Design and Validation of a PLC-Controlled Morbidostat for Investigating Bacterial Drug Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Morbidostat Design and Building
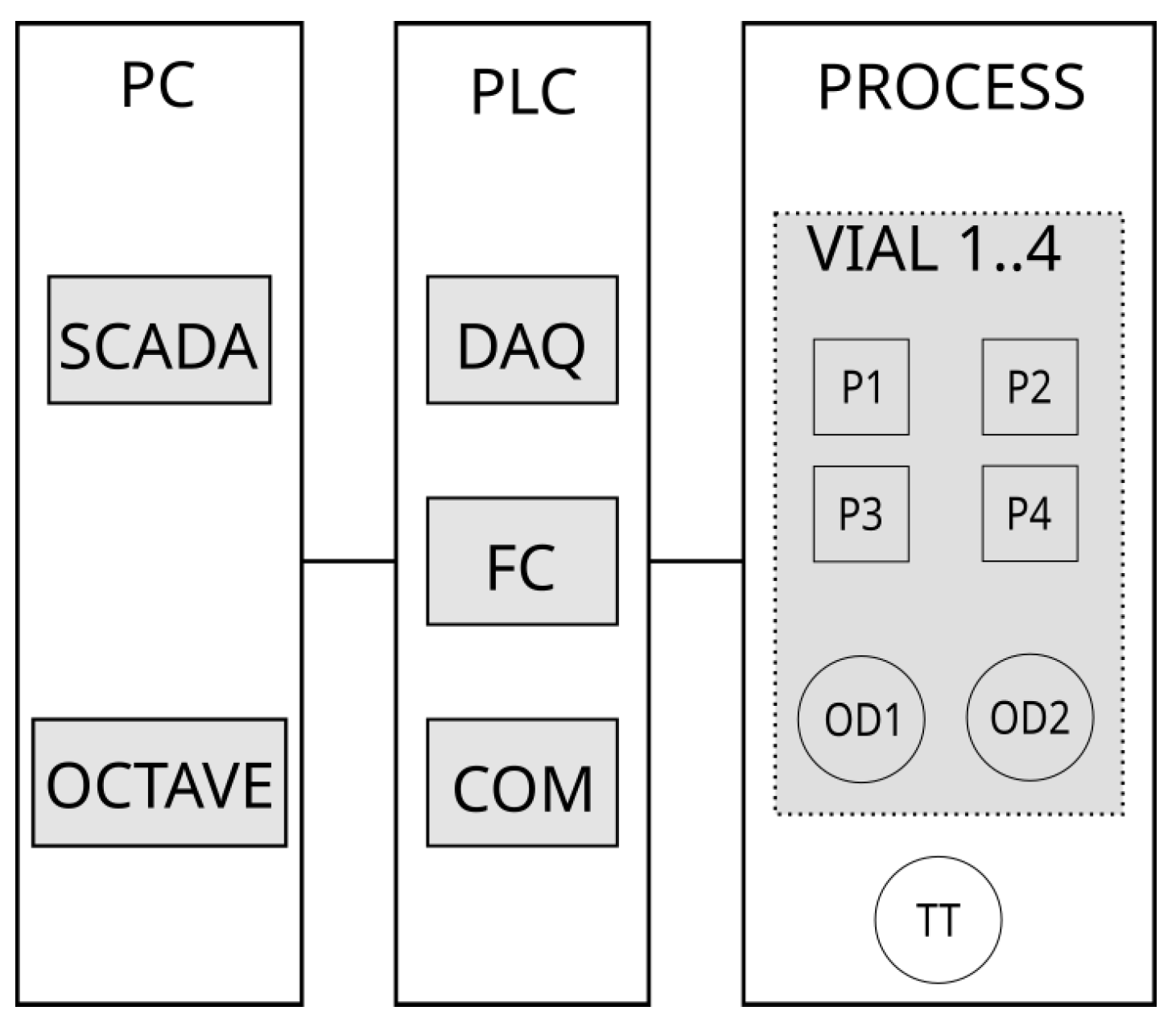
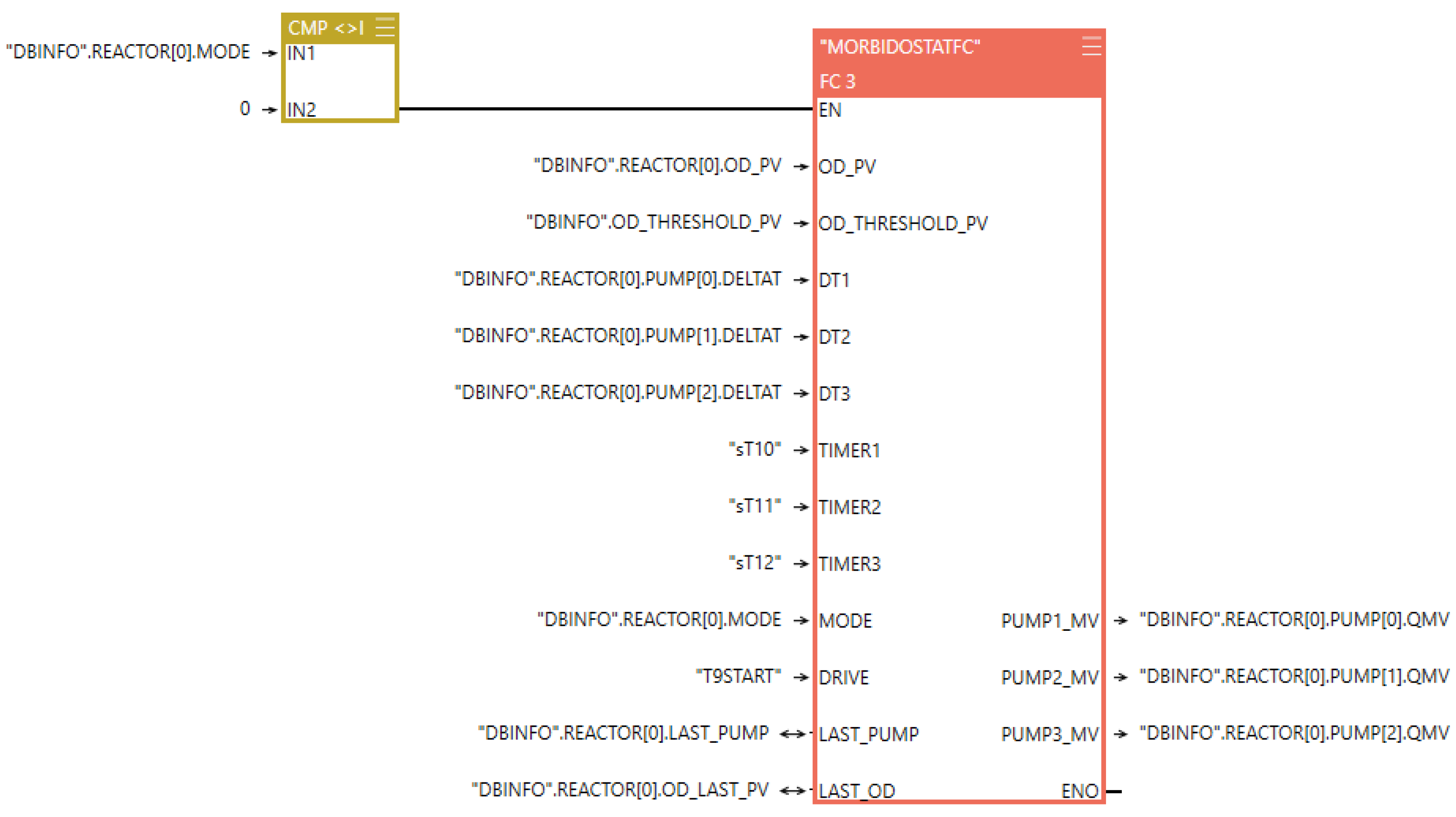
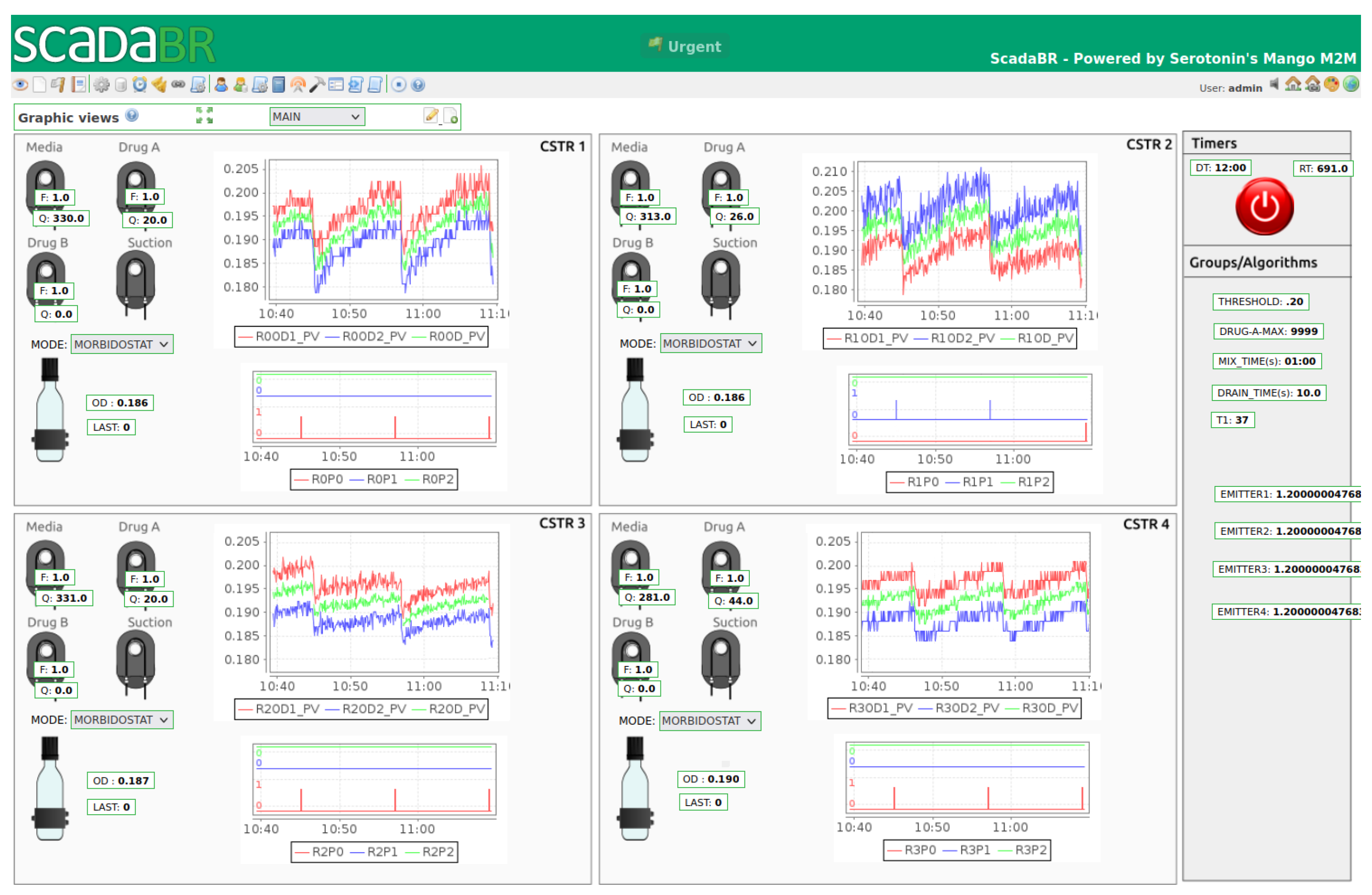
2.1.1. Control System
Processing Unit
Programmable Logic Controller (PLC)
Personal Computer (PC)
- It satisfies real-time, low-latency performance requirements via PLC and ensures that the system will continue operating with the latest available setpoints, even if SCADA or MATLAB become unavailable (crash, network problems, etc.);
- It provides a cost-effective and feature-rich SCADA for data visualization, supervisory control, and data historicization and features web access from other computers if needed;
- Using MODBUS TCP standard industrial protocol, it allows for eventual integration in other, more complex setups.
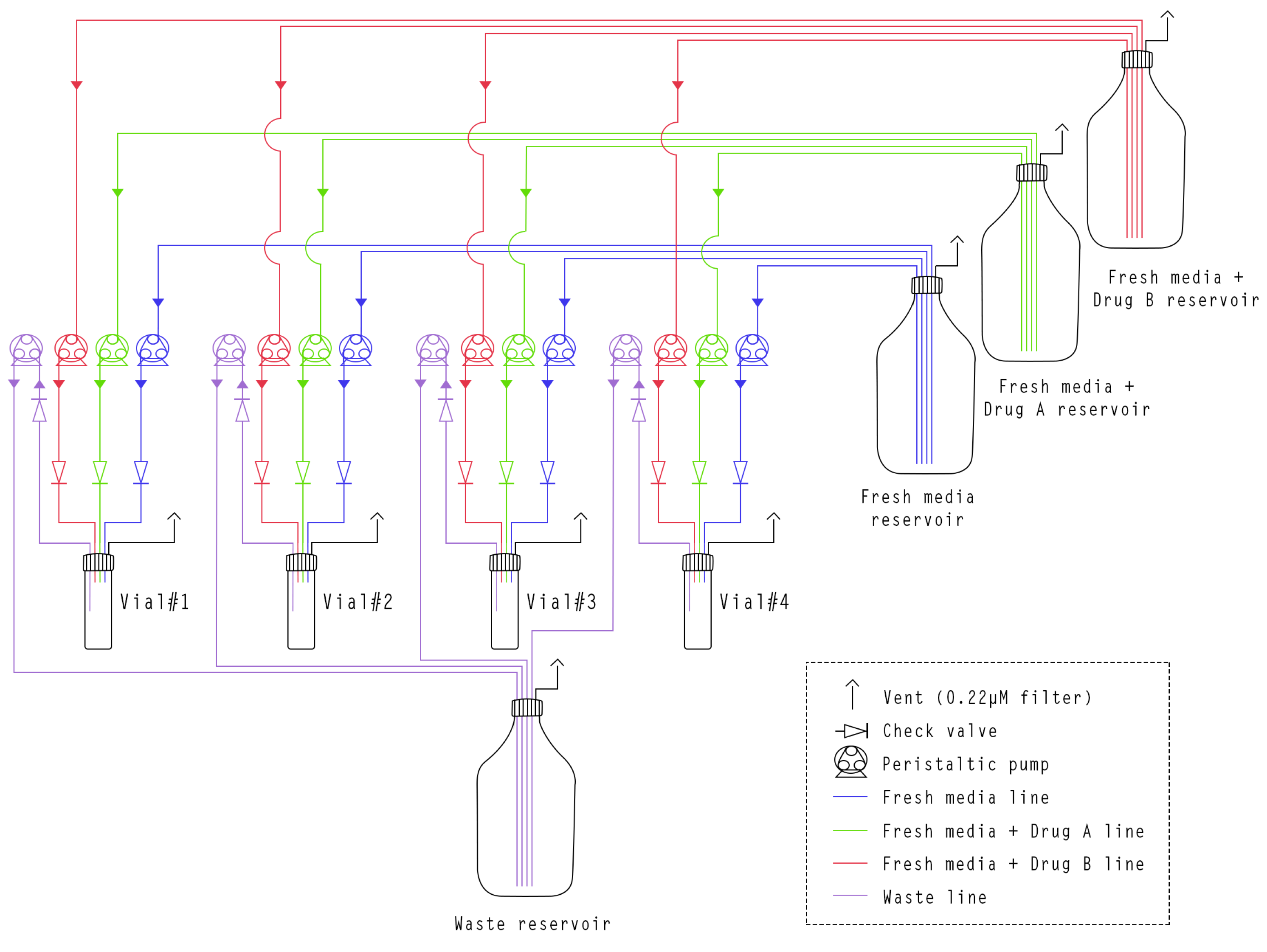
2.1.2. Fluidic System
2.2. Experimental Procedure on Morbidostat
2.2.1. Strain and Antimicrobials
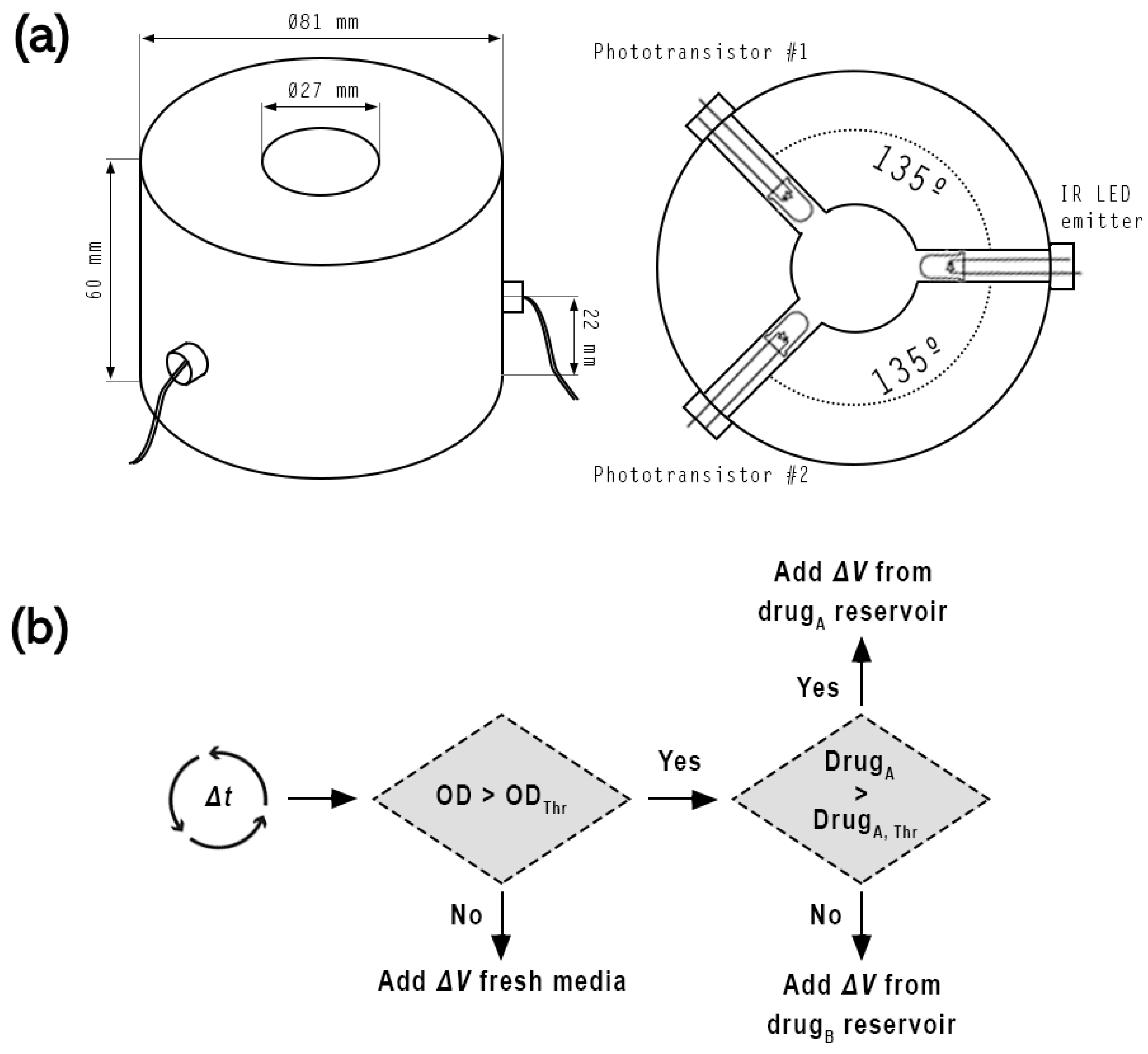
2.2.2. Calibration of the Optical Subsystem
2.2.3. Sterilization of the Fluidic Subsystem
2.2.4. Morbidostat Experiment
2.2.5. Evaluation of Sensitivity to DDAC of the Ancestral and Evolved Populations
3. Results and Discussion
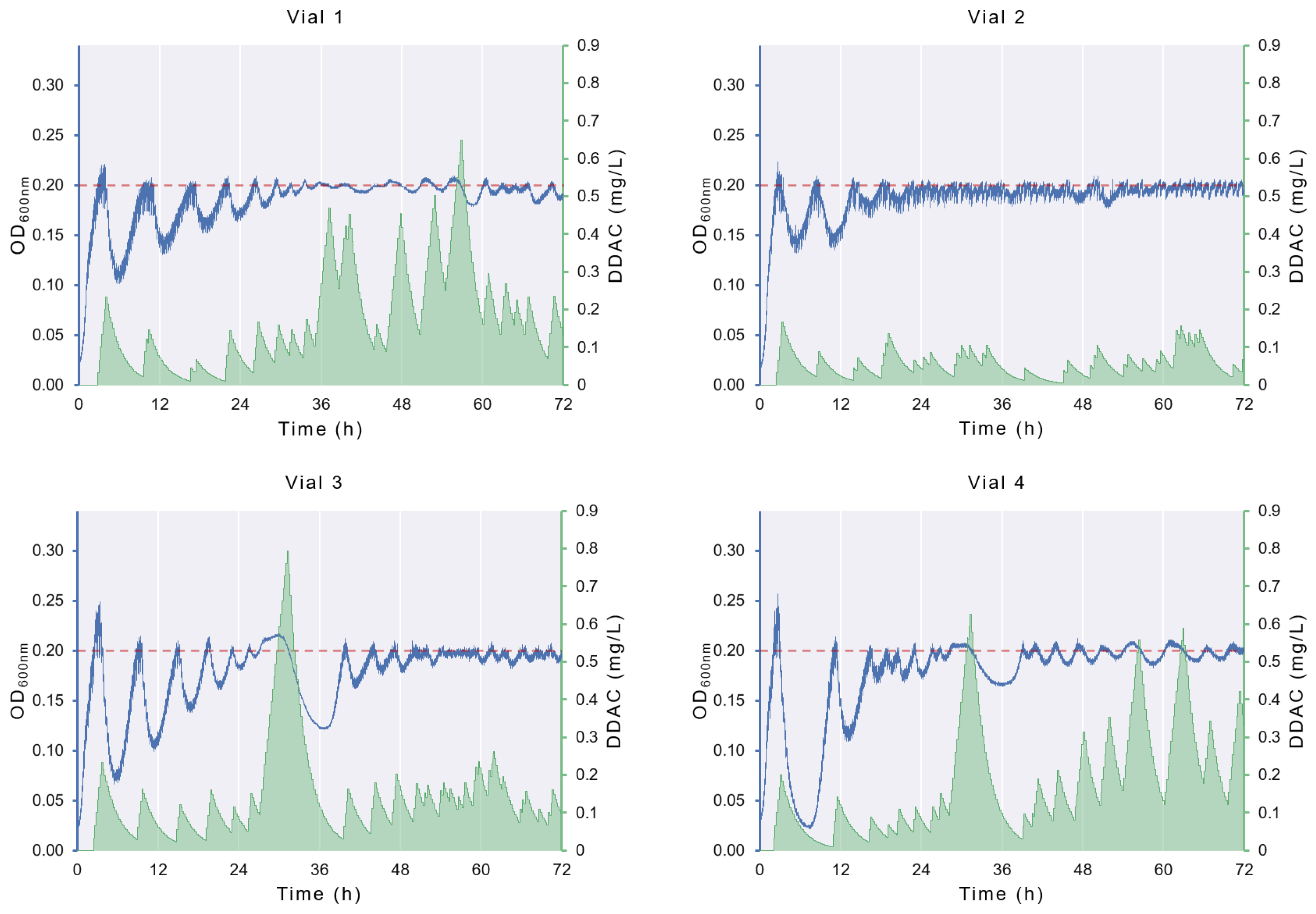
3.1. ALE in Morbidostat
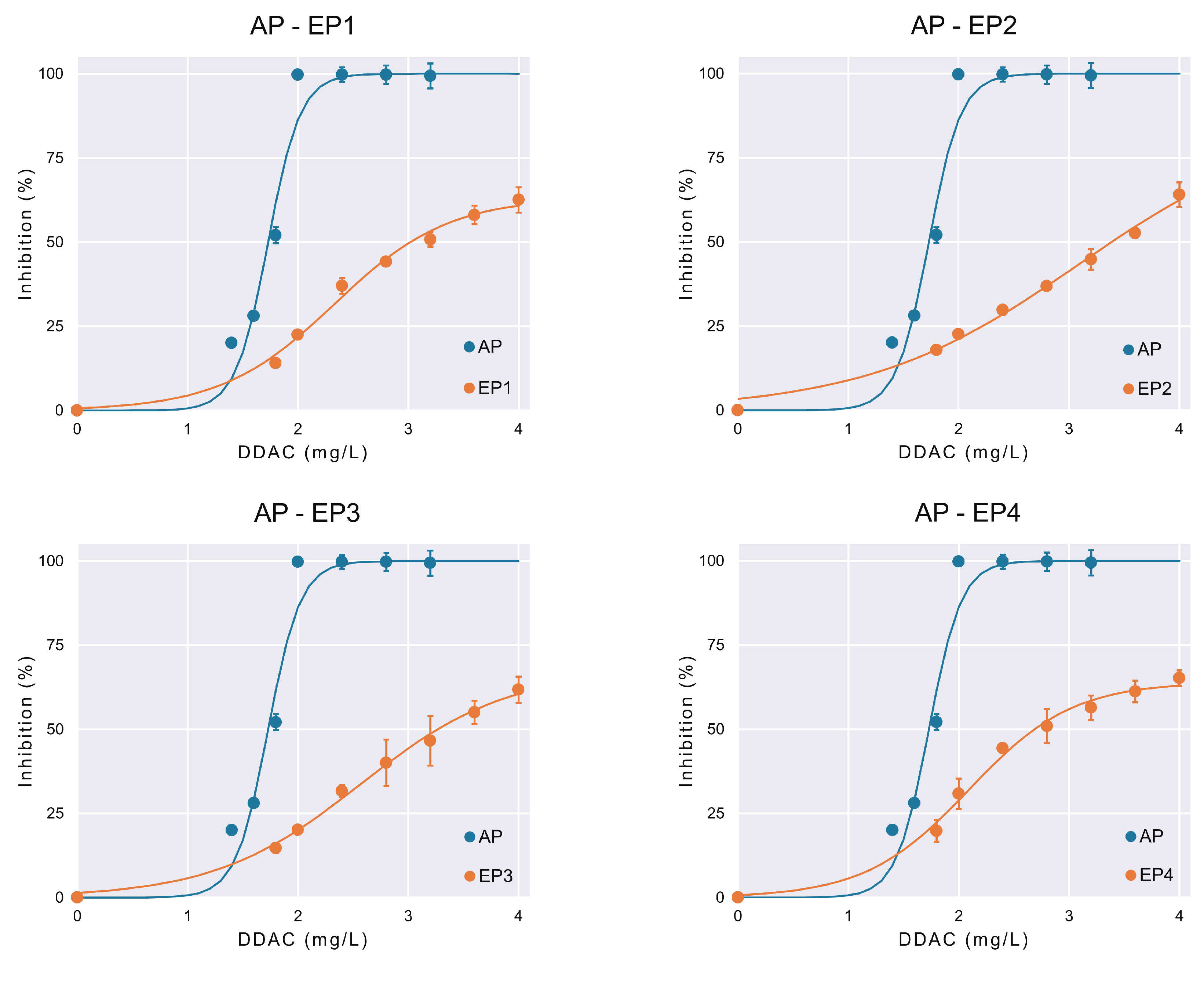
3.2. Changes in DDAC Susceptibility
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALE | Adaptive Laboratory Evolution |
| COM | Communication |
| CFU | Colony-Forming Units |
| DAQ | Data Acquisition |
| DDAC | Didecyldimethylammonium Chloride |
| FBD | Function Block Diagram |
| FC | Flow Control |
| GUI | Graphical User Interface |
| IC50 | Half-Maximal Inhibitory Concentration |
| MIC | Miminium Inhibitory Concentration |
| MPB | Meat-Peptone Broth |
| LED | Light-Emitting Diode |
| OD | Optical Density |
| PLC | Power Line Communications |
| PWM | Pulse Width Modulation |
| SCADA | Supervisory Control And Data Acquisition |
| STL | Standard Template Library |
| TCP | Transmission Control Protocol |
| VDC | Volts Direct Current |
Appendix A

References
- WHO. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Toprak, E.; Veres, A.; Yildiz, S.; Pedraza, J.M.; Chait, R.; Paulsson, J.; Kishony, R. Building a morbidostat: An automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Nat. Protoc. 2013, 8, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Chevereau, G.; Dravecká, M.; Batur, T.; Guvenek, A.; Ayhan, D.H.; Toprak, E.; Bollenbach, T. Quantifying the Determinants of Evolutionary Dynamics Leading to Drug Resistance. PLoS Biol. 2015, 13, e1002299. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, E.; Abdellati, S.; Nys, P.; Laumen, J.; De Baetselier, I.; Crucitti, T.; Kenyon, C. Construction and optimization of a ‘NG Morbidostat’—An automated continuous-culture device for studying the pathways towards antibiotic resistance in Neisseria gonorrhoeae. F1000Research 2020, 8, 560. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.V.; Nelson, E.J.; Vengurlekar, A.; Zhang, R.; White, K.I.; Salinas, V.; Johnson, T.T. Model-based design and analysis of a reconfigurable continuous-culture bioreactor. In Proceedings of the 4th ACM SIGBED International Workshop on Design, Modeling, and Evaluation of Cyber-Physical Systems, Berlin, Germany, 14–17 April 2014; pp. 48–51. [Google Scholar] [CrossRef]
- MathWorks. MATLAB Documentation: Timer Object. 2024. Available online: https://www.mathworks.com/help/matlab/ref/timer.html (accessed on 18 June 2024).
- Dößelmann, B.; Willmann, M.; Steglich, M.; Bunk, B.; Nübel, U.; Peter, S.; Neher, R.A. Rapid and Consistent Evolution of Colistin Resistance in Extensively Drug-Resistant Pseudomonas aeruginosa during Morbidostat Culture. Antimicrob. Agents Chemother. 2017, 61, e00043-17. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.; Ueltzhoeffer, V.; Heinrich, M.; Siegrist, H.J.; Wildermuth, R.; Lorenz, F.R.; Neher, R.A.; Willmann, M. Colistin susceptibility test evaluation of multiple-resistance-level Pseudomonas aeruginosa isolates generated in a morbidostat device. J. Antimicrob. Chemother. 2018, 73, 3368–3374. [Google Scholar] [CrossRef] [PubMed]
- Madden, M.M. Challenges Using Linux as a Real-Time Operating System. In Proceedings of the AIAA Scitech 2019 Forum, San Diego, CA, USA, 7–11 January 2019. [Google Scholar] [CrossRef]
- Ziv, N.; Brandt, N.J.; Gresham, D. The Use of Chemostats in Microbial Systems Biology. JOVE 2013, 80, e50168. [Google Scholar] [CrossRef]
- Martínez-López, N.; Pedreira, A.; García, M.R. Control Predictivo Basado en Modelo Como Alternativa al Modo de Operación Estándar en Morbidostato. 2022. Available online: https://ruc.udc.es/dspace/handle/2183/31472 (accessed on 16 June 2024).
- De Battista, H.; Picó-Marco, E.; Santos-Navarro, F.N.; Picó, J. Output feedback linearization of turbidostats after time scaling. IEEE Trans. Control Syst. Technol. 2018, 27, 1668–1676. [Google Scholar] [CrossRef]
- M07-A10; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015.
- Pedreira, A.; Fernandes, S.; Simões, M.; García, M.R.; Vázquez, J.A. Synergistic Bactericidal Effects of Quaternary Ammonium Compounds with Essential Oil Constituents. Foods 2024, 13, 1831. [Google Scholar] [CrossRef] [PubMed]
- Murado, M.; González, M.P.; Vázquez, J. Dose–response relationships: An overview, a generative model and its application to the verification of descriptive models. Enzym. Microb. Technol. 2002, 31, 439–455. [Google Scholar] [CrossRef]
- De Levie, R. How to Use Excel® in Analytical Chemistry: And in General Scientific Data Analysis; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Laumen, J.G.E.; Van Dijck, C.; Manoharan-Basil, S.S.; Abdellati, S.; De Baetselier, I.; Cuylaerts, V.; De Block, T.; Van den Bossche, D.; Xavier, B.B.; Malhotra-Kumar, S.; et al. Sub-Inhibitory Concentrations of Chlorhexidine Induce Resistance to Chlorhexidine and Decrease Antibiotic Susceptibility in Neisseria gonorrhoeae. Front. Microbiol. 2021, 12, 776909. [Google Scholar] [CrossRef] [PubMed]
- Pioreactor. Wetware Assembly. 2024. Available online: https://docs.pioreactor.com/user-guide/wetware-assembly (accessed on 31 July 2024).
- García, M.R.; Vázquez, J.A.; Teixeira, I.G.; Alonso, A.A. Stochastic individual-based modeling of bacterial growth and division using flow cytometry. Front. Microbiol. 2018, 8, 286497. [Google Scholar] [CrossRef] [PubMed]
- Coates, J.; Park, B.R.; Le, D.; Şimşek, E.; Chaudhry, W.; Kim, M. Antibiotic-induced population fluctuations and stochastic clearance of bacteria. eLife 2018, 7, e32976. [Google Scholar] [CrossRef] [PubMed]
- Leyn, S.A.; Zlamal, J.E.; Kurnasov, O.V.; Li, X.; Elane, M.; Myjak, L.; Godzik, M.; de Crecy, A.; Garcia-Alcalde, F.; Ebeling, M.; et al. Experimental evolution in morbidostat reveals converging genomic trajectories on the path to triclosan resistance. Microb. Genom. 2021, 7, 000553. [Google Scholar] [CrossRef] [PubMed]
- Martínez-López, N.; Vilas, C.; García, M.R. A Birth-Death Model to Understand Bacterial Antimicrobial Heteroresistance from Time-Kill Curves. Preprint 2024. [Google Scholar] [CrossRef]
- Pedreira, A.; Vázquez, J.A.; García, M.R. Kinetics of Bacterial Adaptation, Growth, and Death at Didecyldimethylammonium Chloride sub-MIC Concentrations. Front. Microbiol. 2022, 13, 758237. [Google Scholar] [CrossRef] [PubMed]

| Aspect | Previous Works | Proposed Solution |
|---|---|---|
| Architecture | Monolithic, single process running in MATLAB or the like [2,3,7,17]. | Hybrid, hierarchical design with well-defined functions distributed among the levels. |
| Long operation | Time-critical segments may suffer as the data sets grow throughout the experiment [5]. | Real-time portions and subroutines are executed in the PLC, unaffected by data-acquisition growth. |
| Logic of the controller | Focus on Toprak’s morbidostat control law [2] with two conditions that cannot control the system when the antimicrobial effect has some latency. | Single-condition control law that ensures control even when the antimicrobial has a delayed effect. |
| Temperature control | Incubator chambers [2,3] or in-built heater solutions [18]. Cannot operate below ambient temperature. | Incubator equipped with a heating and cooling system. Stable temperature within a 10–60 °C range. |
| Sterilization of fluidic subsystem | Partially or totally chemical (successive washes with hypochlorite, ethanol, and distilled water) [2,3]. | Totally physical (autoclaving). No risk of chemical residues. |
| Asepsis | Risk of external microbial contamination. Risk of contamination of the culture chambers and reservoirs by backflow [4]. | Check valves reduce the risk of contamination in culture chambers and reservoirs. |
| Parameter | AP | EP1 | EP2 | EP3 | EP4 |
|---|---|---|---|---|---|
| (%) | 100 ± 13.33 | 63.26 ± 7.80 | 86.92 ± 37.37 | 67.49 ± 10.39 | 64.05 ± 5.85 |
| r (L/mg) | 6.83 ± 4.54 | 1.93 ± 0.71 | 1.03 ± 0.44 | 1.51 ± 0.45 | 2.13 ± 0.14 |
| (mg/L) | 1.73 ± 0.11 | 2.33 ± 0.20 | 3.09 ± 0.94 | 2.57 ± 0.27 | 2.09 ± 0.14 |
| 0.971 | 0.992 | 0.993 | 0.995 | 0.993 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedreira, A.; Vázquez, J.A.; Romanenko, A.; García, M.R. Design and Validation of a PLC-Controlled Morbidostat for Investigating Bacterial Drug Resistance. Bioengineering 2024, 11, 815. https://doi.org/10.3390/bioengineering11080815
Pedreira A, Vázquez JA, Romanenko A, García MR. Design and Validation of a PLC-Controlled Morbidostat for Investigating Bacterial Drug Resistance. Bioengineering. 2024; 11(8):815. https://doi.org/10.3390/bioengineering11080815
Chicago/Turabian StylePedreira, Adrián, José A. Vázquez, Andrey Romanenko, and Míriam R. García. 2024. "Design and Validation of a PLC-Controlled Morbidostat for Investigating Bacterial Drug Resistance" Bioengineering 11, no. 8: 815. https://doi.org/10.3390/bioengineering11080815
APA StylePedreira, A., Vázquez, J. A., Romanenko, A., & García, M. R. (2024). Design and Validation of a PLC-Controlled Morbidostat for Investigating Bacterial Drug Resistance. Bioengineering, 11(8), 815. https://doi.org/10.3390/bioengineering11080815








