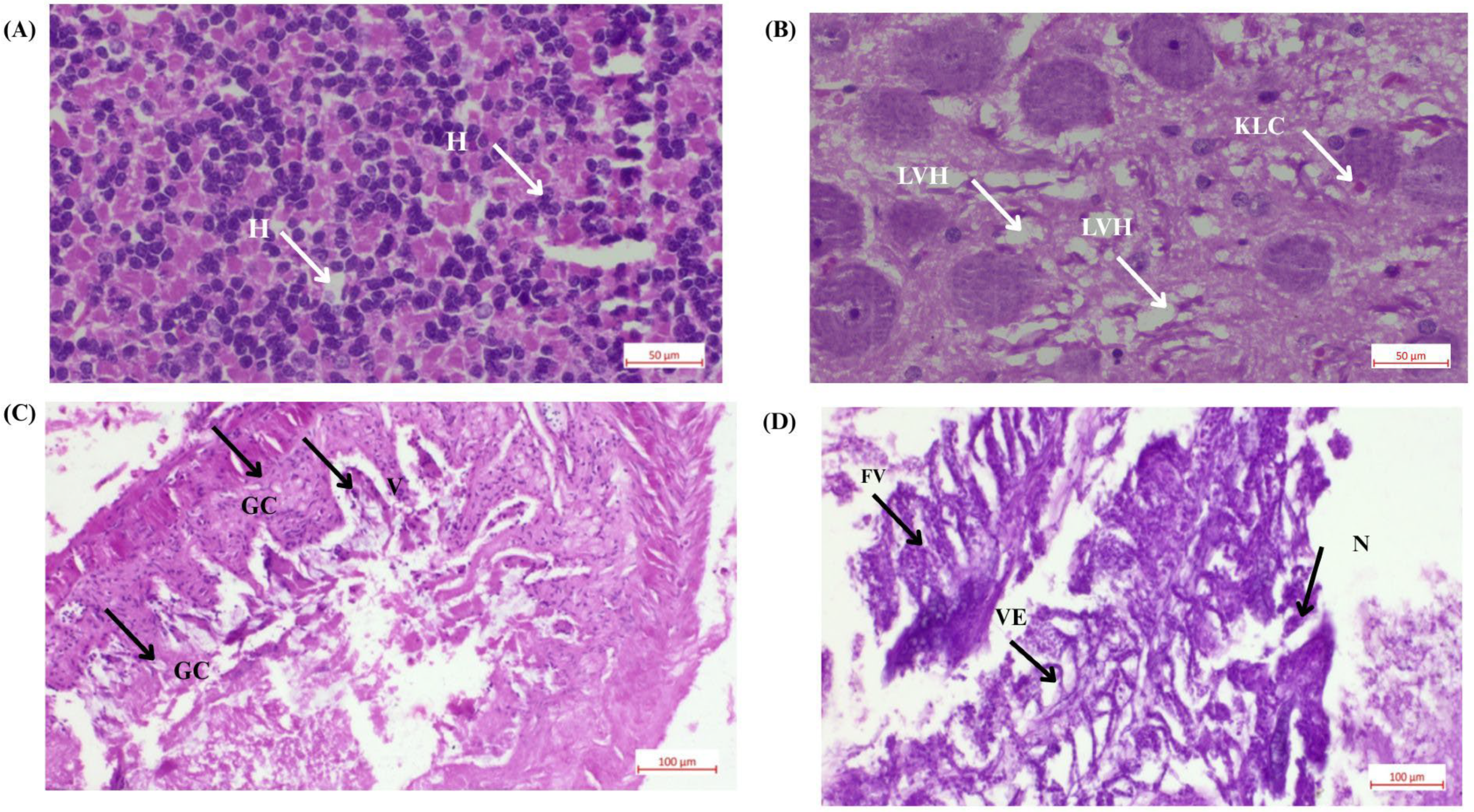
2.8. Histological Test
2.9. CaO-NP Accumulation Analysis Using ICP-OES
2.10. Computational Methods
2.10.1. Selection of Metabolic Pathway, Retrieval of Proteins and Its 3D Structure
2.10.2. Retrieval of CaO-NP 3D Structure
2.10.3. Three-Dimensional Modelling and Verification
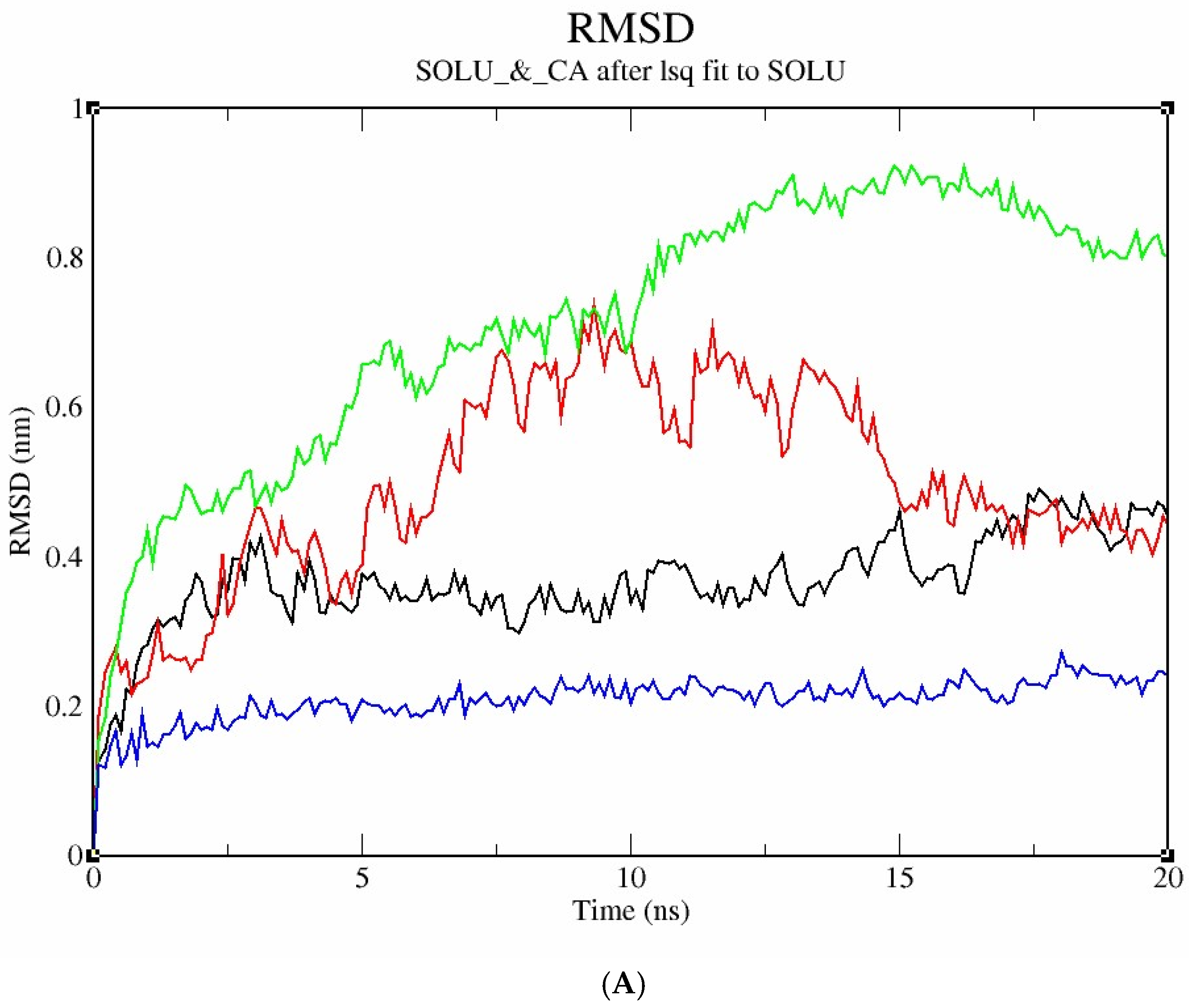
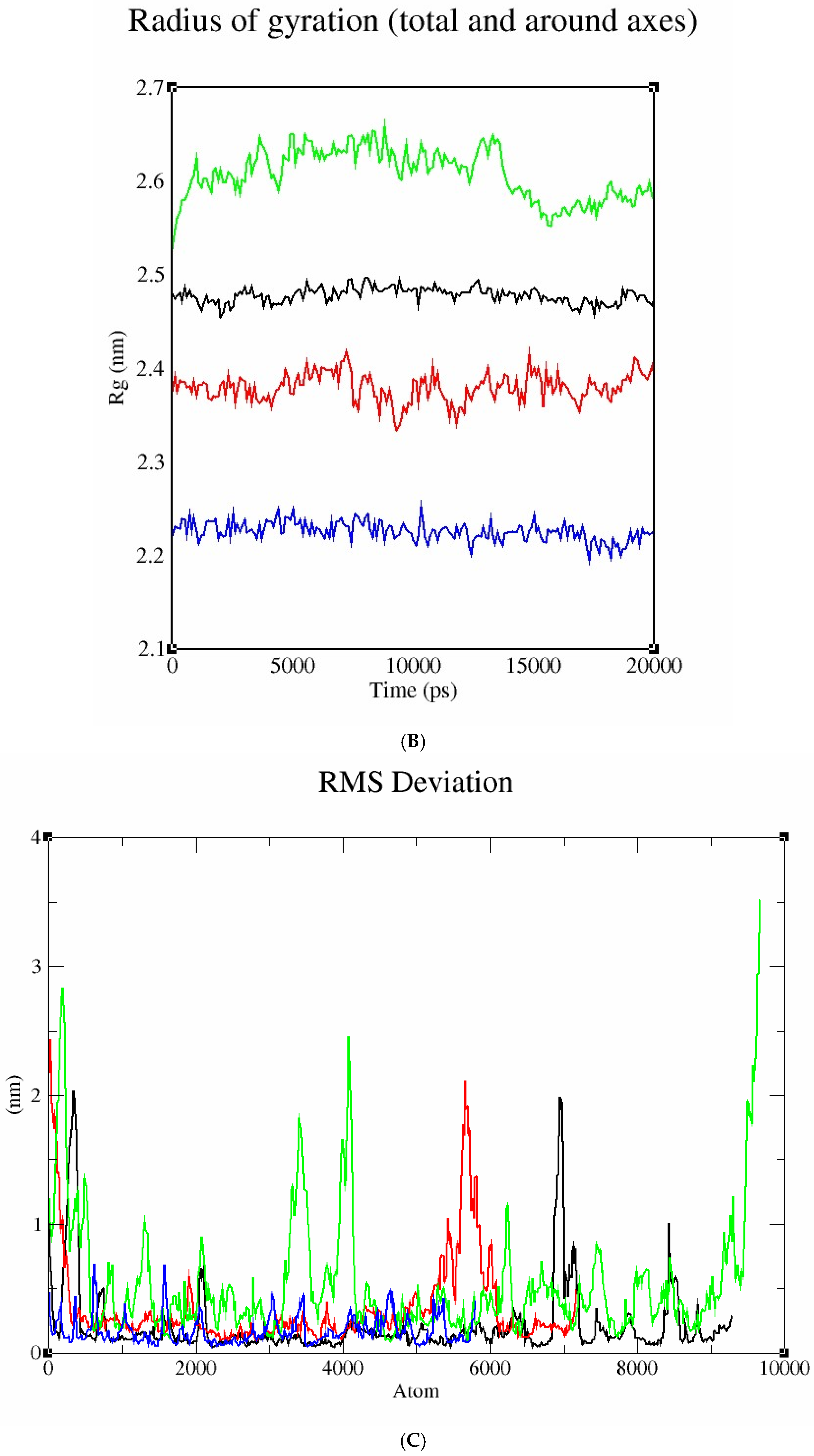
2.10.4. Molecular Dynamics Study
2.10.5. Active Site Prediction, Molecular Docking Analysis of Proteins
3. Results
3.1. CaO-NP Synthesis and Characterization
3.2. Water Sample Physiochemical Characterization
3.3. Histological Analysis
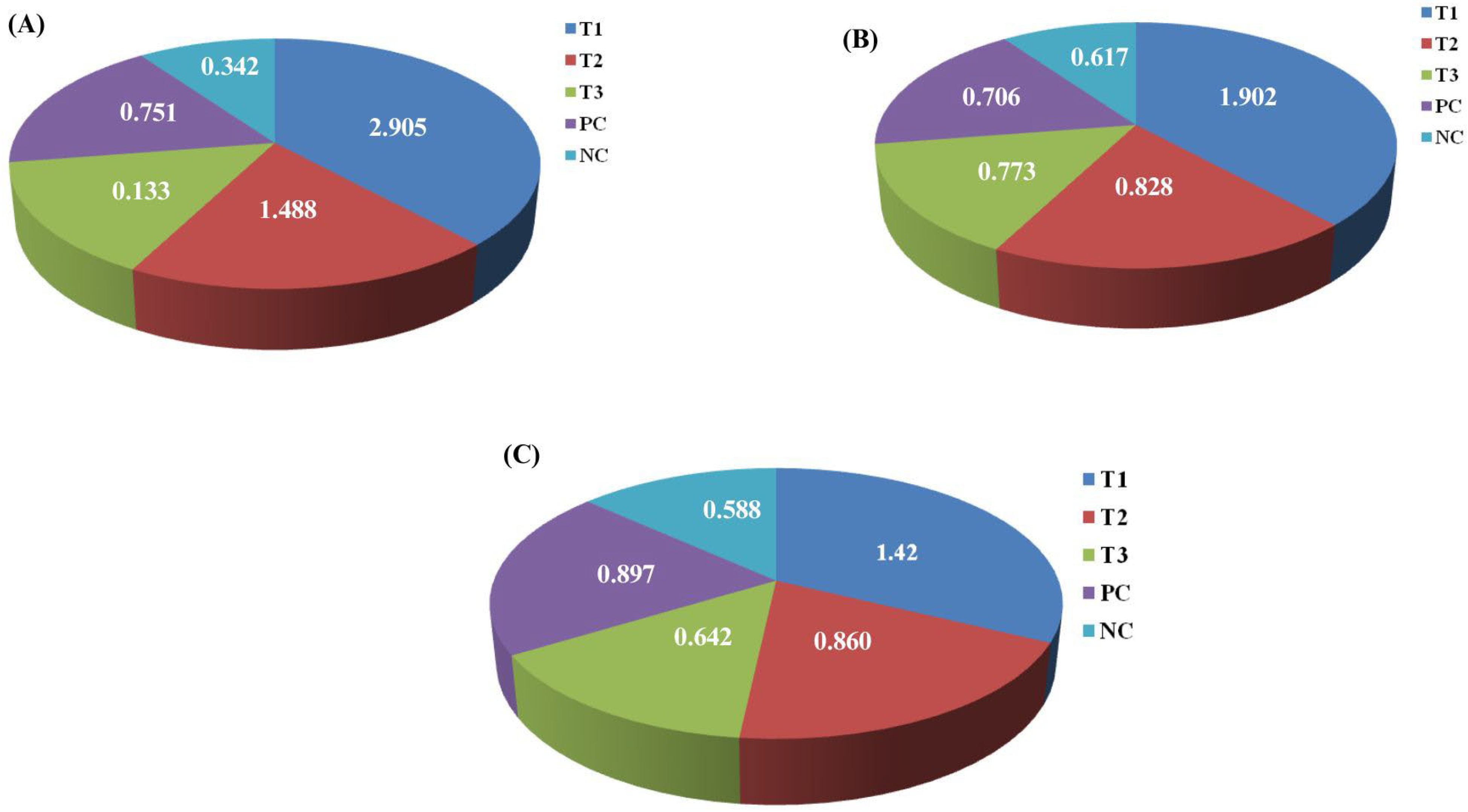
3.4. CaO-NP Accumulation Analysis in D. rerio Tissues Using ICP-OES
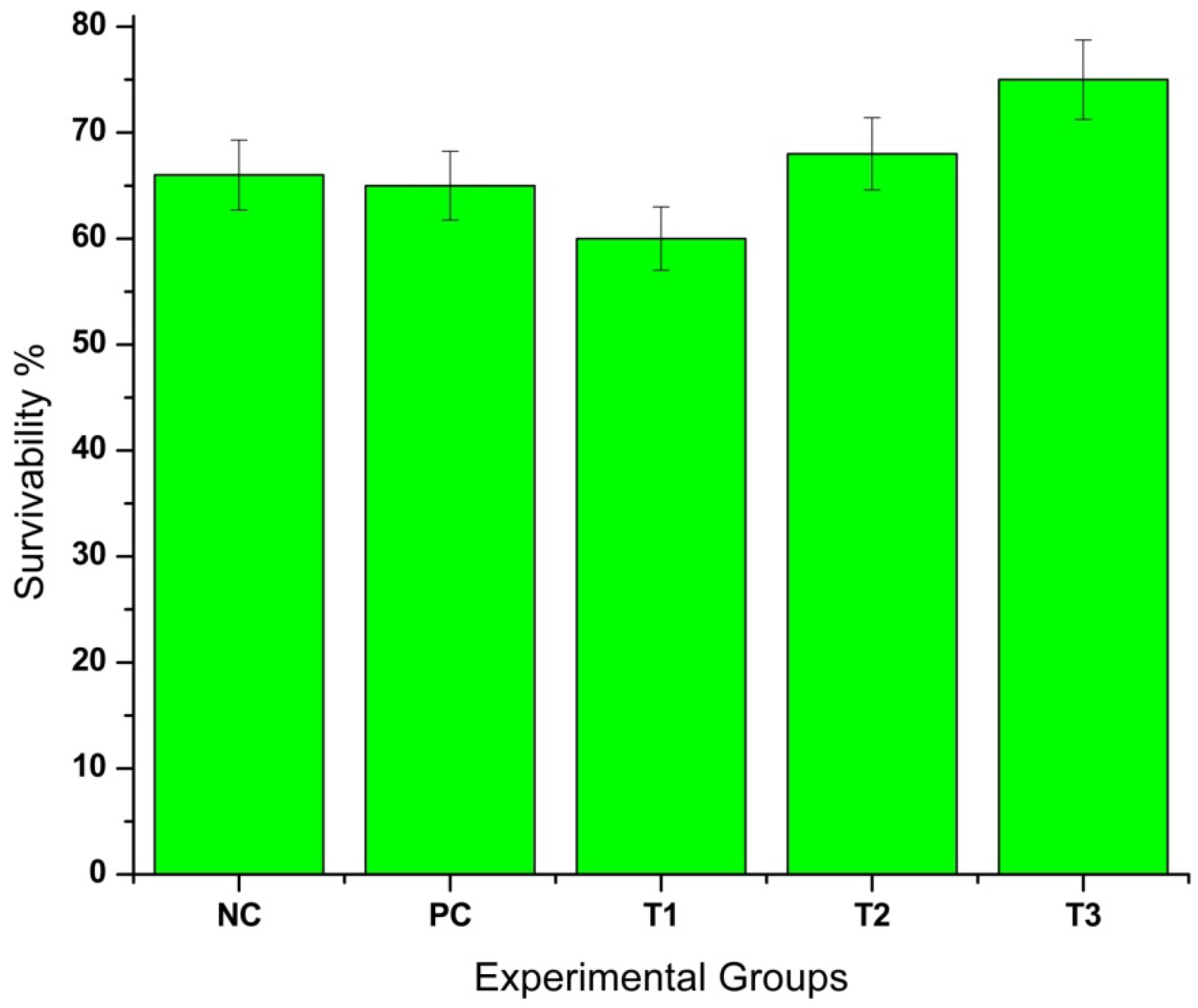
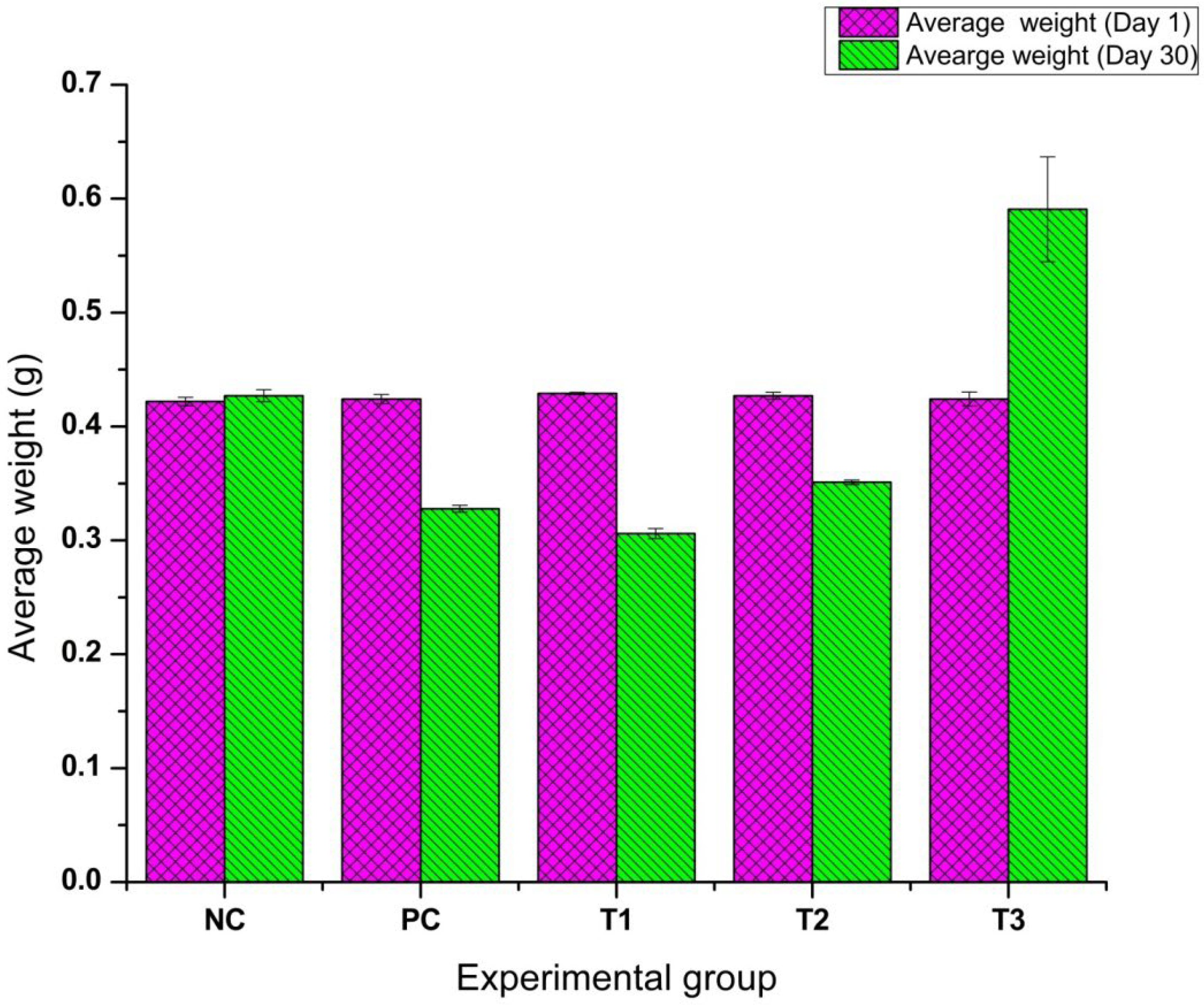
3.5. Feeding, Growth Performance, and Statistical Analysis in D. rerio
3.6. Computational Analysis
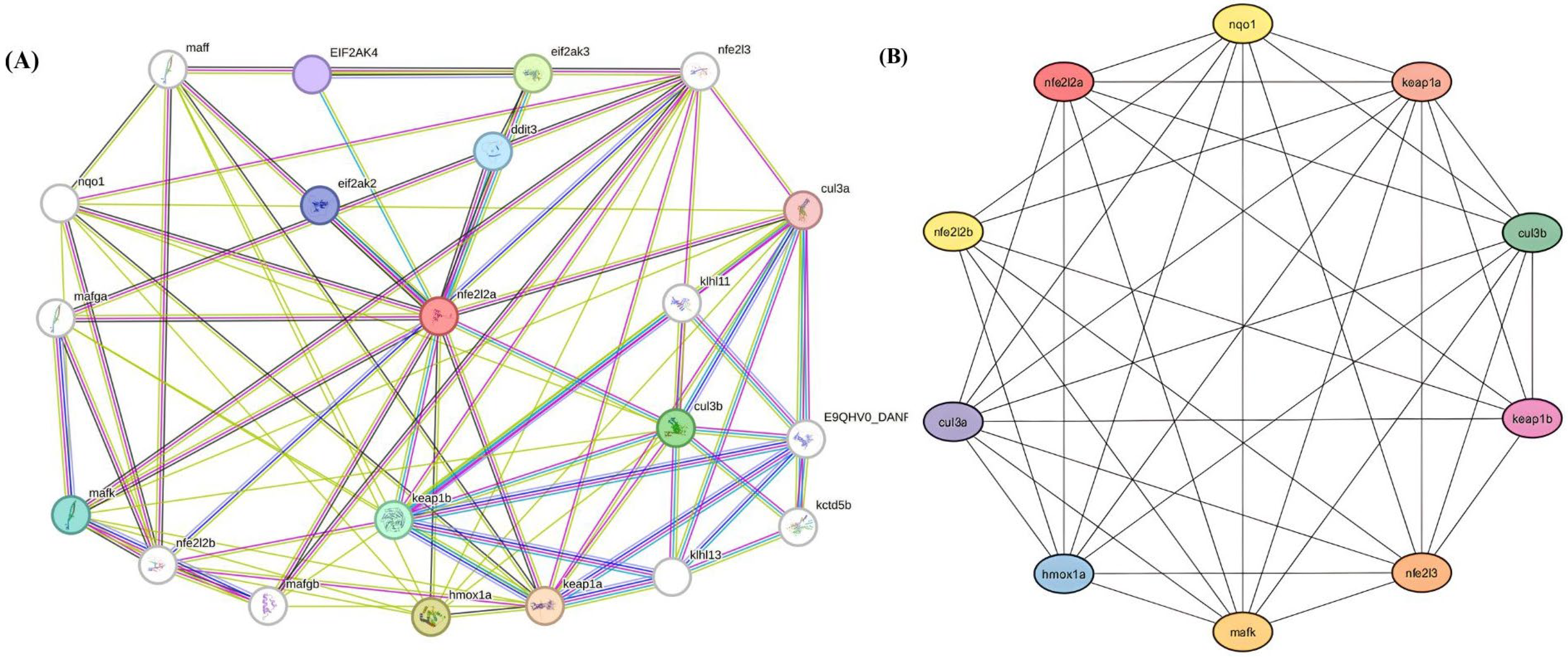
3.6.1. Selection of Metabolic Pathway
3.6.2. Three-Dimensional Protein Modelling and Verification
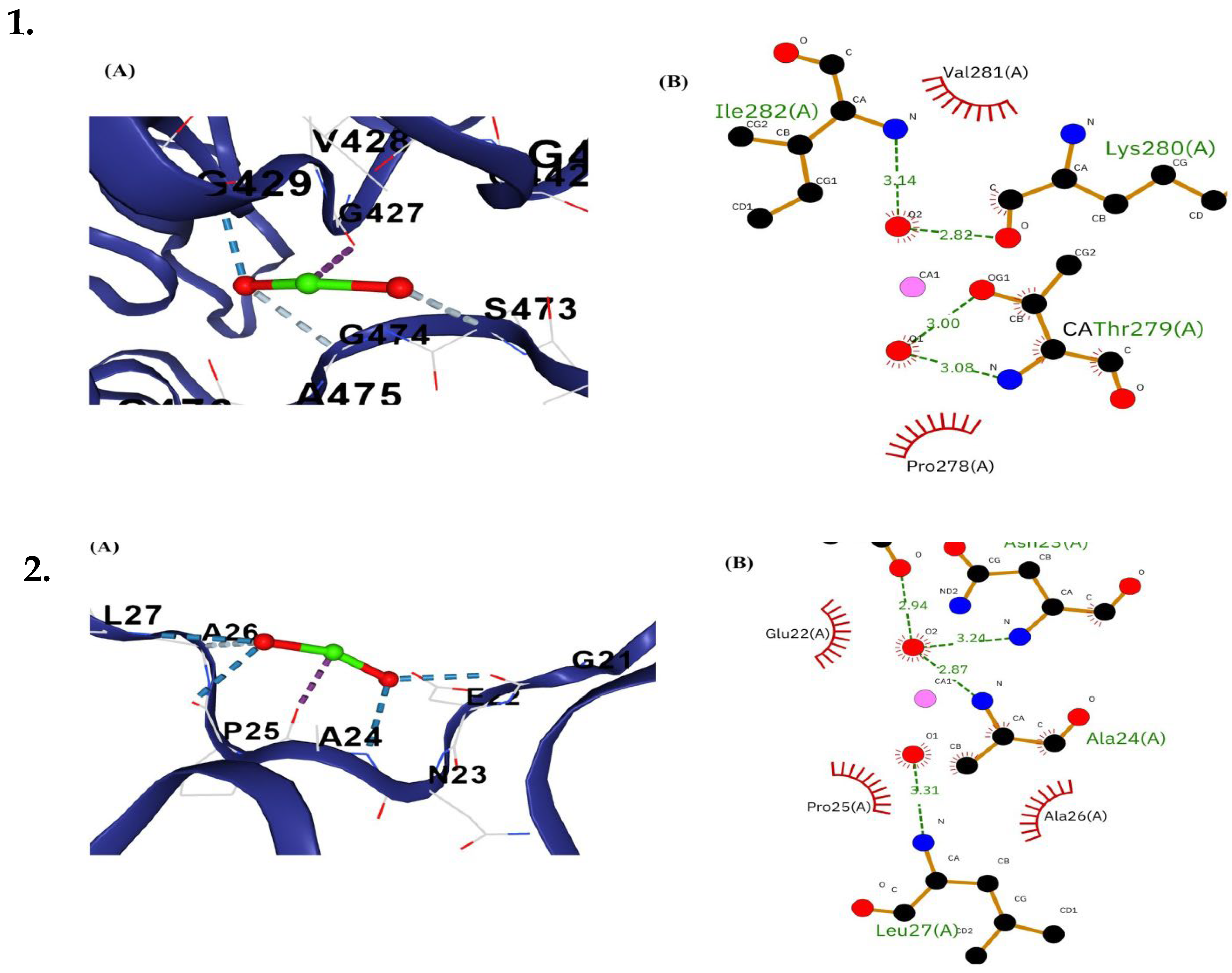
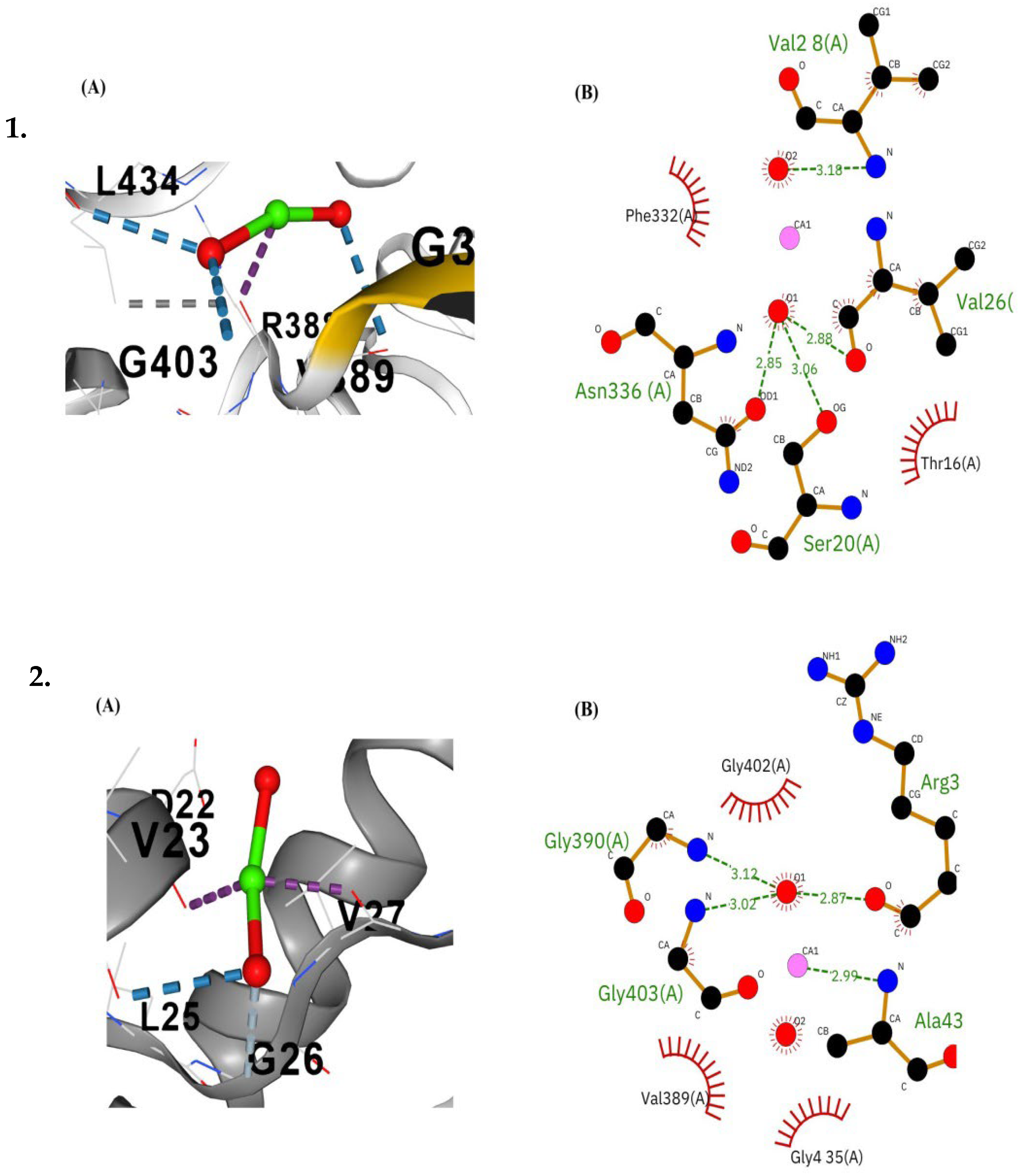
3.6.3. Molecular Docking and Post-Docking Analysis of Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiong, H.; Xu, H. How Current Global Aqua-Nutrition Research Matches the New Industry and Market Demands. Fishes 2024, 9, 259. [Google Scholar] [CrossRef]
- Raghuvaran, N.; Varghese, T.; Jana, P.; Angela, B.R.J.; Sudarshan, S.; Alrashdi, Y.B.; Ibrahim, A.E.; El Deeb, S. Current Status and Global Research Trend Patterns of Insect Meal in Aquaculture From Scientometric Perspective:(2013–2022). Aquac. Nutr. 2024, 22, 5466604. [Google Scholar]
- Tumwesigye, Z.; Tumwesigye, W.; Opio, F.; Kemigabo, C.; Mujuni, B. The effect of water quality on aquaculture productivity in Ibanda District, Uganda. Aquac. J. 2022, 4, 23–36. [Google Scholar] [CrossRef]
- Akpoilih, B.U.; Nwafili, S.A.; Erondu, E.S. Microbial Phytases as Functional Feed Additives in Aquaculture: Impact, Challenges, Recent Developments and Future Opportunities. Sustain. Feed Ingred. Addit. Aquac. Farm. 2024, 19, 521–563. [Google Scholar]
- Sivagurunathan, U.; Izquierdo, M.; Tseng, Y.; Prabhu, P.A.; Zamorano, M.J.; Robaina, L.; Domínguez, D. Effects of the Interaction between Dietary Vitamin D3 and Vitamin K3 on Growth, Skeletal Anomalies, and Expression of Bone and Calcium Metabolism-Related Genes in Juvenile Gilthead Seabream (Sparus aurata). Animals 2024, 14, 2808. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.I.; Eding, E.H.; Verdegem, M.C.; Heinsbroek, L.T.; Schneider, O.; Blancheton, J.P.; d’Orbcastel, E.R.; Verreth, J.A. New developments in recirculating aquaculture systems in Europe: A perspective on environmental sustainability. Aquac. Eng. 2010, 1, 83–93. [Google Scholar] [CrossRef]
- Kumari, M.; Sarkar, B.; Mukherjee, K. Nanoscale calcium oxide and its biomedical applications: A comprehensive review. Biocatal. Agric. Biotechnol. 2023, 47, 102506. [Google Scholar] [CrossRef]
- Goodrich, H.R.; Berry, A.A.; Montgomery, D.W.; Davison, W.G.; Wilson, R.W. Fish feeds supplemented with calcium-based buffering minerals decrease stomach acidity, increase the blood alkaline tide and cost more to digest. Sci. Rep. 2022, 12, 18468. [Google Scholar] [CrossRef]
- Sánchez-Velázquez, J.; Peña-Herrejón, G.A.; Aguirre-Becerra, H. Fish Responses to Alternative Feeding Ingredients under Abiotic Chronic Stress. Animals 2024, 14, 765. [Google Scholar] [CrossRef]
- Kayiira, J.C.; Mi, H.; Liang, H.; Ren, M.; Huang, D.; Zhang, L.; Teng, T. Effect of Dietary Copper on Growth Performance, Antioxidant Capacity, and Immunity in Juvenile Largemouth Bass (Micropterus salmoides). Fishes 2024, 9, 369. [Google Scholar] [CrossRef]
- Wei, X.L.; Hao, Z.W.; Kotzamanis, Y.P.; Zhang, T.H.; Liu, Z.B.; Yang, H.; Luo, Z. Dietary iron oxide (Fe2O3) nanoparticles modulate growth performance, body composition, mineral content and intestinal health of yellow catfish juveniles (Pelteobagrus fulvidraco). Aquac. Rep. 2025, 42, 102739. [Google Scholar] [CrossRef]
- Mahmoud, A.S.; Sayed, A.E.; Mahmoud, U.T.; Mohammed, A.A.; Darwish, M.H. Impact of zinc oxide nanoparticles on the behavior and stress indicators of African catfish (Clarias gariepinus) exposed to heat stress. BMC Vet. Res. 2024, 20, 474. [Google Scholar] [CrossRef]
- Eissa, E.S.; Bazina, W.K.; Abd El-Aziz, Y.M.; Abd Elghany, N.A.; Tawfik, W.A.; Mossa, M.I.; Abd El Megeed, O.H.; Abd El-Hamed, N.N.; El-Saeed, A.F.; El-Haroun, E.; et al. Nano-selenium impacts on growth performance, digestive enzymes, antioxidant, immune resistance and histopathological scores of Nile tilapia, Oreochromis niloticus against Aspergillus flavus infection. Aquac. Int. 2024, 32, 1587–1611. [Google Scholar] [CrossRef]
- Meyers, J.R. Zebrafish: Development of a vertebrate model organism. Curr. Protoc. Essent. Lab. Tech. 2018, 16, 19. [Google Scholar] [CrossRef]
- Süntar, I.; Çetinkaya, S.; Panieri, E.; Saha, S.; Buttari, B.; Profumo, E.; Saso, L. Regulatory role of Nrf2 signaling pathway in wound healing process. Molecules 2021, 26, 2424. [Google Scholar] [CrossRef] [PubMed]
- Emami, M.H.; Sereshki, N.; Malakoutikhah, Z.; Dehkordi, S.A.; Fahim, A.; Mohammadzadeh, S.; Maghool, F. Nrf2 signaling pathway in trace metal carcinogenesis: A cross-talk between oxidative stress and angiogenesis. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2022, 254, 109266. [Google Scholar] [CrossRef] [PubMed]
- Attarilar, S.; Yang, J.; Ebrahimi, M.; Wang, Q.; Liu, J.; Tang, Y.; Yang, J. The toxicity phenomenon and the related occurrence in metal and metal oxide nanoparticles: A brief review from the biomedical perspective. Front. Bioeng. Biotechnol. 2020, 8, 822. [Google Scholar] [CrossRef]
- Kumar, H.; Bhardwaj, K.; Nepovimova, E.; Kuča, K.; Singh Dhanjal, D.; Bhardwaj, S.; Bhatia, S.K.; Verma, R.; Kumar, D. Antioxidant functionalized nanoparticles: A combat against oxidative stress. Nanomaterials 2020, 10, 1334. [Google Scholar] [CrossRef]
- Briggs, J.P. The zebrafish: A new model organism for integrative physiology. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 282, R3–R9. [Google Scholar] [CrossRef]
- Ba-Abbad, M.M.; Benamour, A.; Ewis, D.; Mohammad, A.W.; Mahmoudi, E. Synthesis of Fe3O4 nanoparticles with different shapes through a co-precipitation method and their application. JOM 2018, 74, 3531–3539. [Google Scholar] [CrossRef]
- Inam, E.; Inoh, G.G.; Offiong, N.O.; Etim, B.B. Physicochemical characteristics and health risk assessment of drinking water sources in Okoroette Community, Eastern Coast of Nigeria. Am. J. Water Resour. 2017, 5, 13–23. [Google Scholar] [CrossRef][Green Version]
- Sarkar, B.; Kumar, M.; Rathore, R.M.; Verma, S. Effect of dietary nanosilver on gut proteases and general performance in Zebrafish (Danio rerio). Int. J. Aquat. Biol. 2015, 3, 360–367. [Google Scholar][Green Version]
- Lawrence, C.; Best, J.; James, A.; Maloney, K. The effects of feeding frequency on growth and reproduction in zebrafish (Danio rerio). Aquaculture 2012, 368, 103–108. [Google Scholar] [CrossRef]
- Vulesevic, B.; McNeill, B.; Perry, S.F. Chemoreceptor plasticity and respiratory acclimation in the zebrafish (Danio rerio). J. Exp. Biol. 2006, 209, 1261–1273. [Google Scholar] [CrossRef]
- Wong, P.B.; Wiley, E.O.; Johnson, W.E.; Ryder, O.A.; O’Brien, S.J.; Haussler, D.; Koepfli, K.P.; Houck, M.L.; Perelman, P.; Mastromonaco, G.; et al. Tissue sampling methods and standards for vertebrate genomics. Gigascience 2012, 1, 2047-17X. [Google Scholar] [CrossRef]
- Chen, Y.J.; Liu, Y.J.; Yang, H.J.; Yuan, Y.; Liu, F.J.; Tian, L.X.; Liang, G.Y.; Yuan, R.M. Effect of dietary oxidized fish oil on growth performance, body composition, antioxidant defence mechanism and liver histology of juvenile largemouth bass Micropterus salmoides. Aquac. Nutr. 2012, 18, 321–331. [Google Scholar] [CrossRef]
- Li, J.; Ni, J.; Li, J.; Wang, C.; Li, X.; Wu, S.; Zhang, T.; Yu, Y.; Yan, Q. Comparative study on gastrointestinal microbiota of eight fish species with different feeding habits. J. Appl. Microbiol. 2014, 117, 1750–1760. [Google Scholar] [CrossRef]
- Jin, Q.; Dai, M.; Zhan, X.; Wang, S.; He, Z. Carbon nanotubes and graphene composites used in Cr(VI) detection techniques: A review. J. Alloy. Compd. 2022, 922, 166268. [Google Scholar] [CrossRef]
- Kuo, T.C.; Tian, T.F.; Tseng, Y.J. 3Omics: A web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst. Biol. 2013, 7, 1–5. [Google Scholar] [CrossRef]
- Labena, A.A.; Gao, Y.Z.; Dong, C.; Hua, H.L.; Guo, F.B. Metabolic pathway databases and model repositories. Quant. Biol. 2018, 6, 30–39. [Google Scholar] [CrossRef]
- Zhang, L.; Feng, C.; Zhou, Y.; Zhou, Q. Dysregulated genes targeted by microRNAs and metabolic pathways in bladder cancer revealed by bioinformatics methods. Oncol. Lett. 2018, 15, 9617–9624. [Google Scholar] [CrossRef]
- She, H.; Tan, L.; Zhou, Y.; Zhu, Y.; Ma, C.; Wu, Y.; Du, Y.; Liu, L.; Hu, Y.; Mao, Q.; et al. The landscape of featured metabolism-related genes and imbalanced immune cell subsets in sepsis. Front. Genet. 2022, 13, 821275. [Google Scholar] [CrossRef] [PubMed]
- Vlah, D.; Čok, V.; Urbas, U. VR as a 3D modelling tool in engineering design applications. Appl. Sci. 2021, 11, 7570. [Google Scholar] [CrossRef]
- Pradeepkiran, J.A.; Sainath, S.B.; Balne, P.K.; Bhaskar, M. Computational Modeling and Evaluation of Best Potential Drug Targets Through Comparative Modeling; Academic Press: Cambridge, MA, USA, 2021; Volume 1. [Google Scholar]
- Pardeshi, T.A.; Jobby, A.; Chaudhary, S. Comparative study of homology-based structure prediction and structure validation tools on some proteins from the bHLH family. Int. J. Comput. Biol. 2015, 4, 59–67. [Google Scholar] [CrossRef]
- Dhivya, S.; Suresh, C.K.; Bommuraj, V.; Janarthanam, R.; Chandran, M.; Usha, T.; Middha, S.K. A study of comparative modelling, simulation and molecular dynamics of CXCR3 receptor with lipid bilayer. J. Biomol. Struct. Dyn. 2018, 36, 2361–2372. [Google Scholar] [CrossRef]
- Agrawal, R.; Punarva, H.B.; Heda, G.O.; Vishesh, Y.M.; Karunakar, P. Vina LigGen: A method to generate LigPlots and retrieval of hydrogen and hydrophobic interactions from protein-ligand complexes. J. Biomol. Struct. Dyn. 2024, 42, 12040–12043. [Google Scholar] [CrossRef]
- Mabrouk, M.; Fouad, G.I.; El-Sayed, S.A.; Rizk, M.Z.; Beherei, H.H. Hepatotoxic and neurotoxic potential of iron oxide nanoparticles in Wistar rats: A biochemical and ultrastructural study. Biol. Trace Elem. Res. 2021, 199, 1–28. [Google Scholar] [CrossRef]
- Liang, J.; Cao, C.; Chen, P.; Bharadwaj, A.S.; Wu, X.; Gu, X.; Tao, Q.; Xue, M. Effects of dietary zinc sources and levels on growth performance, tissue zinc retention and antioxidant response of juvenile common carp (Cyprinus carpio) fed diets containing phytic acid. Aquac. Nutr. 2020, 26, 444–455. [Google Scholar] [CrossRef]
- Fadia, B.S.; Mokhtari-Soulimane, N.; Meriem, B.; Wacila, N.; Zouleykha, B.; Karima, R.; Soulimane, T.; Tofail, S.A.; Townley, H.; Thorat, N.D. Histological injury to rat brain, liver, and kidneys by gold nanoparticles is dose-dependent. ACS Omega 2022, 7, 20656–20665. [Google Scholar] [CrossRef]
- Bootz, A.; Vogel, V.; Schubert, D.; Kreuter, J. Comparison of Scanning Electron Microscopy, Dynamic Light Scattering and Analytical Ultracentrifugation for the Sizing of Polyalkylcyanoacrylate Nanoparticles. Eur. J. Pharm. Biopharm. 2004, 57, 369–375. [Google Scholar] [CrossRef]
- Pyrgiotakis, G.; Blattmann, C.O.; Demokritou, P. Real-Time Nanoparticle-Cell Interactions in Physiological Media by Atomic Force Microscopy. ACS Sustain. Chem. Eng. 2014, 2, 1681–1690. [Google Scholar] [CrossRef]
- Piloni, A.; Casalini, T.; Rossi, F.; Bertini, S.; Parnigiani, C.; Pugliese, R.; Montalbano, C.; Mari, L.; Mollica, H.; Monopoli, M.P.; et al. Surface Roughness Influences the Protein Corona Formation of Glycosylated Nanoparticles and Alters Their Cellular Uptake. Nanoscale 2019, 11, 23259–23267. [Google Scholar] [CrossRef]
- Hoseini, S.M.; Khosravi-Darani, K.; Hedayati, A.; Naghshi, H. Comparative toxicity of organic, inorganic and nanoparticulate zinc following dietary exposure to common carp (Cyprinus carpio). Sci. Total Environ. 2019, 656, 1191–1198. [Google Scholar]
- Taheri, S.; Banaee, M.; Haghi, B.N.; Mohiseni, M. Effects of dietary supplementation of zinc oxide nanoparticles on some biochemical biomarkers in common carp (Cyprinus carpio). Int. J. Aquat. Biol. 2017, 5, 286–294. [Google Scholar]
- Chen, G.H.; Song, C.C.; Zhao, T.; Hogstrand, C.; Wei, X.L.; Lv, W.H.; Song, Y.F.; Luo, Z. Mitochondria-dependent oxidative stress mediates ZnO nanoparticle-induced mitophagy and lipotoxicity in freshwater teleost fish. Environ. Sci. Technol. 2022, 56, 2407–2420. [Google Scholar] [CrossRef] [PubMed]
- Johari, S.A.; Kalbassi, M.R.; Yu, I.J.; Lee, J.H. Chronic effect of waterborne silver nanoparticles on rainbow trout (Oncorhynchus mykiss): Histopathology and bioaccumulation. Comp. Clin. Pathol. 2015, 24, 995–1007. [Google Scholar] [CrossRef]
- Koner, D.; Banerjee, B.; Kumari, A.; Lanong, A.S.; Snaitang, R.; Saha, N. Molecular characterization of superoxide dismutase and catalase genes, and the induction of antioxidant genes under the zinc oxide nanoparticle-induced oxidative stress in air-breathing magur catfish (Clarias magur). Fish Physiol. Biochem. 2021, 47, 1909–1932. [Google Scholar] [CrossRef]
- Liu, F.; Wang, Y.; Chen, L.; Bello, B.K.; Zhang, T.; Yang, H.; Li, X.; Pan, E.; Feng, H.; Dong, J. Difenoconazole disrupts the blood-brain barrier and results in neurotoxicity in carp by inhibiting the Nrf2 pathway mediated ROS accumulation. Ecotoxicol. Environ. Saf. 2022, 244, 114081. [Google Scholar] [CrossRef]
- Alhamhoom, Y.; Hani, U.; Bennani, F.E.; Rahman, N.; Rashid, M.A.; Abbas, M.N.; Rastrelli, L. Identification of new drug target in Staphylococcus lugdunensis by subtractive genomics analysis and their inhibitors through molecular docking and molecular dynamic simulation studies. Bioengineering 2022, 9, 451. [Google Scholar] [CrossRef]
- Bodea, F.; Bungau, S.G.; Negru, A.P.; Radu, A.; Tarce, A.G.; Tit, D.M.; Bungau, A.F.; Bustea, C.; Behl, T.; Radu, A.F. Exploring new therapeutic avenues for ophthalmic disorders: Glaucoma-related molecular docking evaluation and bibliometric analysis for improved management of ocular diseases. Bioengineering 2023, 10, 983. [Google Scholar] [CrossRef]
- Ghufran, M.; Ullah, M.; Khan, H.A.; Ghufran, S.; Ayaz, M.; Siddiq, M.; Abbas, S.Q.; Hassan, S.S.U.; Bungau, S. In-silico lead druggable compounds identification against SARS COVID-19 main protease target from in-house, ChemBridge and ZINC databases by structure-based virtual screening, molecular docking and molecular dynamics simulations. Bioengineering 2023, 10, 100. [Google Scholar] [CrossRef]
- Li, M.Y.; Shi, Y.C.; Xu, W.X.; Zhao, L.; Zhang, A.Z. Exploring Cr(VI)-induced blood-brain barrier injury and neurotoxicity in zebrafish and snakehead fish, and inhibiting toxic effects of astaxanthin. Environ. Pollut. 2024, 355, 124280. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.M.; Naeem, E.; Ali, S.; Adrees, M.; Riaz, D.; Paray, B.A.; Naeem, A. Evaluation of growth, nutrient absorption, body composition and blood indices under dietary exposure of iron oxide nanoparticles in common carp (Cyprinus carpio). J. Anim. Physiol. Anim. Nutr. 2024, 108, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Gupta, A.; Sarkar, B.; Gupta, R.; Kumar, M.; Kumari, A.; Foysal, M.J. Pomegranate (Punica granatum) peel extract supplementation in diet influences growth performance, haemato-immunological responses and cytokine expression in pathogen-aggravated Labeo rohita fingerlings. Aquaculture 2023, 562, 738823. [Google Scholar] [CrossRef]
- Alhujaily, M.; Albukhaty, S.; Yusuf, M.; Mohammed, M.K.; Sulaiman, G.M.; Al-Karagoly, H.; Alyamani, A.A.; Albaqami, J.; AlMalki, F.A. Recent advances in plant-mediated zinc oxide nanoparticles with their significant biomedical properties. Bioengineering 2022, 9, 541. [Google Scholar] [CrossRef]
- Baki, A.; Wiekhorst, F.; Bleul, R. Advances in magnetic nanoparticles engineering for biomedical applications—A review. Bioengineering 2021, 8, 134. [Google Scholar] [CrossRef]
- Zhao, P.; Tian, Y.; You, J.; Hu, X.; Liu, Y. Recent advances of calcium carbonate nanoparticles for biomedical applications. Bioengineering 2022, 9, 691. [Google Scholar] [CrossRef]
- Balhaddad, A.A.; Garcia, I.M.; Mokeem, L.; Alsahafi, R.; Collares, F.M.; Sampaio de Melo, M.A. Metal oxide nanoparticles and nanotubes: Ultrasmall nanostructures to engineer antibacterial and improved dental adhesives and composites. Bioengineering 2021, 8, 146. [Google Scholar] [CrossRef]
- Balaji, S.; Mandal, B.K.; Reddy, V.K.; Sen, D. Biogenic ceria nanoparticles (CeO2 NPs) for effective photocatalytic and cytotoxic activity. Bioengineering 2020, 7, 26. [Google Scholar] [CrossRef]

| Organs | Proteins | Cur ID | Vina Score | Cavity Volume (Å3) | Contact Residues | No. of H-Bonds |
|---|---|---|---|---|---|---|
| Polo like kinase 1 | C1 | −3.3 | 1173 | ARG43 LEU45 CYS53 ALA66 LYS68 VAL70 MET84 GLU87 ILE88 HIS91 VAL114 LEU116 GLU117 ILE118 CYS119 ARG120 ARG121 ARG122 SER123 ILE159 HIS160 ARG161 ASP162 LYS164 ASN167 PHE169 LEU170 GLY179 ASP180 PHE181 GLY182 LEU183 ALA184 THR200 PRO201 ASN202 | - | |
| Gills | TRPV6 | C1 | −3.2 | 3387 | GLY194 VAL233 PRO234 ASN235 TYR236 GLY238 ARG247 SER283 ARG284 ASP286 GLU287 HIS288 SER289 GLU292 ILE293 ILE294 THR296 SER297 HIS298 VAL409 ARG413 PHE414 PHE415 GLN417 ALA419 LEU420 GLY421 GLY422 HIS425 TYR466 ARG469 GLN595 VAL596 ALA598 THR599 LEU601 MET602 ARG605 | - |
| N-K ATPase alpha−2 | C1 | −3.2 | 4499 | LYS89 GLN90 LEU91 PHE92 GLY93 GLY94 PHE95 SER96 GLY139 CYS140 GLN145 LYS148 SER149 SER150 ILE152 GLU273 VAL274 GLY275 THR277 LEU332 ALA343 LEU352 VAL353 LYS354 ASN355 LEU356 GLN700 ALA718 LEU719 LYS720 LYS721 ALA722 ASP723 ILE724 GLY725 GLN738 ALA739 ALA740 ASP741 | - | |
| Intestine | Glutaminase A | C1 | −3.1 | 1811 | SER252 LYS255 GLU278 PRO279 SER280 GLY281 LEU282 ARG283 PHE284 ASN285 LYS286 LEU287 PHE288 LEU289 HIS296 ASN297 MET299 VAL300 ASN301 ALA302 ARG353 ASN354 ILE357 TYR380 CYS384 SER428 CYS429 GLY430 MET431 TYR432 ASP433 SER435 LYS447 VAL450 | - |
| Glycogen | C1 | −3.1 | 2601 | GLY65 PHE67 LYS85 ARG96 GLU97 ILE100 HIS179 ARG180 ASP181 LYS183 ASN186 ASP200 PHE201 GLY202 SER203 ALA204 LYS205 ILE217 | - | |
| Isotocin | C1 | −2.6 | 581 | ARG40 GLN41 CYS42 ARG50 GLY51 ARG52 CYS53 PHE54 CYS60 GLY61 GLU62 GLY63 ILE64 GLU87 MET88 SER89 GLY90 ARG98 CYS99 ALA100 ALA101 PRO102 GLY103 VAL104 CYS117 VAL118 ASP119 GLY120 ASP121 ALA122 ALA124 ALA125 ALA126 | - | |
| Dopamine | C1 | −3.1 | 2229 | LE152 PHE153 PHE154 ILE155 ARG156 PRO190 GLU191 SER192 LEU193 HIS194 GLN195 VAL196 PHE200 SER201 ASP202 ARG203 MET284 TRP292TRP294 ASP298 THR300 LYS301 VAL302 TRP303 SER304 GLU307 PHE308 ASP437 ASP438 ASN439 VAL440 THR441 GLN442 VAL443 PHE446 LEU451 ALA454 GLU455 ARG456 ARG458 LEU459 | - | |
| Brain | Acetylcholineeasterase | C1 | −3.1 | 1282 | ALA437 GLN438 THR453 GLY455 TRP456 GLY457 ASN458 SER459 TYR464 ASN465 SER466 GLY467 ASN468 SER469 HIS470 GLY471 ALA472 VAL473 TYR474 GLY535 ASN536 PRO537 GLN553 PHE554 SER555 ALA556 ASN557 | - |
| GST | C1 | −3.2 | 633 | TYR8 VAL11 LYS12 GLY13 ARG14 CYS15 GLY16 ALA17 CYS48 VAL49 PHE50 GLY51 GLN52 LEU53 PRO54 LYS55 PHE64 GLN65 SER66 ASN67 ALA68 LEU70 ASN94 ASP95 VAL97 GLU98 ASP99 ARG101 TYR104 ILE105 TYR109 ASN153 ASP156 LEU157 ASN160 ILE202 ASN203 GLY204 | 2 | |
| Liver | Glutathione synthetase | C1 | −3.3 | 2304 | ARG125 ASP127 GLU144 ILE145 ASN146 THR147 ILE148 ALA149 ALA150 SER151 LEU209 VAL210 GLU211 GLN214 ASN216 HIS220 ARG267 ASN268 TYR270 LYS306 LYS365 PRO366 GLN367 ARG368 GLU369 GLY370 GLY371 GLY372 ASN373 ASN374 TYR376 LEU411 LEU412 ARG413 PRO414 SER421 ARG451 ASP459 | 2 |
| IGF-1 | C1 | −2.7 | 770 | ARG65 TYR68 PHE69 SER70 LYS71 PRO72 THR73 SER82 HIS83 ASN84 ARG85 GLY86 ILE87 VAL88 ASP89 GLU90 ARG100 GLU102 MET103 TYR104 CYS105 ALA106 PRO107 | - | |
| Catalase | C1 | −3.7 | 1442 | ASN5 TYR17 LEU18 THR19 VAL20 LEU21 HIS46 PRO48 ASN60 ILE61 SER62 TYR63 PRO64 LYS65 GLN66 GLU67 LEU69 LEU70 GLU71 VAL72 LYS73 ASP110 ALA111 TRP112 SER113 SER146 PHE147 SER148 THR149 CYS150 ASP151 PRO152 HIS153 ASP154 GLY160 THR161 VAL162 PRO199 PRO201 THR224 THR225 TYR226 ARG325 VAL328 TYR330 ARG340 ASP341 SER342 SER343 GLY344 | 4 | |
| Keap 1a | C1 | −3.5 | 1594 | GLY337 LEU338 GLY339 ALA340 ASN387 ARG388 VAL389 GLY390 VAL391 VAL401 GLY402 GLY403 ARG433 LEU434 GLY435 ALA436 GLY437 VAL438 GLY482 LEU483 GLY484 VAL495 SER528 ALA529 HIS530 GLY531 VAL532 SER575 GLY576 MET577 GLY578 VAL579 | 4 | |
| Mafk | C1 | −2.4 | 439 | GLY21 GLU22 ASN23 ALA24 PRO25 ALA26 LEU27 GLN42 HIS43 LEU44 ARG45 GLY46 LEU47 THR48 GLU50 ASP51 VAL52 ARG54 LEU55 ARG58 | 4 | |
| Cul3a | C2 | −3.1 | 1386 | GLY571 ILE573 ALA586 LEU587 LEU588 THR589 GLY590 SER591 ASN592 THR593 ARG594 LYS595 HIS596 PHE671 THR672 SER673 LYS674 HIS676 | 2 | |
| Cul3b | C2 | −3.0 | 5731 | GLU428 ARG429 LYS432 GLN433 CYS463 GLN464 PHE465 THR466 SER467 LYS468 LEU469 ASP475 ILE478 VAL506 LEU507 THR508 THR509 GLY510 TYR511 TRP512 PRO513 THR514 GLN515 SER516 GLN546 GLU681 PRO684 GLU685 ARG686 GLU688 THR689 LYS692 GLU700 ARG731 | 2 | |
| Nfe2I3 | C2 | −3.1 | 1225 | ILE17 LEU19 SER20 LEU21 ILE22 GLY23 VAL24 ARG25 HIS334 ILE335 ASN336 MET337 LEU338 ASP339 LEU340 PHE570 SER574 PHE578 PRO591 GLU592 TYR594 THR595 LEU596 HIS597 CYS598 GLN607 ARG609 | - | |
| Keap1b | C2 | −3.3 | 943 | GLY329 LEU330 ALA331 ALA332 CYS333 ILE381 GLY382 VAL383 GLY384 GLY427 VAL428 GLY429 VAL430 VAL440 GLY441 GLY442 ARG472 SER473 GLY474 ALA475 GLY476 SER520 ALA521 LEU522 GLY523 VAL524 SER567 GLY568 VAL569 GLY570 VAL571 ALA572 | 4 | |
| Nfe2I2b | C2 | −2.5 | 1003 | LEU143 ASP144 VAL145 LEU146 GLU147 SER148 GLU149 SER150 SER151 SER152 LEU153 GLU156 LYS448 ARG452 TYR455 PRO472 ARG490 | 1 | |
| Nfe2I2a | C1 | −2.3 | 889 | ASN272 PHE275 TYR276 PRO277 GLU278 MET279 MET534 LYS535 GLN536 LEU538 SER539 THR540 TYR542 GLN543 PRO559 ASN560 GLU561 SER563 LEU564 GLN565 HIS566 ARG577 | 2 | |
| NAD(P)H Quinone dehydrogenase 1 | C5 | −3.2 | 219 | MET143 GLN144 ASN145 MET146 TYR147 ASP148 ASN149 TRP184 ASN188 ARG192 PHE237 ALA238 PRO239 SER240 PHE243 LEU245 THR269 HIS272 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, M.; Tiwary, A.; Sheel, R.; Roy Chowdhury, A.; Sarkar, B.; Mukherjee, K.; Maity, D. Evaluating the Molecular Basis of Nanocalcium-Induced Health Regulation in Zebra Fish (Danio rerio). Bioengineering 2025, 12, 1016. https://doi.org/10.3390/bioengineering12101016
Kumari M, Tiwary A, Sheel R, Roy Chowdhury A, Sarkar B, Mukherjee K, Maity D. Evaluating the Molecular Basis of Nanocalcium-Induced Health Regulation in Zebra Fish (Danio rerio). Bioengineering. 2025; 12(10):1016. https://doi.org/10.3390/bioengineering12101016
Chicago/Turabian StyleKumari, Madhubala, Aastha Tiwary, Rishav Sheel, Arnab Roy Chowdhury, Biplab Sarkar, Koel Mukherjee, and Dipak Maity. 2025. "Evaluating the Molecular Basis of Nanocalcium-Induced Health Regulation in Zebra Fish (Danio rerio)" Bioengineering 12, no. 10: 1016. https://doi.org/10.3390/bioengineering12101016
APA StyleKumari, M., Tiwary, A., Sheel, R., Roy Chowdhury, A., Sarkar, B., Mukherjee, K., & Maity, D. (2025). Evaluating the Molecular Basis of Nanocalcium-Induced Health Regulation in Zebra Fish (Danio rerio). Bioengineering, 12(10), 1016. https://doi.org/10.3390/bioengineering12101016










