Evaluation of UNeXt for Automatic Bone Surface Segmentation on Ultrasound Imaging in Image-Guided Pediatric Surgery
Abstract
1. Introduction
2. Materials and Methods
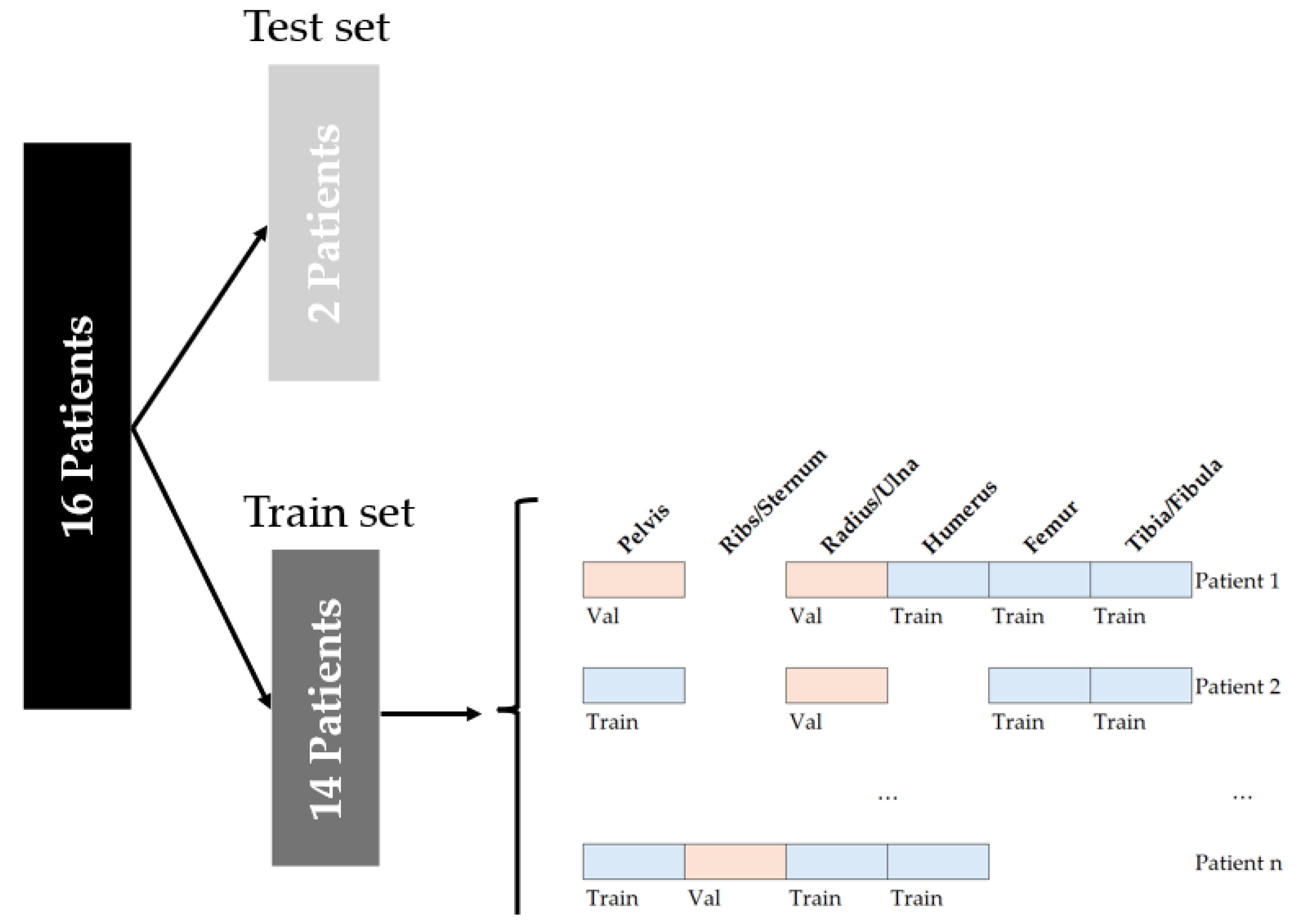
2.1. Patient Population and Data Split
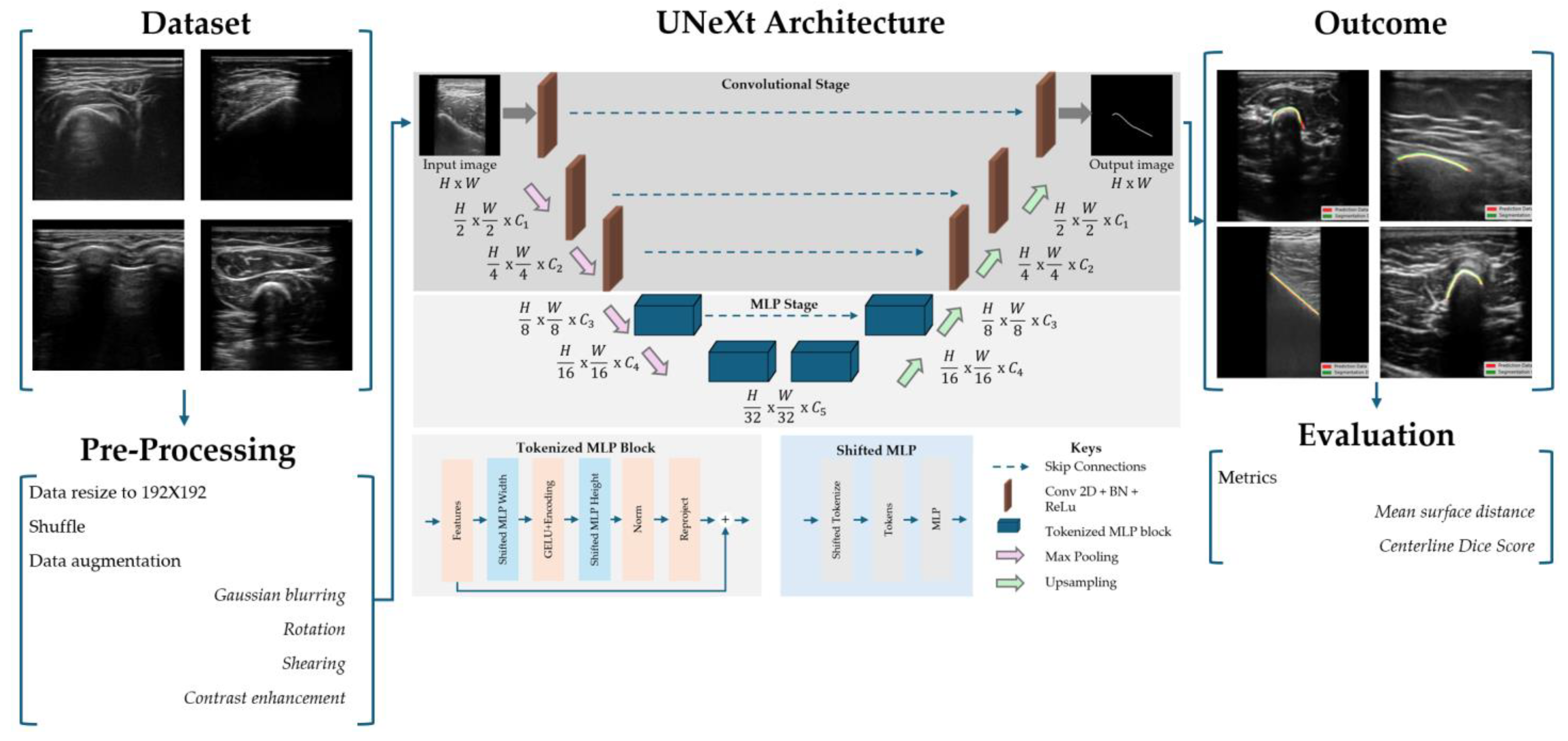
2.2. Data Acquisition and Image Processing
2.3. UNeXt Architecture
2.4. Model Parameters
2.5. Evaluation Metrics
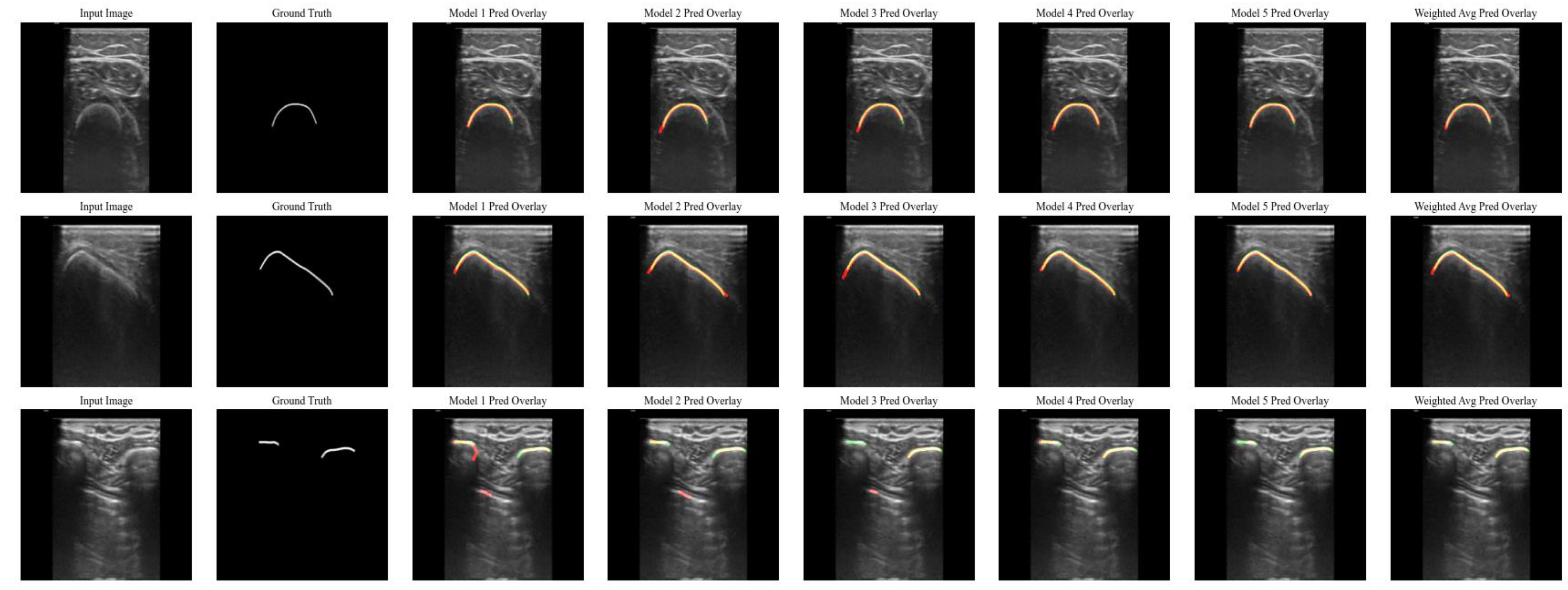
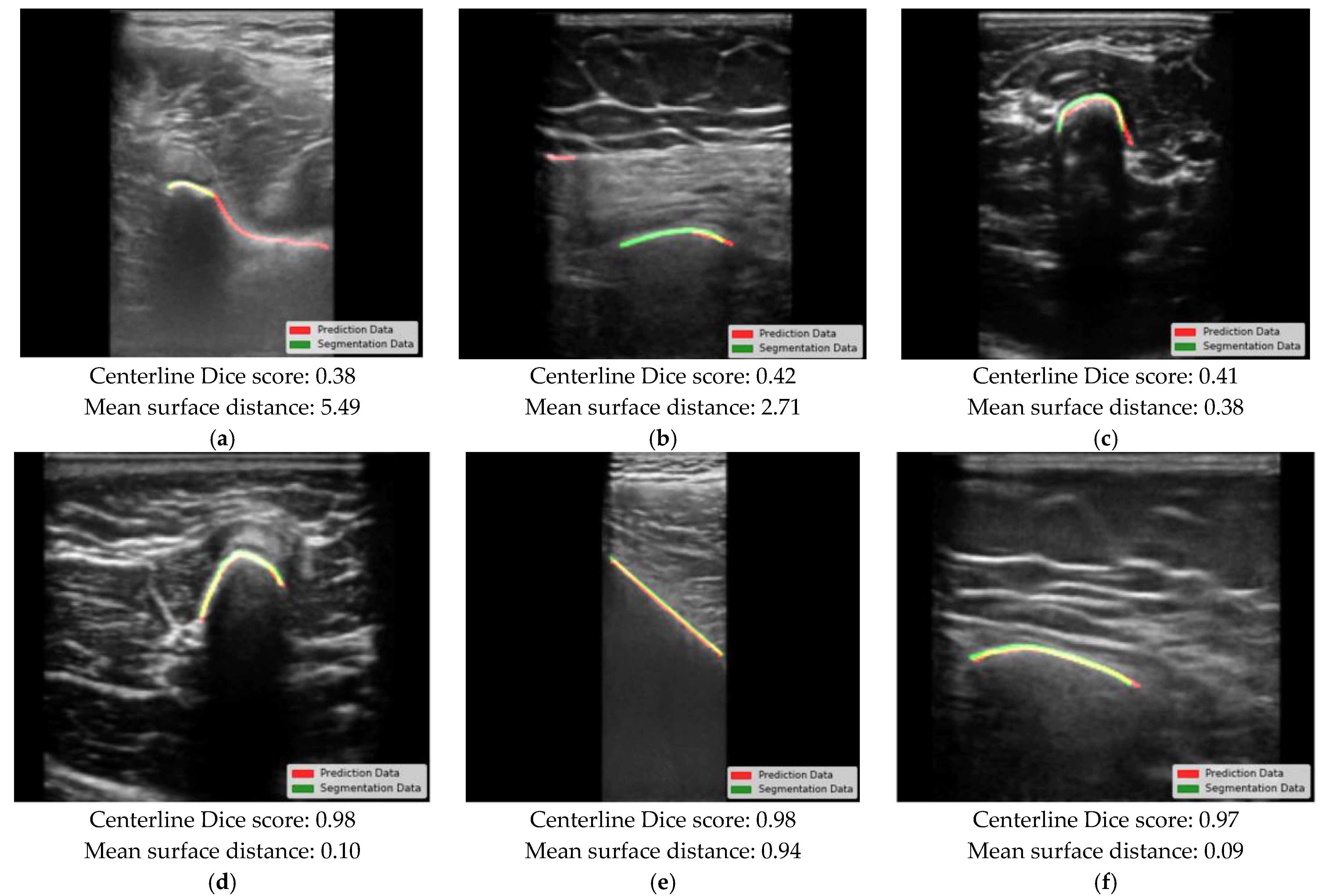
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3D | Three-dimensional |
| IQR | Inter quartile range |
| SD | Standard deviation |
References
- Zheng, G.; Kowal, J.; González Ballester, M.A.; Caversaccio, M.; Nolte, L.-P. (i) Registration techniques for computer navigation. Curr. Orthop. 2007, 21, 170–179. [Google Scholar] [CrossRef]
- Pearce, M.S.; Salotti, J.A.; Little, M.P.; McHugh, K.; Lee, C.; Kim, K.P.; Howe, N.L.; Ronckers, C.M.; Rajaraman, P.; Craft, A.W.; et al. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: A retrospective cohort study. Lancet 2012, 380, 499–505. [Google Scholar] [CrossRef]
- Amin, D.V.; Kanade, T.; Digioia, A.M.; Jaramaz, B. Ultrasound Registration of the Bone Surface for Surgical Navigation. Comput. Aided Surg. 2003, 8, 1–16. [Google Scholar] [CrossRef]
- Yan, C.X.B.; Goulet, B.; Pelletier, J.; Chen, S.J.-S.; Tampieri, D.; Collins, D.L. Towards accurate, robust and practical ultrasound-CT registration of vertebrae for image-guided spine surgery. Int. J. CARS 2011, 6, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Gueziri, H.-E. Toward real-time rigid registration of intra-operative ultrasound with preoperative CT images for lumbar spinal fusion surgery. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1933–1943. [Google Scholar] [CrossRef]
- Gueziri, H.-E.; Georgiopoulos, M.; Santaguida, C.; Collins, D.L. Ultrasound-based navigated pedicle screw insertion without intraoperative radiation: Feasibility study on porcine cadavers. Spine J. 2022, 22, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Schumann, S. State of the Art of Ultrasound-Based Registration in Computer Assisted Orthopedic Interventions. In Computational Radiology for Orthopaedic Interventions; Zheng, G., Li, S., Eds.; Lecture Notes in Computational Vision and Biomechanics; Springer International Publishing: Cham, Switzerland, 2016; Volume 23, pp. 271–297. ISBN 978-3-319-23481-6. [Google Scholar]
- Jia, R.; Mellon, S.; Hansjee, S.; Monk, A.P.; Murray, D.; Noble, J.A. Automatic Bone Segmentation in Ultrasound Images Using Local Phase Features and Dynamic Programming. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1005–1008. [Google Scholar]
- Hacihaliloglu, I. Ultrasound imaging and segmentation of bone surfaces: A review. Technology 2017, 5, 74–80. [Google Scholar] [CrossRef]
- Hiep, M.A.J.; Heerink, W.J.; Groen, H.C.; Saiz, L.A.; Grotenhuis, B.A.; Beets, G.L.; Aalbers, A.G.J.; Kuhlmann, K.F.D.; Ruers, T.J.M. Real-time intraoperative ultrasound registration for accurate surgical navigation in patients with pelvic malignancies. Int. J. CARS 2024, 20, 249–258. [Google Scholar] [CrossRef]
- van Sloun, R.J.; Cohen, R.; Eldar, Y.C. Deep learning in ultrasound imaging. Proc. IEEE 2020, 108, 11–29. [Google Scholar] [CrossRef]
- Noble, J.A.; Boukerroui, D. Ultrasound image segmentation: A survey. IEEE Trans. Med. Imaging 2006, 25, 987–1010. [Google Scholar] [CrossRef]
- Hohlmann, B.; Broessner, P.; Radermacher, K. Ultrasound-based 3D bone modelling in computer assisted orthopedic surgery—A review and future challenges. Comput. Assist. Surg. 2024, 29, 2276055. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.U.; Quader, N.; Guy, P.; Garbi, R.; Hodgson, A.J. Ultrasound Bone Segmentation: A Scoping Review of Techniques and Validation Practices. Ultrasound Med. Biol. 2020, 46, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Munich, Germany, 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Pandey, P. Fast and automatic bone segmentation and registration of 3D ultrasound to CT for the full pelvic anatomy: A comparative study. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1515–1524. [Google Scholar] [CrossRef]
- Jain, A.K.; Taylor, R.H. Understanding bone responses in B-mode ultrasound images and automatic bone surface extraction using a Bayesian probabilistic framework. In Medical Imaging 2004; Walker, W.F., Emelianov, S.Y., Eds.; SPIE: San Diego, CA, USA, 2004; p. 131. [Google Scholar]
- Quader, N.; Hodgson, A.J.; Mulpuri, K.; Schaeffer, E.; Abugharbieh, R. Automatic Evaluation of Scan Adequacy and Dysplasia Metrics in 2-D Ultrasound Images of the Neonatal Hip. Ultrasound Med. Biol. 2017, 43, 1252–1262. [Google Scholar] [CrossRef] [PubMed]
- El-Hariri, H.; Mulpuri, K.; Hodgson, A.; Garbi, R. Comparative evaluation of Hand-Engineered and Deep-Learned features for neonatal hip bone segmentation in ultrasound. In Medical Image Computing and Computer Assisted Intervention; Shen, D., Ed.; Springer International Publishing: Shenzhen, China, 2019; pp. 12–20. [Google Scholar]
- Van Der Woude, R.; Fitski, M.; Van Der Zee, J.M.; Van De Ven, C.P.; Bökkerink, G.M.J.; Wijnen, M.H.W.A.; Meulstee, J.W.; Van Doormaal, T.P.C.; Siepel, F.J.; Van Der Steeg, A.F.W. Clinical Application and Further Development of Augmented Reality Guidance for the Surgical Localization of Pediatric Chest Wall Tumors. J. Pediatr. Surg. 2024, 59, 1549–1555. [Google Scholar] [CrossRef]
- Guillerman, R.P. Marrow: Red, yellow and bad. Pediatr. Radiol. 2013, 43, 181–192. [Google Scholar] [CrossRef]
- Bronner, F.; Farach-Carson, M.C.; Roach, H.I. (Eds.) Bone and Development; Springer: London, UK, 2010; ISBN 978-1-84882-821-6. [Google Scholar]
- Valanarasu, J.M.J.; Patel, V.M. UNeXt: MLP-based Rapid Medical Image Segmentation Network. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer Nature: Cham, Switzerland, 2022; pp. 23–33. [Google Scholar]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Ungi, T.; Lasso, A.; Fichtinger, G. Open-source platforms for navigated image-guided interventions. Med. Image Anal. 2016, 33, 181–186. [Google Scholar] [CrossRef]
- Zaman, A.; Park, S.H.; Bang, H.; Park, C.; Park, I.; Joung, S. Generative approach for data augmentation for deep learning-based bone surface segmentation from ultrasound images. Int. J. CARS 2020, 15, 931–941. [Google Scholar] [CrossRef]
- Shit, S.; Paetzold, J.C.; Sekuboyina, A.; Ezhov, I.; Unger, A.; Zhylka, A.; Pluim, J.P.W.; Bauer, U.; Menze, B.H. clDice—A Novel Topology-Preserving Loss Function for Tubular Structure Segmentation. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 16555–16564. [Google Scholar]
- Gebhardt, C.; Göttling, L.; Buchberger, L.; Ziegler, C.; Endres, F.; Wuermeling, Q.; Holzapfel, B.M.; Wein, W.; Wagner, F.; Zettinig, O. Femur reconstruction in 3D ultrasound for orthopedic surgery planning. Int. J. Comput. Assist. Radiol. Surg. 2023, 18, 1001–1008. [Google Scholar] [CrossRef]
- Sørensen, T.J. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species Content and Its Application to Analyses of the Vegetation on Danish Commons; Kongelige Danske Videnskabernes Selskab: Kobenhaven, Denmark, 1948. [Google Scholar]
- Hers, B.; Bonta, M.; Du, S.; Mulpuri, K.; Schaeffer, E.K.; Hodgson, A.J.; Garbi, R. SegFormer3D: Improving the Robustness of Deep Learning Model-Based Image Segmentation in Ultrasound Volumes of the Pediatric Hip. Ultrasound Med. Biol. 2025, 51, 751–758. [Google Scholar] [CrossRef]
- Laursen, C.B.; Clive, A.; Hallifax, R.; Pietersen, P.I.; Asciak, R.; Davidsen, J.R.; Bhatnagar, R.; Bedawi, E.O.; Jacobsen, N.; Coleman, C.; et al. European Respiratory Society statement on thoracic ultrasound. Eur. Respir. J. 2021, 57, 2001519. [Google Scholar] [CrossRef]
- Buser, M.A.D.; Van Der Rest, J.K.; Wijnen, M.H.W.A.; De Krijger, R.R.; Van Der Steeg, A.F.W.; Van Den Heuvel-Eibrink, M.M.; Reismann, M.; Veldhoen, S.; Pio, L.; Markel, M. Deep Learning and Multidisciplinary Imaging in Pediatric Surgical Oncology: A Scoping Review. Cancer Med. 2025, 14, e70574. [Google Scholar] [CrossRef]
- Wu, L.; Cavalcanti, N.A.; Seibold, M.; Loggia, G.; Reissner, L.; Hein, J.; Beeler, S.; Viehöfer, A.; Wirth, S.; Calvet, L.; et al. UltraBones100k: A reliable automated labeling method and large-scale dataset for ultrasound-based bone surface extraction. Comput. Biol. Med. 2025, 194, 110435. [Google Scholar] [CrossRef]
- van der Zee, J.M. Tracked ultrasound registration for intraoperative navigation during pediatric bone tumor resections with soft tissue components: A porcine cadaver study. Int. J. Comput. Assist. Radiol. Surg. 2023, 19, 297–302. [Google Scholar] [CrossRef]
| Osseous Structure | 0–9 Years (n = 7) | 10–19 Years (n = 9) | Total (n = 16) |
|---|---|---|---|
| Mean Age: 3.5 +/− 2.7 Years | Mean Age: 14.7 +/− 2.0 Years | Mean Age: 9.8 +/− 6.0 Years | |
| Pelvis | 699 | 491 | 1190 |
| Ribs/Sternum | 420 | 277 | 697 |
| Radius/Ulna | 154 | 126 | 280 |
| Humerus | 365 | 378 | 743 |
| Femur | 176 | 386 | 562 |
| Tibia/Fibula | 442 | 395 | 837 |
| Total | 2256 | 2053 | 4309 + 363 empty labels = 4672 |
| Layer | Type | Number of Parameters |
|---|---|---|
| Encoder1_Conv2D | Conv2d | 160 |
| Encoder1_BatchNorm | BatchNorm2d | 32 |
| Encoder2_Conv2D | Conv2d | 4640 |
| Encoder2_BatchNorm | BatchNorm2d | 64 |
| Encoder3_Conv2D | Conv2d | 36,992 |
| Encoder3_BatchNorm | BatchNorm2d | 256 |
| PatchEmbed3_Conv2D | Conv2d | 184,480 |
| PatchEmbed4_Conv2D | Conv2d | 368,896 |
| ShiftMLP1_fc1 | Linear | 25,760 |
| ShiftMLP1_fc2 | Linear | 25,760 |
| ShiftMLP2_fc1 | Linear | 65,792 |
| ShiftMLP2_fc2 | Linear | 65,792 |
| Decoder1_Conv2D | Conv2d | 368,800 |
| Decoder2_Conv2D | Conv2d | 184,448 |
| Decoder3_Conv2D | Conv2d | 36,896 |
| Decoder4_Conv2D | Conv2d | 4624 |
| Decoder5_Conv2D | Conv2d | 2320 |
| Final_1 × 1_Conv | Conv2d (1 × 1) | 17 |
| Total: 1,375,712 parameters |
| Model | Centerline Dice Score (Mean ± SD) | Mean Surface Distance (Mean ± SD), Direction of Segmentation [mm] | Computation Time Per Million Pixels | ||||
|---|---|---|---|---|---|---|---|
| All Ages | <10 Years | >10 Years | All Ages [mm] | <10 Years [mm] | >10 Years [mm] | All Ages [ms] | |
| 1 | 0.80 ± 0.15 | 0.77 ± 0.17 | 0.83 ± 0.12 | 0.86 ± 1.04, 0.00 | 0.93 ± 1.35, −0.17 | 0.77 ± 0.69, +0.13 | 1.21 |
| 2 | 0.81 ± 0.14 | 0.78 ± 0.17 | 0.84 ± 0.10 | 0.82 ± 1.00, +0.07 | 0.98 ± 1.37, −0.07 | 0.69 ± 0.52, +0.18 | 0.79 |
| 3 | 0.82 ± 0.14 | 0.79 ± 0.17 | 0.84 ± 0.11 | 0.91 ± 1.08, +0.11 | 1.09 ± 1.46, +0.05 | 0.77 ± 0.61, +0.16 | 0.84 |
| 4 | 0.84 ± 0.14 | 0.81 ± 0.17 | 0.86 ± 0.10 | 0.48 ± 1.30, +0.21 | 0.74 ± 1.89, +0.19 | 0.28 ± 0.33, +0.21 | 1.12 |
| 5 | 0.84 ± 0.14 | 0.83 ± 0.16 | 0.85 ± 0.11 | 0.51 ± 0.93, +0.07 | 0.65 ± 1.23, +0.09 | 0.40 ± 0.48, +0.05 | 0.85 |
| Weighted model | 0.85 ± 0.13 | 0.82 ± 0.16 | 0.87 ± 0.09 | 0.78 ± 1.15, +0.05 | 1.0 ± 1.63, +0.07 | 0.61 ± 0.46, +0.03 | Not specified |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zee, J.M.v.d.; Rahman, A.M.; Gunnewiek, K.K.; Hiep, M.A.J.; Fitski, M.; Hacihaliloglu, I.; Alsinan, A.Z.; Patel, V.M.; Littooij, A.S.; Steeg, A.F.W.v.d. Evaluation of UNeXt for Automatic Bone Surface Segmentation on Ultrasound Imaging in Image-Guided Pediatric Surgery. Bioengineering 2025, 12, 1008. https://doi.org/10.3390/bioengineering12101008
Zee JMvd, Rahman AM, Gunnewiek KK, Hiep MAJ, Fitski M, Hacihaliloglu I, Alsinan AZ, Patel VM, Littooij AS, Steeg AFWvd. Evaluation of UNeXt for Automatic Bone Surface Segmentation on Ultrasound Imaging in Image-Guided Pediatric Surgery. Bioengineering. 2025; 12(10):1008. https://doi.org/10.3390/bioengineering12101008
Chicago/Turabian StyleZee, Jasper M. van der, Aimon M. Rahman, Kevin Klein Gunnewiek, Marijn A. J. Hiep, Matthijs Fitski, Ilker Hacihaliloglu, Ahmed Z. Alsinan, Vishal M. Patel, Annemieke S. Littooij, and Alida F. W. van der Steeg. 2025. "Evaluation of UNeXt for Automatic Bone Surface Segmentation on Ultrasound Imaging in Image-Guided Pediatric Surgery" Bioengineering 12, no. 10: 1008. https://doi.org/10.3390/bioengineering12101008
APA StyleZee, J. M. v. d., Rahman, A. M., Gunnewiek, K. K., Hiep, M. A. J., Fitski, M., Hacihaliloglu, I., Alsinan, A. Z., Patel, V. M., Littooij, A. S., & Steeg, A. F. W. v. d. (2025). Evaluation of UNeXt for Automatic Bone Surface Segmentation on Ultrasound Imaging in Image-Guided Pediatric Surgery. Bioengineering, 12(10), 1008. https://doi.org/10.3390/bioengineering12101008





