Machine Learning Algorithm for Predicting Distant Metastasis of T1 and T2 Gallbladder Cancer Based on SEER Database
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Model Construction
3. Results
3.1. Analysis of Patient Information
3.2. Analysis of Results Obtained from Machine Learning Algorithms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ji, Z.; Ren, L.; Liu, F.; Liu, L.; Song, J.; Zhu, J.; Ji, G.; Huang, G. Effect of different surgical options on the long-term survival of stage I gallbladder cancer: A retrospective study based on SEER database and Chinese Multi-institutional Registry. J. Cancer Res. Clin. Oncol. 2023, 149, 12297–12313. [Google Scholar] [CrossRef]
- Huang, J.; Patel, H.K.; Boakye, D.; Chandrasekar, V.T.; Koulaouzidis, A.; Lucero-Prisno, D.E., III; Ngai, C.H.; Pun, C.N.; Bai, Y.; Lok, V.; et al. Worldwide distribution, associated factors, and trends of gallbladder cancer: A global country-level analysis. Cancer Lett. 2021, 521, 238–251. [Google Scholar] [CrossRef]
- Torres, O.J.M.; Alikhanov, R.; Li, J.; Serrablo, A.; Chan, A.C.; de Souza, M.F.E. Extended liver surgery for gallbladder cancer revisited: Is there a role for hepatopancreatoduodenectomy? Int. J. Surg. 2020, 82, 82–86. [Google Scholar] [CrossRef]
- Lim, H.; Seo, D.W.; Park, D.H.; Lee, S.S.; Lee, S.K.; Kim, M.H.; Hwang, S. Prognostic factors in patients with gallbladder cancer after surgical resection: Analysis of 279 operated patients. J. Clin. Gastroenterol. 2013, 47, 443–448. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, K.L.; Gupta, A.; Yadav, A.; Kumar, A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J. Gastroenterol. 2017, 23, 3978–3998. [Google Scholar] [CrossRef]
- Cai, Y.L.; Lin, Y.X.; Jiang, L.S.; Ye, H.; Li, F.Y.; Cheng, N.S. A Novel Nomogram Predicting Distant Metastasis in T1 and T2 Gallbladder Cancer: A SEER-based Study. Int. J. Med. Sci. 2020, 17, 1704–1712. [Google Scholar] [CrossRef]
- Yang, Y.; Tu, Z.; Ye, C.; Cai, H.; Yang, S.; Chen, X.; Tu, J. Site-specific metastases of gallbladder adenocarcinoma and their prognostic value for survival: A SEER-based study. BMC Surg. 2021, 21, 59. [Google Scholar] [CrossRef]
- Wang, X.; Yu, G.Y.; Chen, M.; Wei, R.; Chen, J.; Wang, Z. Pattern of distant metastases in primary extrahepatic bile-duct cancer: A SEER-based study. Cancer Med. 2018, 7, 5006–5014. [Google Scholar] [CrossRef]
- Mady, M.; Prasai, K.; Tella, S.H.; Yadav, S.; Hallemeier, C.L.; Rakshit, S.; Roberts, L.; Borad, M.; Mahipal, A. Neutrophil to lymphocyte ratio as a prognostic marker in metastatic gallbladder cancer. HPB 2020, 22, 1490–1495. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, X.; Hu, X.; Ren, H.; Wu, S.; Wu, J.; Wu, G.; Si, X.; Wang, B. Survival analysis of patients with primary gallbladder cancer from 2010 to 2015: A retrospective study based on SEER data. Medicine 2020, 99, e22292. [Google Scholar] [CrossRef]
- Zhong, X.; Lin, Y.; Zhang, W.; Bi, Q. Predicting diagnosis and survival of bone metastasis in breast cancer using machine learning. Sci. Rep. 2023, 13, 18301. [Google Scholar] [CrossRef]
- Liu, W.C.; Li, Z.Q.; Luo, Z.W.; Liao, W.J.; Liu, Z.L.; Liu, J.M. Machine learning for the prediction of bone metastasis in patients with newly diagnosed thyroid cancer. Cancer Med. 2021, 10, 2802–2811. [Google Scholar] [CrossRef]
- Mao, Y.; Lan, H.; Lin, W.; Liang, J.; Huang, H.; Li, L.; Wen, J.; Chen, G. Machine learning algorithms are comparable to conventional regression models in predicting distant metastasis of follicular thyroid carcinoma. Clin. Endocrinol. 2023, 98, 98–109. [Google Scholar] [CrossRef]
- Wernberg, J.A.; Lucarelli, D.D. Gallbladder cancer. Surg. Clin. N. Am. 2014, 94, 343–360. [Google Scholar] [CrossRef]
- Zhong, Y.; Wu, X.; Li, Q.; Ge, X.; Wang, F.; Wu, P.; Deng, X.; Miao, L. Long noncoding RNAs as potential biomarkers and therapeutic targets in gallbladder cancer: A systematic review and meta-analysis. Cancer Cell Int. 2019, 19, 169. [Google Scholar] [CrossRef]
- Shen, H.; He, M.; Lin, R.; Zhan, M.; Xu, S.; Huang, X.; Xu, C.; Chen, W.; Yao, Y.; Mohan, M.; et al. PLEK2 promotes gallbladder cancer invasion and metastasis through EGFR/CCL2 pathway. J. Exp. Clin. Cancer Res. CR 2019, 38, 247. [Google Scholar] [CrossRef]
- Hundal, R.; Shaffer, E.A. Gallbladder cancer: Epidemiology and outcome. Clin. Epidemiol. 2014, 6, 99–109. [Google Scholar] [CrossRef]
- Shindoh, J.; de Aretxabala, X.; Aloia, T.A.; Roa, J.C.; Roa, I.; Zimmitti, G.; Javle, M.; Conrad, C.; Maru, D.M.; Aoki, T.; et al. Tumor location is a strong predictor of tumor progression and survival in T2 gallbladder cancer: An international multicenter study. Ann. Surg. 2015, 261, 733–739. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, Z.; Sa, B. Construction and validation of the predictive model for gallbladder cancer liver metastasis patients: A SEER-based study. Eur. J. Gastroenterol. Hepatol. 2024, 36, 129–134. [Google Scholar] [CrossRef]
- Fang, C.; Li, W.; Wang, Q.; Wang, R.; Dong, H.; Chen, J.; Chen, Y. Risk factors and prognosis of liver metastasis in gallbladder cancer patients: A SEER-based study. Front. Surg. 2022, 9, 899896. [Google Scholar] [CrossRef]
- Leonard, G.; South, C.; Balentine, C.; Porembka, M.; Mansour, J.; Wang, S.; Yopp, A.; Polanco, P.; Zeh, H.; Augustine, M. Machine Learning Improves Prediction Over Logistic Regression on Resected Colon Cancer Patients. J. Surg. Res. 2022, 275, 181–193. [Google Scholar] [CrossRef]
- Guo, Z.T.; Tian, K.; Xie, X.Y.; Zhang, Y.H.; Fang, D.B. Machine Learning for Predicting Distant Metastasis of Medullary Thyroid Carcinoma Using the SEER Database. Int. J. Endocrinol. 2023, 2023, 9965578. [Google Scholar] [CrossRef]
- Han, T.; Zhu, J.; Chen, X.; Chen, R.; Jiang, Y.; Wang, S.; Xu, D.; Shen, G.; Zheng, J.; Xu, C. Application of artificial intelligence in a real-world research for predicting the risk of liver metastasis in T1 colorectal cancer. Cancer Cell Int. 2022, 22, 28. [Google Scholar] [CrossRef]
- Ahn, J.H.; Kwak, M.S.; Lee, H.H.; Cha, J.M.; Shin, H.P.; Jeon, J.W.; Yoon, J.Y. Development of a Novel Prognostic Model for Predicting Lymph Node Metastasis in Early Colorectal Cancer: Analysis Based on the Surveillance, Epidemiology, and End Results Database. Front. Oncol. 2021, 11, 614398. [Google Scholar] [CrossRef]
- Osório, F.M.; Vidigal, P.V.; Ferrari, T.C.; Lima, A.S.; Lauar, G.M.; Couto, C.A. Histologic Grade and Mitotic Index as Predictors of Microvascular Invasion in Hepatocellular Carcinoma. Exp. Clin. Transplant. 2015, 13, 421–425. [Google Scholar] [CrossRef]
- Butte, J.M.; Gönen, M.; Allen, P.J.; D’Angelica, M.I.; Kingham, T.P.; Fong, Y.; Dematteo, R.P.; Blumgart, L.; Jarnagin, W.R. The role of laparoscopic staging in patients with incidental gallbladder cancer. HPB 2011, 13, 463–472. [Google Scholar] [CrossRef]
- Shirai, Y.; Sakata, J.; Wakai, T.; Ohashi, T.; Ajioka, Y.; Hatakeyama, K. Assessment of lymph node status in gallbladder cancer: Location, number, or ratio of positive nodes. World J. Surg. Oncol. 2012, 10, 87. [Google Scholar] [CrossRef]
- Qiu, B.; Su, X.H.; Qin, X.; Wang, Q. Application of machine learning techniques in real-world research to predict the risk of liver metastasis in rectal cancer. Front. Oncol. 2022, 12, 1065468. [Google Scholar] [CrossRef]
- Sakata, J.; Shirai, Y.; Wakai, T.; Ajioka, Y.; Hatakeyama, K. Number of positive lymph nodes independently determines the prognosis after resection in patients with gallbladder carcinoma. Ann. Surg. Oncol. 2010, 17, 1831–1840. [Google Scholar] [CrossRef]
- Negi, S.S.; Singh, A.; Chaudhary, A. Lymph nodal involvement as prognostic factor in gallbladder cancer: Location, count or ratio? J. Gastrointest. Surg. 2011, 15, 1017–1025. [Google Scholar] [CrossRef]
- Feng, S.; Wang, J.; Wang, L.; Qiu, Q.; Chen, D.; Su, H.; Li, X.; Xiao, Y.; Lin, C. Current Status and Analysis of Machine Learning in Hepatocellular Carcinoma. J. Clin. Transl. Hepatol. 2023, 11, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef] [PubMed]

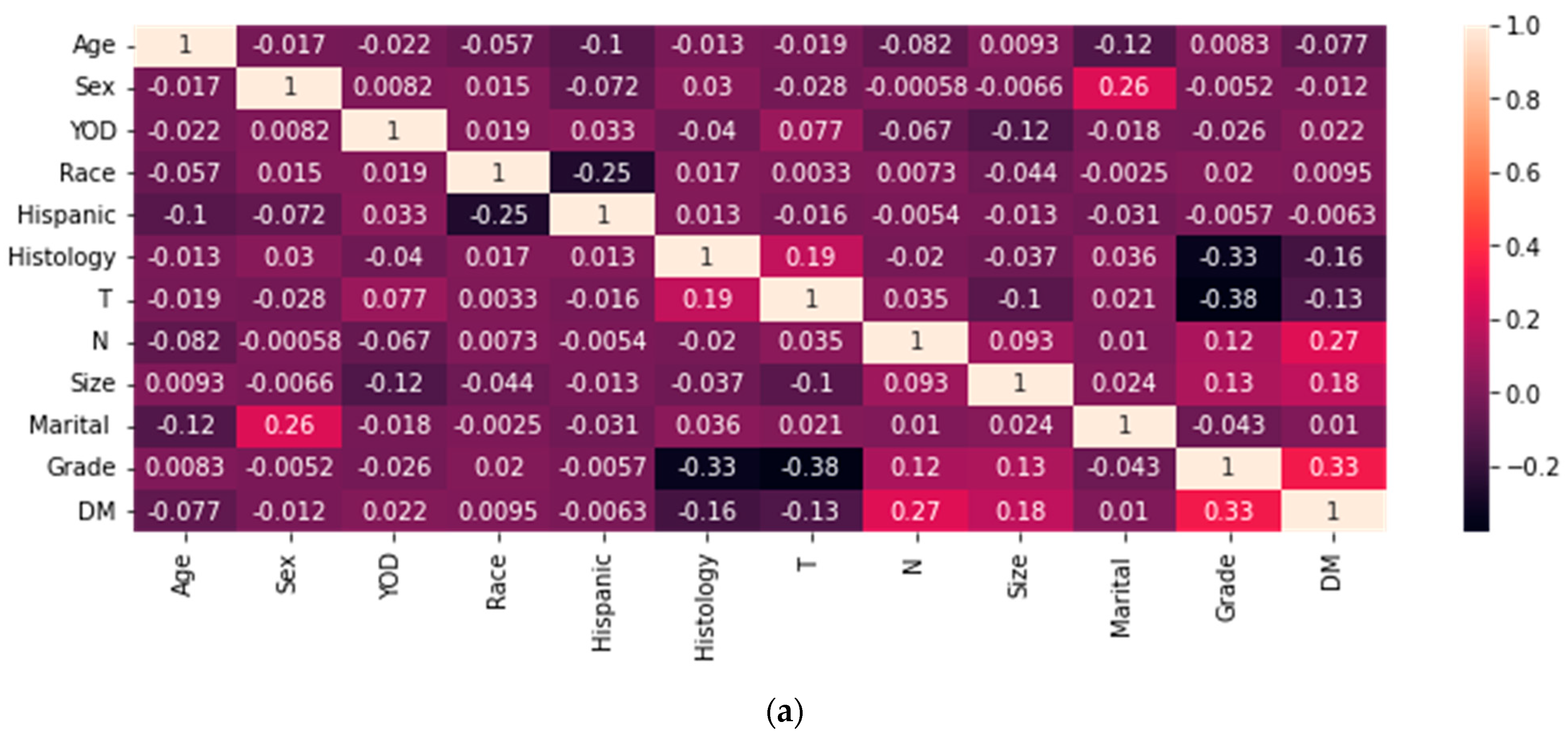


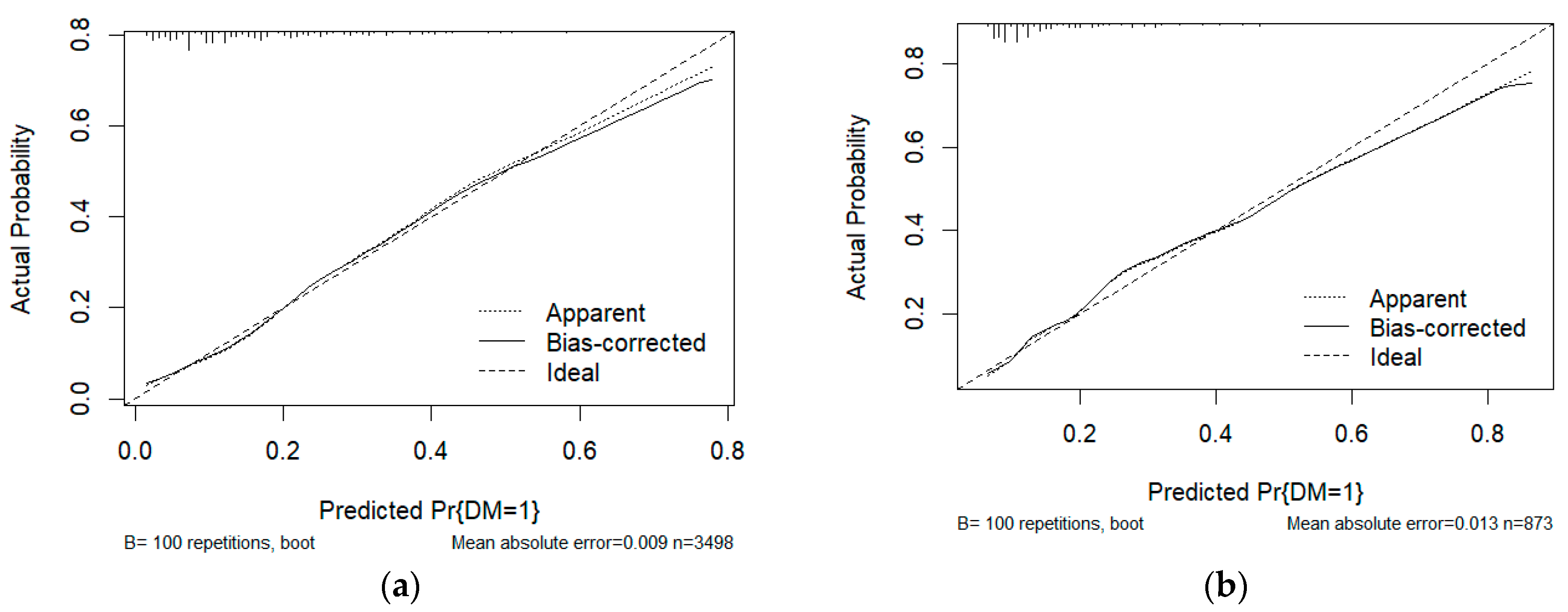


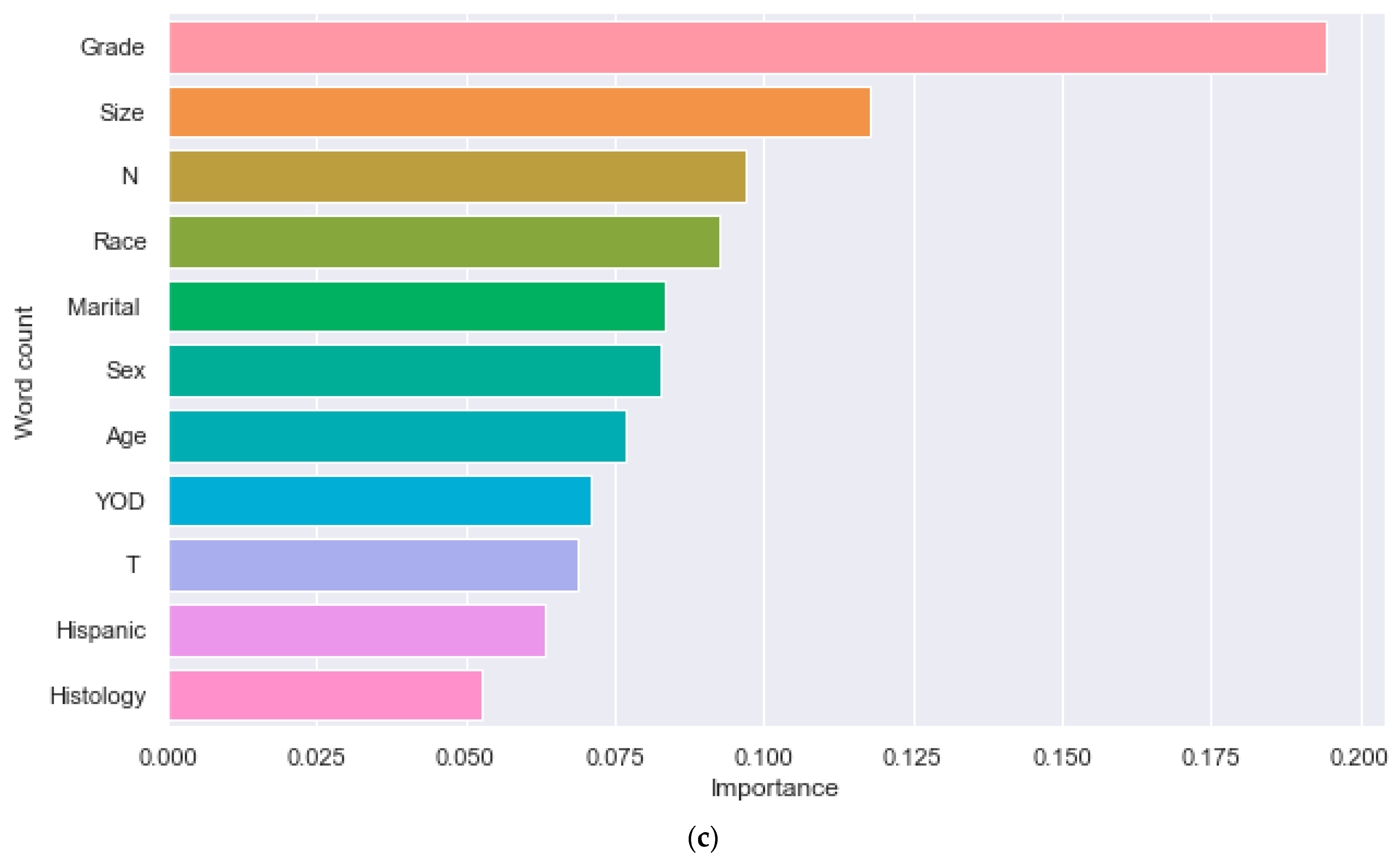
| Characteristic | Without DM (N = 3607) | With DM (N = 764) | p-Value |
|---|---|---|---|
| Age (Year) | <0.001 | ||
| <70 | 1508 (41.8%) | 374 (49.0%) | |
| ≥70 | 2099 (58.2%) | 390 (51.0%) | |
| Gender | 0.181 | ||
| Female | 2523 (69.9%) | 553 (72.4%) | |
| Male | 1084 (30.1%) | 211 (27.6%) | |
| Race | 0.599 | ||
| White | 2770 (76.8%) | 578 (75.7%) | |
| Black | 400 (11.1%) | 97 (12.7%) | |
| Other | 437 (12.1%) | 89 (11.6%) | |
| Hispanic | 0.572 | ||
| YES | 808 (22.4%) | 164 (21.5%) | |
| NO | 2799 (77.6) | 600 (78.5%) | |
| Histology | <0.001 | ||
| Adenocarcinoma | 3308 (91.7%) | 611 (80.0%) | |
| Others | 299 (8.3%) | 153 (20.0%) | |
| Year of Diagnosis | 0.262 | ||
| 2004–2009 | 1624 (45.0%) | 327 (42.8%) | |
| 2010–2015 | 1983 (55.0%) | 437 (57.2%) | |
| Tumor Size(cm) | <0.001 | ||
| <2 | 838 (23.2%) | 81 (10.6%) | |
| ≥2 | 1543 (42.8%) | 321 (42%) | |
| Unknown | 1226 (34%) | 362(47.4%) | |
| T Stage | <0.001 | ||
| T1 | 1259 (34.9%) | 361 (47.3%) | |
| T2 | 2348 (65.1%) | 403 (52.7%) | |
| N stage | <0.001 | ||
| N0 | 2871 (79.6%) | 422 (55.2%) | |
| N1 | 644 (17.8%) | 257 (33.7%) | |
| NX | 92 (2.6%) | 85 (11.1%) | |
| Marital Status | 0.531 | ||
| Single | 1839 (51.0%) | 380 (49.7%) | |
| Married | 1768 (49.0%) | 384 (50.3%) | |
| Grade | <0.001. | ||
| Grade I | 737 (20.4%) | 39 (5.1%) | |
| Grade II | 1536 (42.6%) | 219 (28.6%) | |
| Grade III | 894 (24.8%) | 255 (33.4%) | |
| Grade IV | 55 (1.5%) | 18 (2.4%) | |
| Unknown | 385 (10.7) | 233 (30.5%) |
| Characteristic | Training Set (N = 3498) | Test Set (N = 873) | Total Set (N = 4371) | p-Value |
|---|---|---|---|---|
| Age (Year) | 1.000 | |||
| <70 | 1506 (43.1%) | 376 (43.1%) | 1882 (43.1%) | |
| ≥70 | 1992 (56.9%) | 497 (56.9%) | 2489 (56.9%) | |
| Gender | 0.386 | |||
| Female | 2445 (69.9%) | 631 (72.3%) | 3076 (70.4%) | |
| Male | 1053 (30.1%) | 242 (27.7%) | 1295 (29.6%) | |
| Race | 0.883 | |||
| White | 2686 (76.8%) | 662 (75.8%) | 3348 (76.6%) | |
| Black | 390 (11.1%) | 107 (12.3%) | 497 (1134%) | |
| Other | 422 (12.1%) | 104 (11.9%) | 526 (12.0%) | |
| Hispanic | 0.256 | |||
| YES | 796 (22.8%) | 176 (20.2%) | 972 (22.2%) | |
| NO | 2702 (77.2) | 697 (79.8%) | 3399 (77.8%) | |
| Histology | 0.207 | |||
| Adenocarcinoma | 3122 (89.3%) | 797 (91.3%) | 3919 (89.7%) | |
| Others | 376 (10.7%) | 76 (8.7%) | 452 (10.3%) | |
| Year of Diagnosis | 0.844 | |||
| 2004–2009 | 1569 (44.9%) | 382 (43.8%) | 1951 (44.6%) | |
| 2010–2015 | 1929 (55.1%) | 491 (56.2%) | 2420 (55.4%) | |
| Tumor size(cm) | 0.969 | |||
| <2 | 735 (21%) | 184 (21.1%) | 919 (21.1%) | |
| ≥2 | 1488(42.5%) | 376 (43%) | 1864 (42.6%) | |
| Unknown | 1275(36.4%) | 313 (35.9%) | 1588 (36.3%) | |
| T Stage | 0.756 | |||
| T1 | 1306 (37.3%) | 314 (36%) | 1620 (37.1%) | |
| T2 | 2192 (62.7%) | 559 (64%) | 2751 (62.9%) | |
| N stage | 0.104 | |||
| N0 | 2611 (74.6%) | 682 (78.1%) | 3293 (75.3%) | |
| N1 | 741 (21.2%) | 160 (18.3%) | 901 (20.6%) | |
| NX | 146 (4.2%) | 31 (3.6%) | 177 (4.1%) | |
| Marital Status | 0.998 | |||
| Single | 1775 (50.7%) | 444 (50.9%) | 2219 (50.8%) | |
| Married | 1723 (49.3%) | 429 (49.1%) | 2152 (49.2%) | |
| Grade | 0.620 | |||
| Grade I | 619 (17.7%) | 157 (18.0%) | 776 (17.7%) | |
| Grade II | 1384 (39.6%) | 371 (42.5%) | 1755 (40.2%) | |
| Grade III | 949 (27.1%) | 200 (22.9%) | 1149 (26.3%) | |
| Grade IV | 51 (1.5%) | 22 (2.5%) | 73 (1.7%) | |
| Unknown | 495 (14.1%) | 123 (14.1%) | 618 (14.1%) | |
| Distant Metastasis | 0.998 | |||
| YES | 612 (17.5%) | 152 (17.4%) | 764 (17.5%) | |
| NO | 2886 (82.5%) | 721 (82.6%) | 3607 (82.5%) |
| Univariable Analysis | Multivariable Analysis | |||||
|---|---|---|---|---|---|---|
| OR | 95%CI | p Value | OR | 95%CI | p Value | |
| Age (Year) | ||||||
| <70 | Ref | Ref | ||||
| ≥70 | 0.723 | 0.607–0.861 | <0.001 | 0.705 | 0.583–0.852 | <0.001 |
| Gender | ||||||
| Female | Ref | |||||
| Male | 0.881 | 0.726–1.069 | 0.200 | |||
| Race | ||||||
| White | Ref | |||||
| Black | 1.116 | 0.850–1.464 | 0.431 | |||
| Other | 0.980 | 0.746–1.287 | 0.885 | |||
| Hispanic | ||||||
| YES | 0.997 | 0.810–1.228 | 0.977 | |||
| NO | Ref | |||||
| Histology | ||||||
| Adenocarcinoma | 0.345 | 0.274–0.436 | <0.001 | 0.595 | 0.456–0.777 | <0.001 |
| Others | Ref | Ref | ||||
| Year of Diagnosis | ||||||
| 2004–2009 | Ref | |||||
| 2010–2015 | 1.151 | 0.965–1.374 | 0.117 | |||
| Tumor Size(cm) | ||||||
| <2 | Ref | Ref | ||||
| ≥2 | 1.916 | 1.449–2.534 | <0.001 | 1.507 | 1.121–2.027 | 0.007 |
| Unknown | 2.729 | 2.067–3.602 | <0.001 | 2.023 | 1.509–2.714 | <0.001 |
| T Stage | ||||||
| T1 | Ref | |||||
| T2 | 0.594 | 0.498–0.708 | <0.001 | 0.679 | 0.547–0.843 | <0.001 |
| N Stage | ||||||
| N0 | Ref | Ref | ||||
| N1 | 2.656 | 2.155–3.197 | <0.000 | 2.377 | 1.920–2.944 | <0.001 |
| NX | 6.067 | 4.299–8.563 | <0.000 | 4.913 | 3.398–7.105 | <0.001 |
| Marital Status | ||||||
| Single | Ref | |||||
| Married | 1.096 | 0.920–1.305 | 0.304 | |||
| Grade | ||||||
| Grade I | Ref | Ref | ||||
| Grade II | 3.507 | 2.281–5.391 | <0.001 | 3.236 | 2.090–5.010 | <0.001 |
| Grade III | 6.835 | 4.453–10.489 | <0.001 | 5.776 | 3.721–8.966 | <0.001 |
| Grade IV | 8.990 | 4.316–18.725 | <0.001 | 6.316 | 2.932–13.605 | <0.001 |
| Unknown | 13.936 | 8.977–21.635 | <0.001 | 8.684 | 5.475–13.774 | <0.001 |
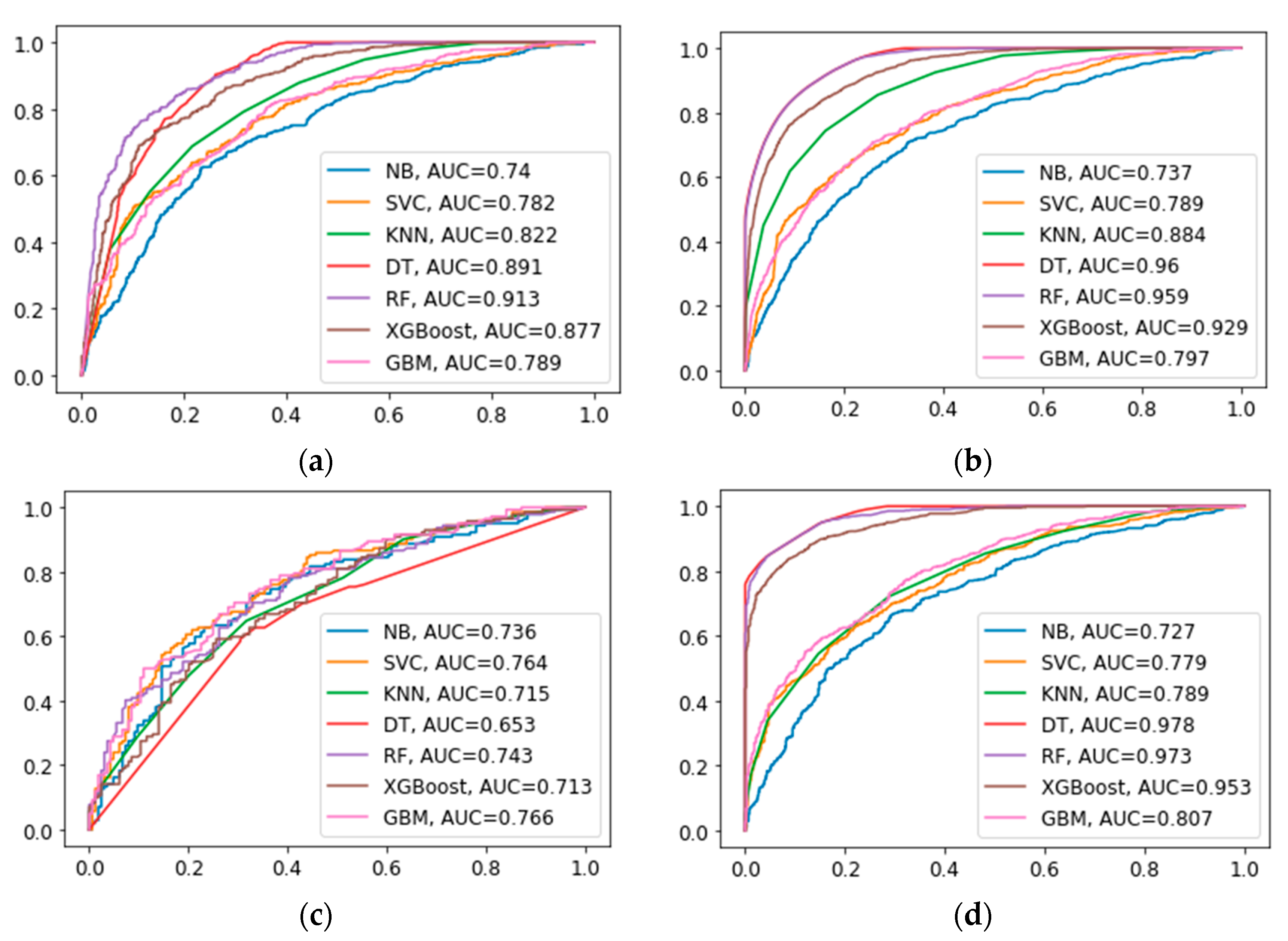
| Model | Accuracy | AUC | Precision | Recall Rate | F1-Score |
|---|---|---|---|---|---|
| NB | 0.681 | 0.740 | 0.734 | 0.587 | 0.652 |
| SVC | 0.707 | 0.782 | 0.722 | 0.690 | 0.706 |
| KNN | 0.738 | 0.822 | 0.721 | 0.791 | 0.761 |
| DT | 0.681 | 0.891 | 0.686 | 0.688 | 0.687 |
| RF | 0.828 | 0.913 | 0.811 | 0.862 | 0.836 |
| XGBoost | 0.784 | 0.877 | 0.781 | 0.799 | 0.790 |
| GBM | 0.704 | 0.789 | 0.711 | 0.704 | 0.707 |
| Model | Accuracy | AUC | Precision | Recall Rate | F1-Score |
|---|---|---|---|---|---|
| NB | 0.689 | 0.736 | 0.715 | 0.549 | 0.621 |
| SVC | 0.702 | 0.764 | 0.691 | 0.647 | 0.669 |
| KNN | 0.604 | 0.715 | 0.562 | 0.661 | 0.687 |
| DT | 0.699 | 0.653 | 0.676 | 0.676 | 0.676 |
| RF | 0.686 | 0.743 | 0.643 | 0.725 | 0.682 |
| XGBoost | 0.656 | 0.713 | 0.624 | 0.654 | 0.639 |
| GBM | 0.702 | 0.766 | 0.683 | 0.669 | 0.676 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.; Zhang, Z.; Liu, L.; Zhao, Y.; Liu, Z.; Zhang, C.; Qi, H.; Feng, J.; Yao, P.; Yuan, H. Machine Learning Algorithm for Predicting Distant Metastasis of T1 and T2 Gallbladder Cancer Based on SEER Database. Bioengineering 2024, 11, 927. https://doi.org/10.3390/bioengineering11090927
Guo Z, Zhang Z, Liu L, Zhao Y, Liu Z, Zhang C, Qi H, Feng J, Yao P, Yuan H. Machine Learning Algorithm for Predicting Distant Metastasis of T1 and T2 Gallbladder Cancer Based on SEER Database. Bioengineering. 2024; 11(9):927. https://doi.org/10.3390/bioengineering11090927
Chicago/Turabian StyleGuo, Zhentian, Zongming Zhang, Limin Liu, Yue Zhao, Zhuo Liu, Chong Zhang, Hui Qi, Jinqiu Feng, Peijie Yao, and Haiming Yuan. 2024. "Machine Learning Algorithm for Predicting Distant Metastasis of T1 and T2 Gallbladder Cancer Based on SEER Database" Bioengineering 11, no. 9: 927. https://doi.org/10.3390/bioengineering11090927
APA StyleGuo, Z., Zhang, Z., Liu, L., Zhao, Y., Liu, Z., Zhang, C., Qi, H., Feng, J., Yao, P., & Yuan, H. (2024). Machine Learning Algorithm for Predicting Distant Metastasis of T1 and T2 Gallbladder Cancer Based on SEER Database. Bioengineering, 11(9), 927. https://doi.org/10.3390/bioengineering11090927







