AI-Powered Telemedicine for Automatic Scoring of Neuromuscular Examinations
Abstract
1. Introduction
- (1)
- The quantitation of neurological deficits is an improvement of the existing outcome measures that are subject to human evaluation with variability among examiners, are at best categorical in nature and not continuous metrics, and are dependent on the availability of examiners. We are also able to capture deficits that are better detected by our technology, which human assessments may potentially miss [11].
- (2)
- Since measures can be performed remotely, a central evaluation unit with a limited number of highly trained, research coordinators can perform evaluations in a more uniform fashion [12].
- (3)
- Inteleclinic allows more complex data collection in a remote location; therefore, research subjects do not need to travel to clinic sites.
- (4)
- The solution has the potential to broaden clinical trial participation to poorly represented groups, including international participation, because of reduced barriers such as excessive travel time, conflicting employment requirements, or the subject’s own disability [10]. We have constructed the computational framework in [11,13] to assess MG-CE scores digitally. To validate our algorithms, we applied them to a large cohort of patients with MG and a diverse control group. The results indicate that our approach yields outcomes that are consistently comparable to those of expert clinicians for the majority of examination metrics under standard clinical conditions [13]. We also determined which parts of the examination were not reliable and revised our understanding of how certain sections of the MG-CE should be interpreted [9].
2. Materials and Methods
2.1. Subjects and Video Recording
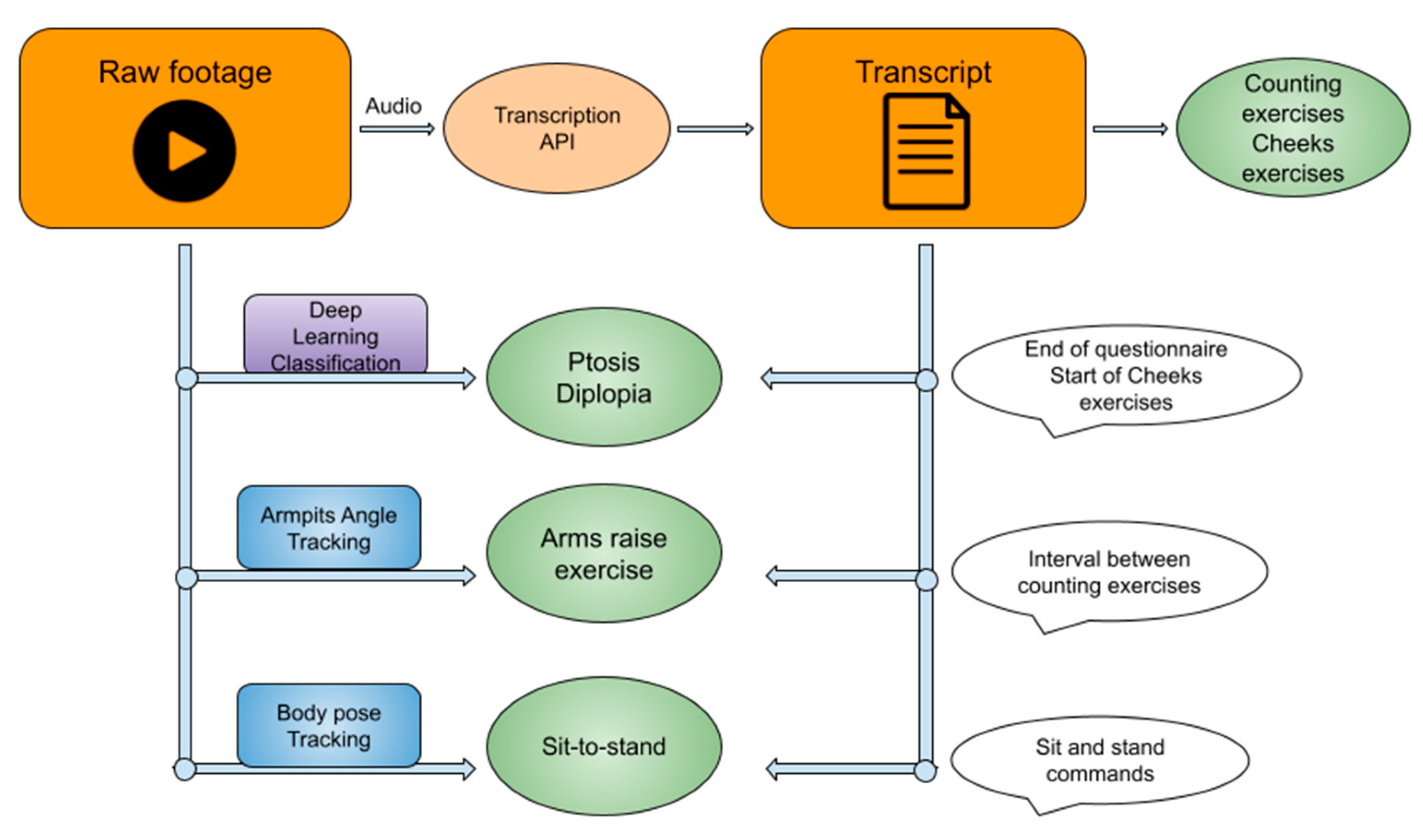
2.2. Overall Algorithm Approach
2.3. General NLP Method
2.4. Counting Exercises
2.5. Cheek Puff and Tongue-to-Cheek Exercises
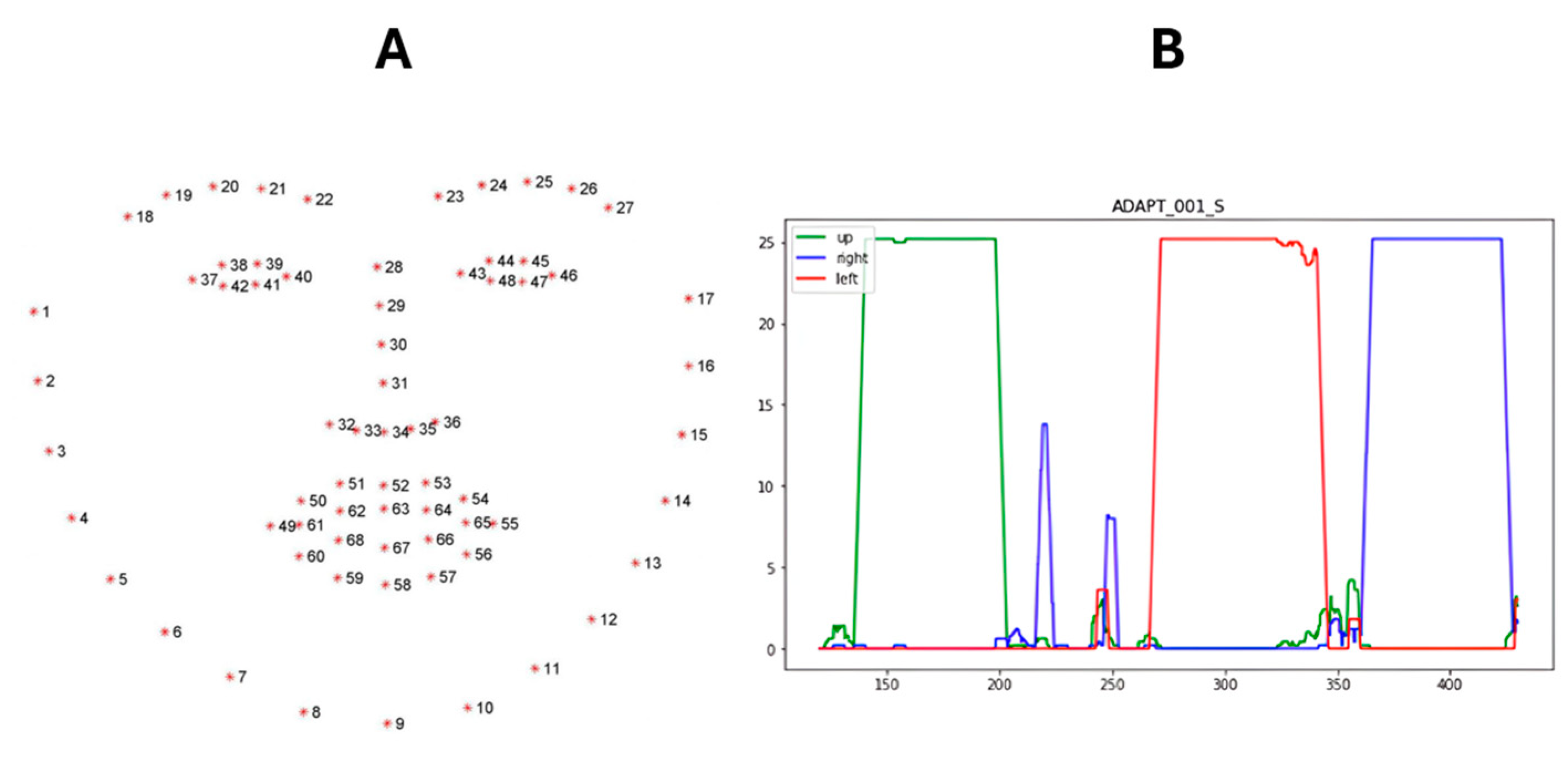
2.6. Ocular Exercises
- Pseudo Code Ocular Exercises Search
- Input:
- Video File
- Transcript Dictionary
- Process:
- 1.
- Identify Ocular Phase (using Transcript Dictionary):
- ○
- Search for the end of the “Questionnaire” section in the transcript dictionary.
- ○
- Search for the start of the “Cheek Puff” exercise in the transcript dictionary.
- ○
- Return the start and end timestamps of the identified ocular phase window.
- 2.
- Process Video Frames (using Computer Vision):
- ○
- For each frame in the video,
- ■
- If no face is detected:
- ■
- Return “No Value”
- ■
- If a face is detected:
- ■
- Use Dlib to place landmarks on facial features.
- ■
- Crop the frame to the region containing both eyes based on the landmarks.
- ■
- Classify the cropped eye region using the CNN.
- ■
- If the classification confidence is greater than 0.95,
- ■
- Record the classified gaze direction (up, left, right, or neutral).
- ■
- Otherwise:
- ■
- Record “No Value”
- ○
- Return a list of gaze directions and their corresponding timestamps.
- 3.
- Analyze Gaze Directions:
- ○
- For each gaze direction in the list,
- ■
- Calculate a 5-s rolling window density curve for that direction.
- ■
- Identify the timestamps of the longest segment in the density curve exceeding 60% of the maximum density.
- ○
- Return a list of significant timestamps corresponding to the detected exercises.
- Output:
- Ptosis timestamps (identified as the longest “up” gaze segment)
- Diplopia Right timestamps (identified as the longest “right” gaze segment)
- Diplopia Left timestamps (identified as the longest “left” gaze segment)
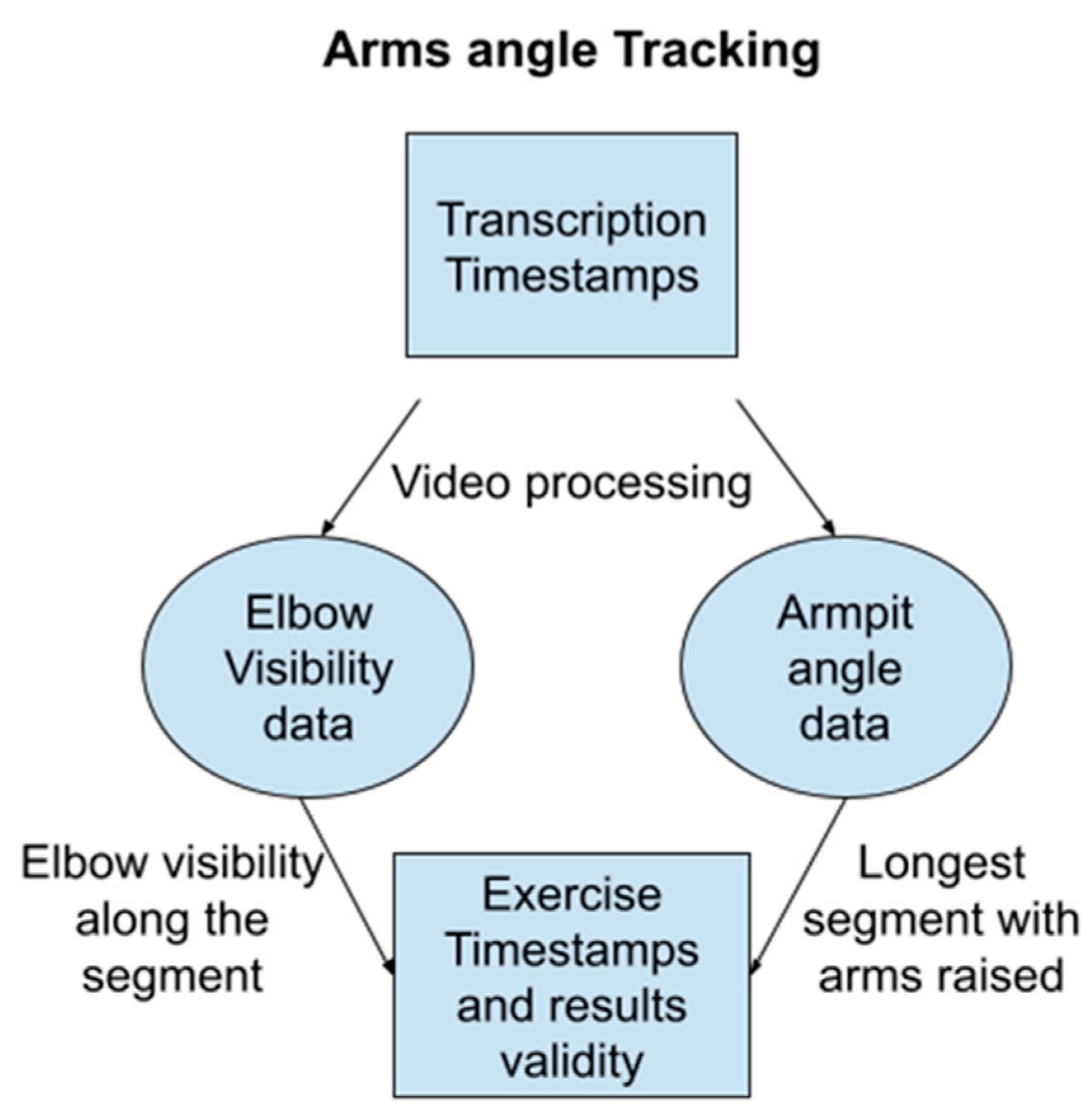
2.7. Full Body Exercises
- Pseudo Code: Arm Raise Search
- Input:
- Video File
- Process:
- 1.
- Iterate Through Video Frames:
- ○
- For each frame in the video:
- ■
- Body Detection:
- ■
- If a body is not detected using MediaPipe, return “No Value”.
- ■
- If a body is detected, proceed to the next steps.
- ■
- Landmark Placement: Place landmarks on the body using MediaPipe.
- ■
- Arm Visibility Metric: Record a metric representing arm visibility (e.g., percentage of arm area visible).
- ■
- Armpit Angle Calculation: Calculate the angles of both armpits based on the landmarks.
- ■
- Return Values: Return a list of arm visibility metrics, a list of timestamps corresponding to each frame, and a list of armpit angles.
- 2.
- Segment Identification:
- ○
- Longest Segment Search: Identify the longest continuous segment where
- ■
- Arm visibility metric is greater than 0.9 (e.g., at least 90% of the arm is visible).
- ■
- Armpit angle is greater than 45 degrees (indicating raised arms).
- ■
- This step can be accomplished using techniques like dynamic programming or a sliding window.
- 3.
- Exercise Timestamps: Return the timestamps corresponding to the beginning and end of the identified segment as the arm exercise timestamps.
- Output:
- Arm Exercise Timestamps (start and end)
- Pseudo Code: Sit-To-Stand Search
- Input:
- Transcript Dictionary
- List of Validation Words (words confirming sit-to-stand execution)
- List of Explanation Words (words preceding sit-to-stand instructions)
- Process:
- Keyword Search in Transcript:
- ○
- Search the transcript dictionary for occurrences of both Validation Words and Explanation Words.
- Start of Exercise:
- ○
- Identify the start of the exercise as the first occurrence of two Validation Words within a 5-s window in the transcript.
- Segment Building with Chains
- ○
- For each Explanation Word found in the transcript:
- ■
- Start a 20-s chain.
- ■
- If any Validation Word is found within the current chain, extend the chain duration by 10 s.
- ○
- The end of the segment is identified as the last word within the most recently extended chain.
- Output Timestamps: Return the timestamps corresponding to the start and end of the identified segment as the sit-to-stand exercise timestamps.
- Output:
- Sit-To-Stand Timestamps (start and end)
- -
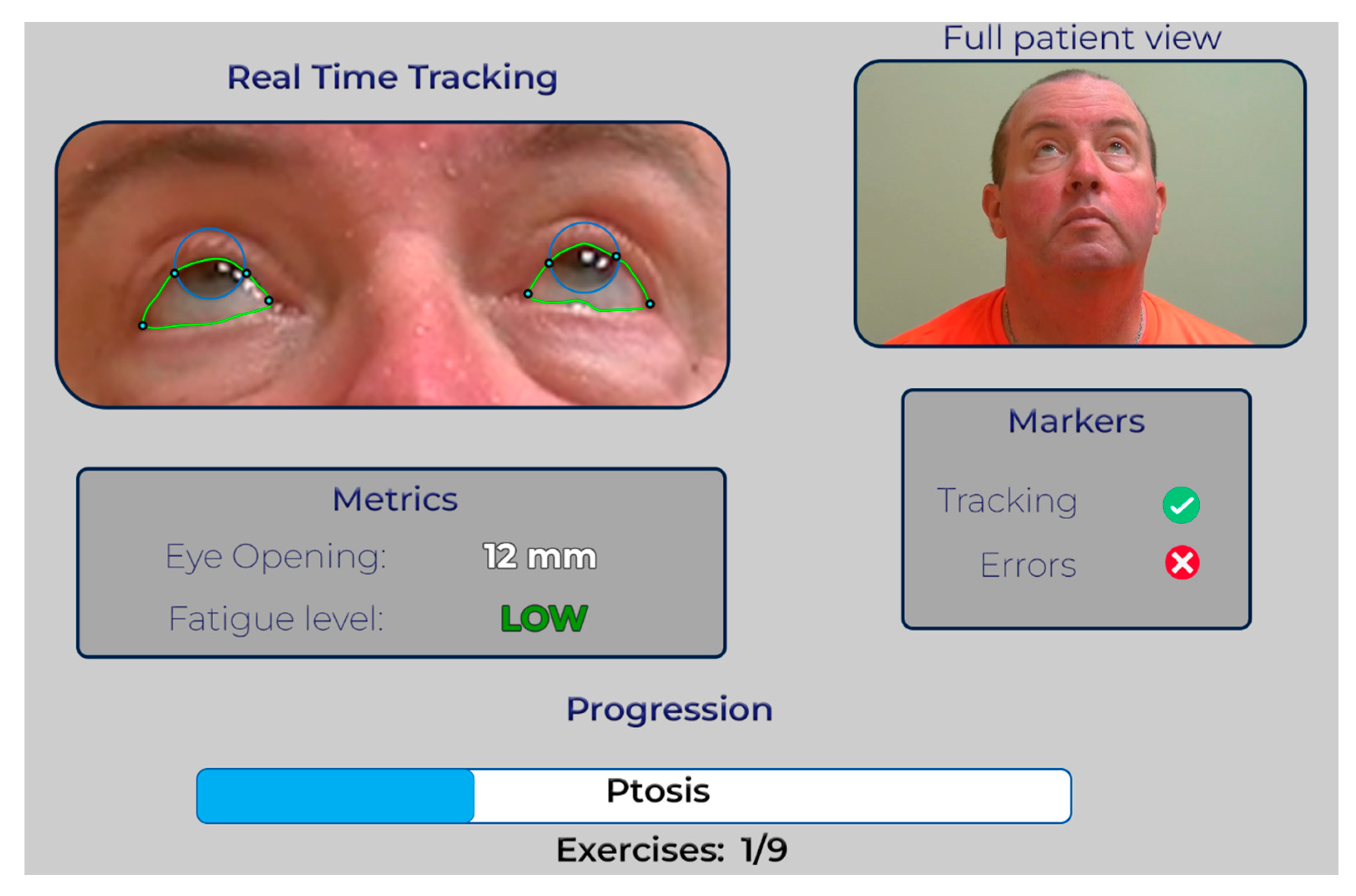
- Efficient Video Review: The GUI allows rapid playback of the patient’s video, focusing solely on the test segments the patient performed.
- -
- Annotated Landmarks: The video is overlaid with graphical annotations highlighting relevant anatomical landmarks during each test. This provides visual cues for the neurologist to assess the scoring accuracy.
- -
- Error Identification: In scenarios where our algorithms encounter difficulties, such as tracking the iris or eyelid during the ptosis exercise, the annotated video will clearly indicate potential scoring errors. This allows the neurologist to quickly identify issues and make informed decisions.
- -
- Fast Forwarding Non-Exercise Segments: During periods when the patient is not actively performing exercises, the video playback speeds up by a factor of 10 to minimize review time.
- -
- Slow Motion for Exercises: During exercise segments, the video playback slows down significantly, allowing the neurologist to clearly see the annotated landmarks and confirm scoring accuracy. This balance ensures an efficient review while maintaining sufficient detail.
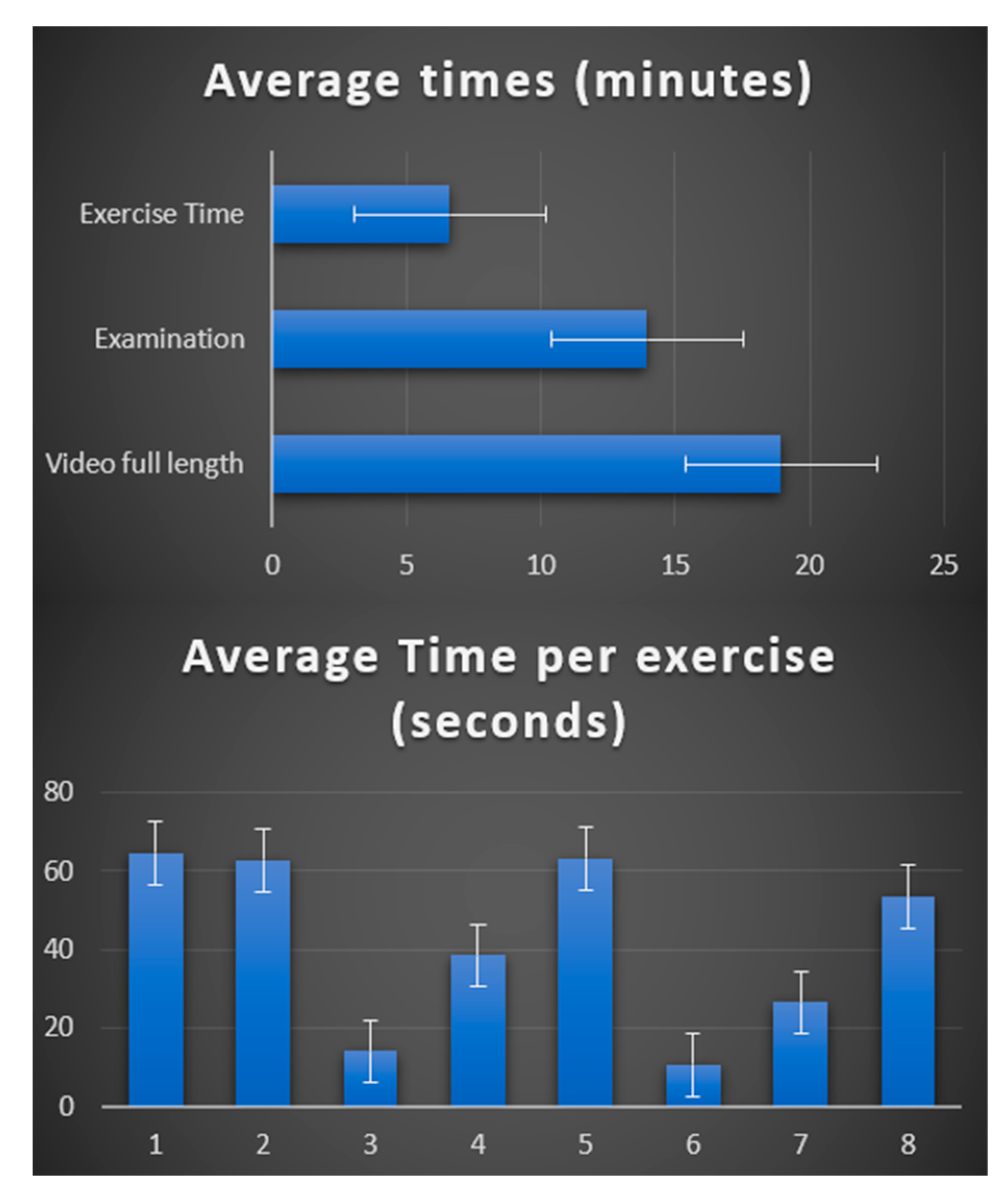
3. Results
- The MGNet Project: This dataset comprises 86 videos of neurological clinical examinations for 51 myasthenia gravis (MG) patients across five different centers, performed by eight certified neurologists.
- Control Subjects: To account for potential variability in clinical examinations, we also processed a dataset of 24 videos featuring healthy control subjects who underwent neurological examinations under strict protocol adherence [15].
3.1. Ocular Exercises
3.2. Speech Test
3.3. Cheek Puff and Tongue-to-Cheek
3.4. Arm Extension
3.5. Sit-to-Stand
4. Discussion
- Video Segmentation: Individual test segments are extracted from the raw clinical examination video.
- Automated Scoring: Specialized software computes the MG score for each segmented test.
- Clinician Review: A Graphical User Interface (GUI) facilitates clinician review of the results and validation of the scoring.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Phase | Keywords |
|---|---|
| Cheek Puff | Cheek puff |
| Cheeks | |
| Puff your cheeks out | |
| Puff your cheek out | |
| Puff out your cheeks | |
| Expanding your cheeks | |
| Tongue-to-Cheek | Tongue-to-cheek |
| Tongue | |
| Tongue push | |
| Tongue in cheek | |
| Tongue into your cheek | |
| Place your tongue | |
| Start of ocular phase | Drooping eyelids |
| Look up | |
| At the ceiling | |
| Double vision | |
| End of ocular phase | Cheek puff |
| Tongue-to-cheek | |
| Cheek/Cheeks | |
| Tongue | |
| Sit-to-stand | Cross your arms |
| Stand up | |
| Sit-to-stand | |
| Sit back down | |
| Sit down | |
| Stand | |
| Validation words and expressions | Good |
| Perfect | |
| All right | |
| Alright | |
| Great | |
| Excellent | |
| Awesome |
Appendix B. Reasons for Removal of Video from Analysis
- General
- Alternating views from examiner view to patient view during test by itself disrupt the possible digital processing of the patient view for 12 videos out of 99 visits.
- Ptosis Test
- Face not included in the image frame, i.e., patient too close to camera
- Face too far from camera leading to lower resolution
- Subject holding head back
- Poor light conditions
- Frontalis Activation
- Target subject is looking at is not high enough
- Diplopia Test
- In addition to Ptosis is test
- Subject’s head turned in gaze direction
- Arm Extension
- Subject too close to camera (Hand and portion of arm not visible)
- Loose garment hides arm contour, especially portion of arm closest to the ground
- Sit-to-Stand test
- Patient too close to camera with either or both
- ○
- Head disappearing during stand up
- ○
- Hip missing during stand up
- Counting to 50 and counting on one breath
- Volume is too low
- External noise perturbs the record.
References
- Giannotta, M.; Petrelli, C.; Pini, A. Telemedicine Applied to Neuromuscular Disorders: Focus on the COVID-19 Pandemic Era. Acta Myol. 2022, 41, 30. [Google Scholar] [CrossRef]
- Spina, E.; Trojsi, F.; Tozza, S.; Iovino, A.; Iodice, R.; Passaniti, C.; Abbadessa, G.; Bonavita, S.; Leocani, L.; Tedeschi, G.; et al. How to Manage with Telemedicine People with Neuromuscular Diseases? Neurol. Sci. 2021, 42, 3553–3559. [Google Scholar] [CrossRef] [PubMed]
- Hooshmand, S.; Cho, J.; Singh, S.; Govindarajan, R. Satisfaction of Telehealth in Patients with Established Neuromuscular Disorders. Front. Neurol. 2021, 12, 667813. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, D.; Casagrande, S.; Iodice, F.; Orlando, B.; Trojsi, F.; Cirillo, G.; Clerico, M.; Bozzali, M.; Leocani, L.; Abbadessa, G.; et al. Myasthenia Gravis and Telemedicine: A Lesson from COVID-19 Pandemic. Neurol. Sci. 2021, 42, 4889–4892. [Google Scholar] [CrossRef] [PubMed]
- Dresser, L.; Wlodarski, R.; Rezania, K.; Soliven, B. Myasthenia Gravis: Epidemiology, Pathophysiology and Clinical Manifestations. J. Clin. Med. 2021, 10, 2235. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Benatar, M.; Cutter, G.; Kaminski, H.J. The best and worst of times in therapy development for myasthenia gravis. Muscle Nerve 2023, 67, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Benatar, M.; Sanders, D.B.; Burns, T.M.; Cutter, G.R.; Guptill, J.T.; Baggi, F.; Kaminski, H.J.; Mantegazza, R.; Meriggioli, M.N.; Quan, J.; et al. Recommendations for myasthenia gravis clinical trials. Muscle Nerve 2012, 45, 909–917. [Google Scholar] [CrossRef]
- Guptill, J.T.; Benatar, M.; Granit, V.; Habib, A.A.; Howard, J.F.; Barnett-Tapia, C.; Nowak, R.J.; Lee, I.; Ruzhansky, K.; Dimachkie, M.M.; et al. Addressing Outcome Measure Variability in Myasthenia Gravis Clinical Trials. Neurology 2023, 101, 442–451. [Google Scholar] [CrossRef]
- Guidon, A.C.; Muppidi, S.; Nowak, R.J.; Guptill, J.T.; Hehir, M.K.; Ruzhansky, K.; Burton, L.B.; Post, D.; Cutter, G.; Conwit, R.; et al. Telemedicine Visits in Myasthenia Gravis: Expert Guidance and the Myasthenia Gravis Core Exam (MG-CE). Muscle Nerve 2021, 64, 270–276. [Google Scholar] [CrossRef]
- Carter-Edwards, L.; Hidalgo, B.; Lewis-Hall, F.; Nguyen, T.; Rutter, J. Diversity, equity, inclusion, and access are necessary for clinical trial site readiness. J. Clin. Transl. Sci. 2023, 7, e268. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Garbey, M.; Joerger, G.; Lesport, Q.; Girma, H.; McNett, S.; Abu-Rub, M.; Kaminski, H. A Digital Telehealth System to Compute Myasthenia Gravis Core Examination Metrics: Exploratory Cohort Study. JMIR Neurotechnol. 2023, 2, e43387. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, V.; Sullivan, J. One Millisecond Face Alignment with an Ensemble of Regression Trees. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1867–1874. [Google Scholar] [CrossRef]
- Lesport, Q.; Joerger, G.; Kaminski, H.J.; Girma, H.; McNett, S.; Abu-Rub, M.; Garbey, M. Eye Segmentation Method for Telehealth: Application to the Myasthenia Gravis Physical Examination. Sensors 2023, 23, 7744. [Google Scholar] [CrossRef] [PubMed]
- Garbey, M.; Lesport, Q.; Girma, H.; Oztosun, G.; Abu-Rub, M.; Guidon, A.C.; Juel, V.; Nowak, R.; Soliven, B.; Aban, I.; et al. Application of Digital Tools and Artificial Intelligence to the Myasthenia Gravis Core Examination. medRxiv 2024. [Google Scholar] [CrossRef]
- Garbey, M. A Quantitative Study of Factors Influencing Myasthenia Gravis Telehealth Examination Score. medRxiv 2024. [Google Scholar] [CrossRef]
- AssemblyAI. Available online: https://www.assemblyai.com/ (accessed on 30 September 2023).
- OpenCV Haar Cascade Eye Detector. Available online: https://github.com/opencv/opencv/blob/master/data/haarcascades/haarcascade_eye.xml (accessed on 30 March 2022).
- Bazarevsky, V.; Grishchenko, I.; Raveendran, K.; Zhu, T.; Zhang, F.; Grundmann, M. BlazePose: On-device Real-time Body Pose Tracking. arXiv 2020, arXiv:2006.10204. Available online: http://arxiv.org/abs/2006.10204 (accessed on 20 October 2023).
- Aristidou, A.; Jena, R.; Topol, E.J. Bridging the chasm between AI and clinical implementation. Lancet 2022, 399, 620. [Google Scholar] [CrossRef]
- Acosta, J.N.; Falcone, G.J.; Rajpurkar, P.; Topol, E.J. Multimodal biomedical AI. Nat. Med. 2022, 28, 1773–1784. [Google Scholar] [CrossRef]
- Tawa, N.; Rhoda, A.; Diener, I. Accuracy of clinical neurological examination in diagnosing lumbo-sacral radiculopathy: A systematic literature review. BMC Musculoskelet. Disord. 2017, 18, 93. [Google Scholar] [CrossRef]
- Jellinger, K.A.; Logroscino, G.; Rizzo, G.; Copetti, M.; Arcuti, S.; Martino, D.; Fontana, A. Accuracy of clinical diagnosis of Parkinson disease: A systematic review and meta-analysis. Neurology 2016, 86, 566–576. [Google Scholar] [CrossRef]
- Heneghan, C.; Goldacre, B.; Mahtani, K.R. Why clinical trial outcomes fail to translate into benefits for patients. Trials 2017, 18, 122. [Google Scholar] [CrossRef]
- Lee, M.; Kang, D.; Joi, Y.; Yoon, J.; Kim, Y.; Kim, J.; Kang, M.; Oh, D.; Shin, S.Y.; Cho, J. Graphical user interface design to improve understanding of the patient-reported outcome symptom response. PLoS ONE 2023, 18, e0278465. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]


| MG-CE Scores | Normal | Mild | Moderate | Severe | Item Score |
|---|---|---|---|---|---|
| Ptosis (61 s upgaze) | 0 no ptosis | 1 Eyelid above pupil | 2 Eyelid at pupil | 3 Eyelid below pupil | |
| Diplopia ** | 0 No diplopia with 61 s sustained gaze | 1 Diplopia with 11–60 s sustained gaze | 2 Diplopia 1–10 s but not immediate | 3 Immediate diplopia with primary or lateral gaze | |
| Cheek puff | 0 Normal seal | 1 Transverse pucker | 2 Opposes lips, but air escapes | 3 Cannot oppose lips or puff cheeks | |
| Tongue-to-cheek ** | 0 Full convex deformity in cheeks | 1 Partial convex deformity in cheeks | 2 Able to move tongue-to-cheek but not deform | 3 Unable to move tongue into cheek at all | |
| Counting to 50 | 0 No dysarthria at 50 | 1 Dysarthria at 30–49 | 2 Dysarthria at 10–29 | 3 Dysarthria at 1–9 | |
| Arm strength ** (shoulder abduction) | 0 No drift during >120 s | 1 Drift 90–120 s | 2 Drift 10–89 s | 3 Drif 0–9 s | |
| Single breath count | 0 SBC > 30 | 1 SBC 25–29 | 2 SBC 20–24 | 3 SBC < 20 | |
| Sit-to-stand | 0 Sit-to-stand with arms crossed, no difficulty | 1 Slow/extra effort | 2 Uses arms/hands | 3 Unable to stand unassisted | |
| ** When the test is performed on the right and left, score the more severe side | Total score: /24 | ||||
| Class | Up | Left | Right |
|---|---|---|---|
| Images in the training set | 42,493 | 30,930 | 30,254 |
| Images in the testing set | 10,057 | 4289 | 5119 |
| MGNet Ocular Test | Ptosis | Diplopia (Left) | Diplopia (Right) |
|---|---|---|---|
| Total videos | 86 | 86 | 86 |
| No Score | 25 | 41 | 43 |
| Score Partially identifiable | 10 | 15 | 12 |
| Score Fully identifiable | 51 | 30 | 31 |
| Control Ocular Test | Ptosis | Diplopia Left | Diplopia Right |
|---|---|---|---|
| Total videos | 24 | 24 | 24 |
| No Score | 6 | 3 | 0 |
| Score Partially identifiable | 1 | 4 | 2 |
| Score Fully identifiable | 17 | 17 | 22 |
| MGNet Test | Count to 50 | Single Breath Count |
|---|---|---|
| Total number of videos | 86 | 86 |
| Number of successes Difference < 10 s | 75 | 62 |
| Control Test | Count to 50 | Single Breath Count |
|---|---|---|
| Total number of videos | 24 | 24 |
| Number of successes Difference < 10 s | 23 | 22 |
| Total Number of Videos | 86 |
| Number of successes Difference < 10 s | 82 |
| Total Number of Videos | 24 |
| Number of successes Difference < 10 s | 23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lesport, Q.; Palmie, D.; Öztosun, G.; Kaminski, H.J.; Garbey, M. AI-Powered Telemedicine for Automatic Scoring of Neuromuscular Examinations. Bioengineering 2024, 11, 942. https://doi.org/10.3390/bioengineering11090942
Lesport Q, Palmie D, Öztosun G, Kaminski HJ, Garbey M. AI-Powered Telemedicine for Automatic Scoring of Neuromuscular Examinations. Bioengineering. 2024; 11(9):942. https://doi.org/10.3390/bioengineering11090942
Chicago/Turabian StyleLesport, Quentin, Davis Palmie, Gülşen Öztosun, Henry J. Kaminski, and Marc Garbey. 2024. "AI-Powered Telemedicine for Automatic Scoring of Neuromuscular Examinations" Bioengineering 11, no. 9: 942. https://doi.org/10.3390/bioengineering11090942
APA StyleLesport, Q., Palmie, D., Öztosun, G., Kaminski, H. J., & Garbey, M. (2024). AI-Powered Telemedicine for Automatic Scoring of Neuromuscular Examinations. Bioengineering, 11(9), 942. https://doi.org/10.3390/bioengineering11090942






