1. Introduction
Detection and interpretation of bioacoustic events are vital in wildlife monitoring, marine life health monitoring, multimedia indexing, and audio surveillance [
1]. These tasks are challenging due to the scarcity of annotated data, as well as class imbalances, noisy environments, and the need for specialized skills to annotate the data. Deep learning has succeeded in recent years by utilizing a vast amount of labeled data for training. Many approaches have been introduced, resulting in good accuracy. However, accuracy can suffer from insufficient amounts of labeled data. Humans can recognize a new class by looking at only a few samples, or even just one. For instance, children can generalize about pandas based on a single image, or will hear about pandas from other children. From that perspective, few-shot learning (FSL) is a promising strategy that learns from small amounts of labeled data. It has emerged as a solution to the aforementioned problems [
2,
3], and is analogous to the way the human brain functions [
4].
In the current age of information technology, there is a lot of unstructured data available, but it can be hard to find annotated data [
5]. Bioacoustic event detection determines whether (and when) a specific animal vocalization occurs in an audio recording [
6]. To start, it can be challenging to record vocalizations of some species; furthermore, only people with specialized skills can annotate the obtained data, which is a labor-intensive process. In that case, few-shot (FS) bioacoustic event detection has arisen as a novel field of study into annotating lengthy recordings [
7]. Additionally, widely recognized methods build on standard supervised learning with regard to effectiveness in other sound event detection (SED) scenarios, which might not deliver the same performance in this particular task. It is reasonable to claim that FS bioacoustic event detection is interesting for research because it can meet the demand.
1.1. Related Work
FS bioacoustic event detection is a task that involves identifying the onset and termination of acoustic events in a series of audio recordings obtained from natural environments. In recent studies, the acoustic characteristics of advanced core structures have been shown to significantly impact the vibroacoustic performance of sandwich systems enhancing the overall sound transmission loss (STL) of the system [
8]. Researchers have investigated how bioacoustics presents unique challenges that make it a particularly demanding application domain for SED [
9,
10]. The bioacoustic environment is surprising and varied. Species, noises, and recording circumstances can affect acoustic signatures considerably. Earlier methods focused on manually extracting features and using basic signal processing methods, which worked well in controlled situations [
11]. However, those methods had trouble with the complexity and variability of bioacoustic environments in the real world. With the emergence of deep learning, there has been a significant change in the way we approach data analysis. Convolutional neural networks (CNNs) and recurrent neural networks (RNNs) have proven highly effective in detecting complex patterns in bioacoustic data [
12]. Nevertheless, these methods often need large quantities of labeled data, thus posing a significant challenge to the time-consuming process of obtaining annotated bioacoustic recordings and to the poor generalizations that result from a lack of supervised data [
10,
11,
12,
13,
14,
15,
16,
17].
Researchers have proposed FSL as a solution to the problem of training with small amounts of labeled data [
18]. An FSL classifier can potentially detect novel classes not present in the training set with just a few samples from each new class. It has been observed that SED only locates the onset and termination of certain sounds of interest, but FSL can identify a new class with just a few labeled samples by fusing the concept with SED [
19,
20]. N-way K-shot classification is typically used in FSL studies [
20], where N is the number of classes, and K is the number of known samples for each class. The problem of bioacoustic event detection has received a lot of attention, and datasets have been released every year from the Detection and Classification of Acoustic Scenes and Events (DCASE) community. Different methods have been used to handle FSL problems. Some compute embeddings (learned representation spaces) to help distinguish between classes not seen before by using prior information about similarities between sounds [
21,
22]. A baseline system uses a prototypical network for FS SED detection, which works on computing Euclidean distance between class prototypes [
18]. But due to background noise and acoustic interference, the class prototypes obtained from such a support set may not precisely represent the class center.
Recent studies demonstrated that it is still insufficient to use a simple prototypical network for a small-scale support set to correctly imply the class center [
23]. Environmental audio recordings contain a lot of background noise; per-channel energy normalization (PCEN) techniques are mostly applied to improve the quality of the data [
24]. This technique involves normalizing the energy of each channel, Gaussianizing the magnitude, and de-correlating the frequency band. However, the most essential factor in the performance of FS bioacoustic event detection is feature extraction. In this process, the most suitable audio features for detection of acoustic events are the time–frequency representation known as a Mel spectrogram and extracted from raw audio data [
25,
26]. Furthermore, in most of the previous research, only Mel spectrograms were utilized or a fine-tuned model overfits the support set. At the same time, other vital features were not investigated [
27]. Additionally, the potential of semi-supervised techniques and various loss functions remains untapped. Overall, all the issues regarding data scarcity, environmental variability, environmental conservation, and overfitting challenges provide significant motivation to explore and present a non-intrusive, scalable, diverse, and comprehensive bioacoustic event detection approach.
1.2. Motivation
The motivation for using transductive inference for sound event detection is that it more effectively represents class prototypes in a few-shot setting compared to other methods, such as inductive learning. More specifically, transductive inference uses a large amount of unlabeled data that is often available in many real-world applications to improve the model’s performance. To create new training examples, data augmentation is a process that transforms the original data. It helps a model learn more robust representations, and improves generalizations about new and unseen data. The combination of data augmentation and transductive inference was motivated by the fact that they complement each other and can lead to even better performance in SED tasks with a limited amount of labeled data. Data augmentation increases diversity in the labeled data, while transductive inference helps the model better utilize the unlabeled data to improve representations of class prototypes. A Mel spectrogram with PCEN is adopted as an input feature due to its excellent performance in most audio processing tasks.
1.3. Contribution
This article details the problems of FS bioacoustic event detection and proposes an effective method for solving them. The major contributions of this research are outlined below:
We investigate, examine, and comprehend audio feature extraction. We use an efficient and unique FSL technique to detect the onset and termination of a bioacoustic event in the DCASE-2022 and DCASE-2021 audio datasets. Moreover, a Mel spectrogram with PCEN is applied to the audio features, which yields higher performance in acoustic event detection.
We calculate cross-entropy loss between predicted and actual labels, and then use back-propagation to update model parameters. The frozen backbone model is utilized as the feature extractor. Then, a classifier with parameterized prototypes for each class in the support set is built, which leads to greater accuracy than from the baseline system. Hence, we provide an innovative approach to fine-tune the feature extractor and make it task-specific.
The subsequent sections of this paper are structured in the following manner:
Section 2 illustrates the proposed methodology regarding few-shot bioacoustic event detection. It includes all significant mathematical modeling and computations related to the proposed technique.
Section 3 describes the experimental setup, the datasets, and the performance metrics to illustrate the proposed methodology more effectively. It presents the results used to validate the simulation and analysis, as well as to compare the suggested method to other advanced approaches. Finally, conclusions are drawn in
Section 4.
2. Our Methodology
This section elaborates on the FS bioacoustic event detection method employing transductive inference with data augmentation.
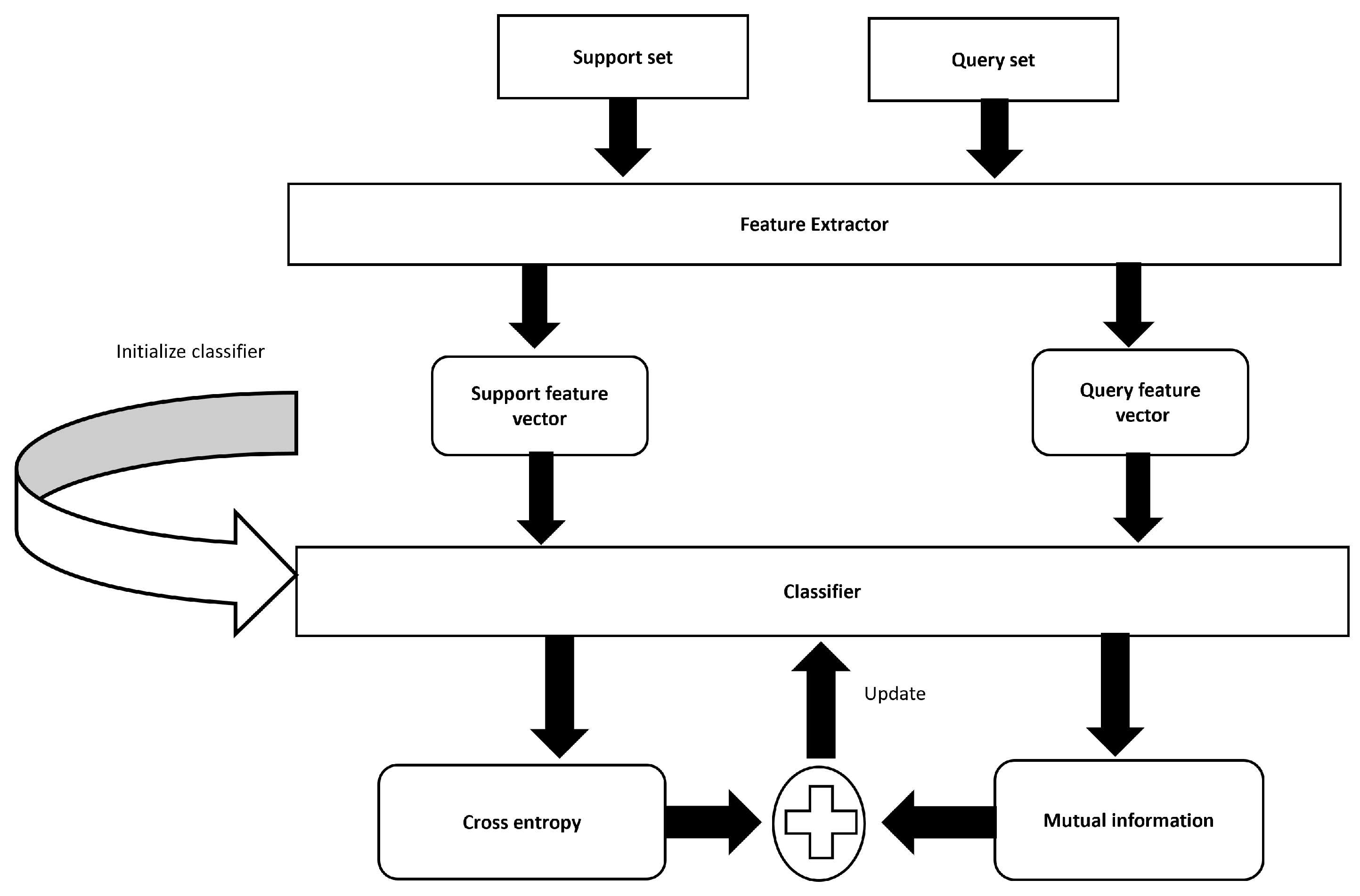
Figure 1 illustrates our proposed method that employs transductive inference by leveraging both labeled support sets and unlabeled query sets. The Mel spectrogram is used as an input feature that gives our model access to sound data. PCEN is then applied to the Mel spectrogram, which reduces the dynamic range and normalizes the energy level across channels. Moreover, PCEN reduces background noise and improves robustness to channel distortion. The transformed features are then fed into a transductive learning framework, which increases the mutual information between the features of the labeled training data and unlabeled test data. The stages involved in the suggested method are subdivided into their individual components below.
- (a)
Features Extraction
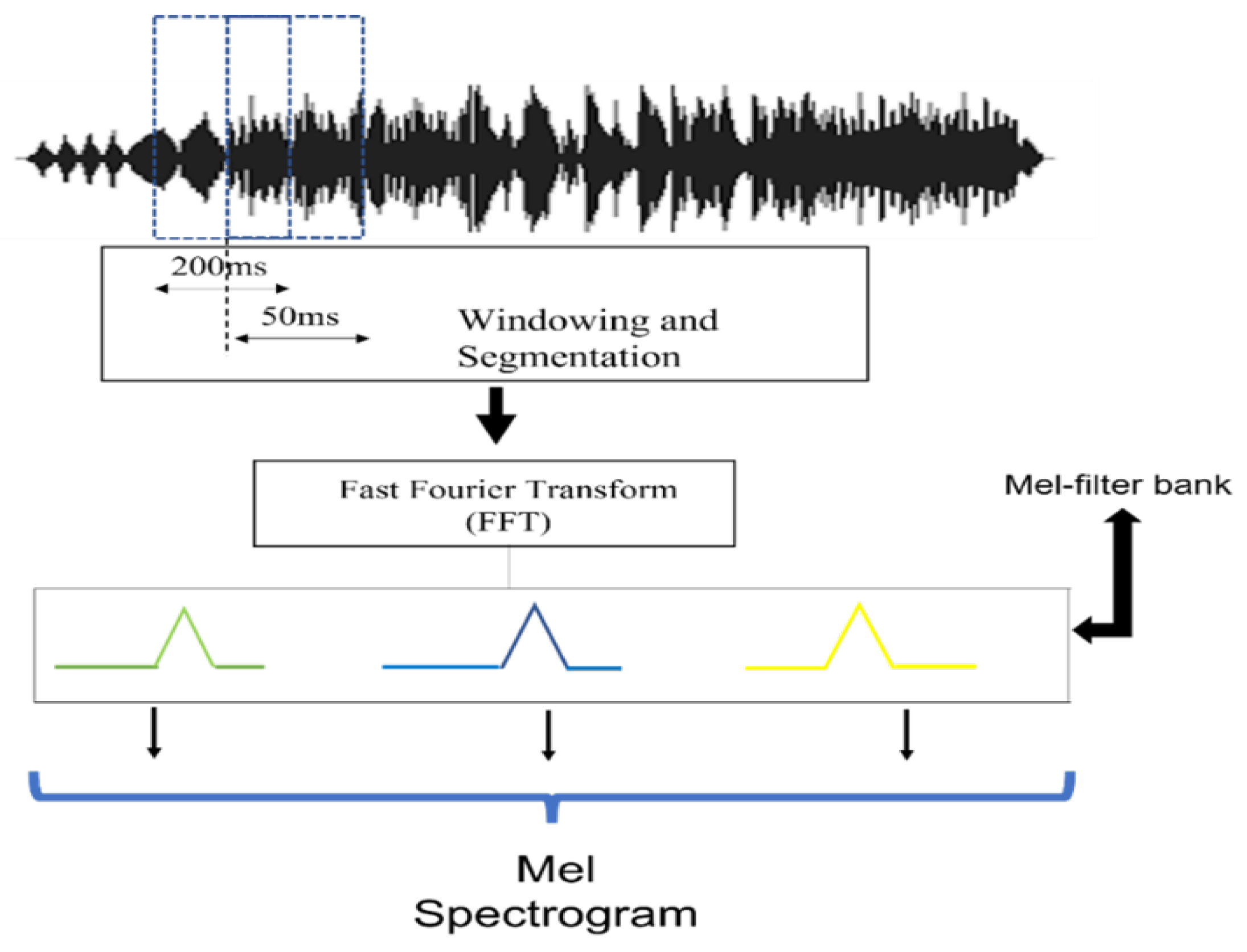
This segment takes a different path compared to traditional methodologies. We concentrate on extracting raw audio features, incorporating contextual and temporal characteristics. This enriches the FSL feature set because a careful approach is essential in bioacoustics, where the intricacies of sound can reflect important ecological events or behaviors. Here, the Mel spectrogram with PCEN is used and features are extracted using Python’s librosa package. The function librosa.load puts the audio signal into memory and returns it as a two-dimensional NumPy array along with the sample rate. After that, computing a short-time Fourier transform (STFT) of the audio signal results in the NumPy array representing the audio signal spectrogram. The resulting complex valued spectrogram is then converted to the Mel frequency domain by applying a Mel filterbank, which maps the linear frequency bins to Mel frequency bins.
Table 1 lists the parameters in this research to extract a Mel spectrogram. After the Mel spectrogram is created, PCEN is applied to reduce background noise and improve robustness to channel distortion, which upgrades the performance of the acoustic event detection task.
Figure 2 shows the Mel spectrogram feature extraction process in this research.
The original datasets were resampled to a fixed sample rate of 22.05 kHz, and the major parameters are listed in
Table 1. Segment length is the length of each audio segment processed to extract the Mel spectrogram. Hop size, known as stride or step size, determines the overlap between consecutive frames or segments of a signal. The size of the fast Fourier transform (FFT) applied to the audio segments is denoted by n_fft and n_Mel is the number of Mel frequency bins used to represent the audio signal. The Mel scale is a non-linear frequency scale designed to better reflect the human perception of sound. The size of the hops in the Mel frequency bins is denoted with hop_Mel and eps denotes the value used as a small constant to prevent division by zero when calculating PCEN, which helps maintain a consistent and non-zero energy level across different audio segments, even in the presence of low-level noise. Thus, eps serves as the lower bound of the Mel spectrum to prevent log 0.
After feature extraction, it is necessary to carefully examine the shape of the input features fed into the backbone network for training. There were 101,702 and 17,168 training samples taken from the DCASE-2022 and DCASE-2021 datasets, respectively, and 392,182 and 284,136 samples, respectively, for evaluation.
- (b)
Data Augmentation
There are two types of data augmentation techniques, i.e., raw data augmentation and SpecAugment [
28,
29]. Raw data augmentation applies transformations such as pitch shift and noise injection to the audio waveform. Meanwhile, SpecAugment operates on the spectrogram representation of the audio data to enhance the target model’s robustness and provide better class prototype representation. SpecAugment is effective in acoustic event detection tasks by introducing variations and enhancing the diversity of the training data. The operations in this method include time masking, frequency masking, and time warping, shown in
Figure 3.
Time Masking: Time masking entails choosing a random time segment in the spectrogram and setting the values to zero or another constant. The parameter t is generated randomly from a discrete uniform distribution between 0 and a maximum time mask parameter . This makes the model insensitive to minor changes in time and thus enhances the model’s ability to handle abrupt changes.
Frequency Masking: Frequency masking is similar to time masking but is applied along the frequency axis and is used to reduce high-frequency noise. The random frequency band of is selected and its values are concealed. The function f is derived randomly from a uniform distribution over the range of 0 and a maximum frequency mask parameter . This leads to a model that is less sensitive to small shifts in frequency, thus improving the model’s ability to generalize.
Time Warping: Time warping involves stretching or compressing the time axis of the spectrogram. A random anchor point is selected along the time axis, and a random warp factor is applied to the left and right of this point. The warping process displaces the spectrogram values to emulate minor speed changes in the audio to assist the model in recognizing temporal distortions. These techniques in aggregate increase the raw number of inputs seen by the model and make the training data more varied, thereby increasing the model’s stability and applicability.
- (c)
Backbone Building
In FSL, the goal is to learn a model that can quickly adapt to new tasks with only a few labeled examples. A common approach in FSL is to use a backbone model pre-trained on a large dataset and then fine-tune it on the FSL task [
30]. Choosing a backbone model for the FSL task significantly impacts the model’s performance [
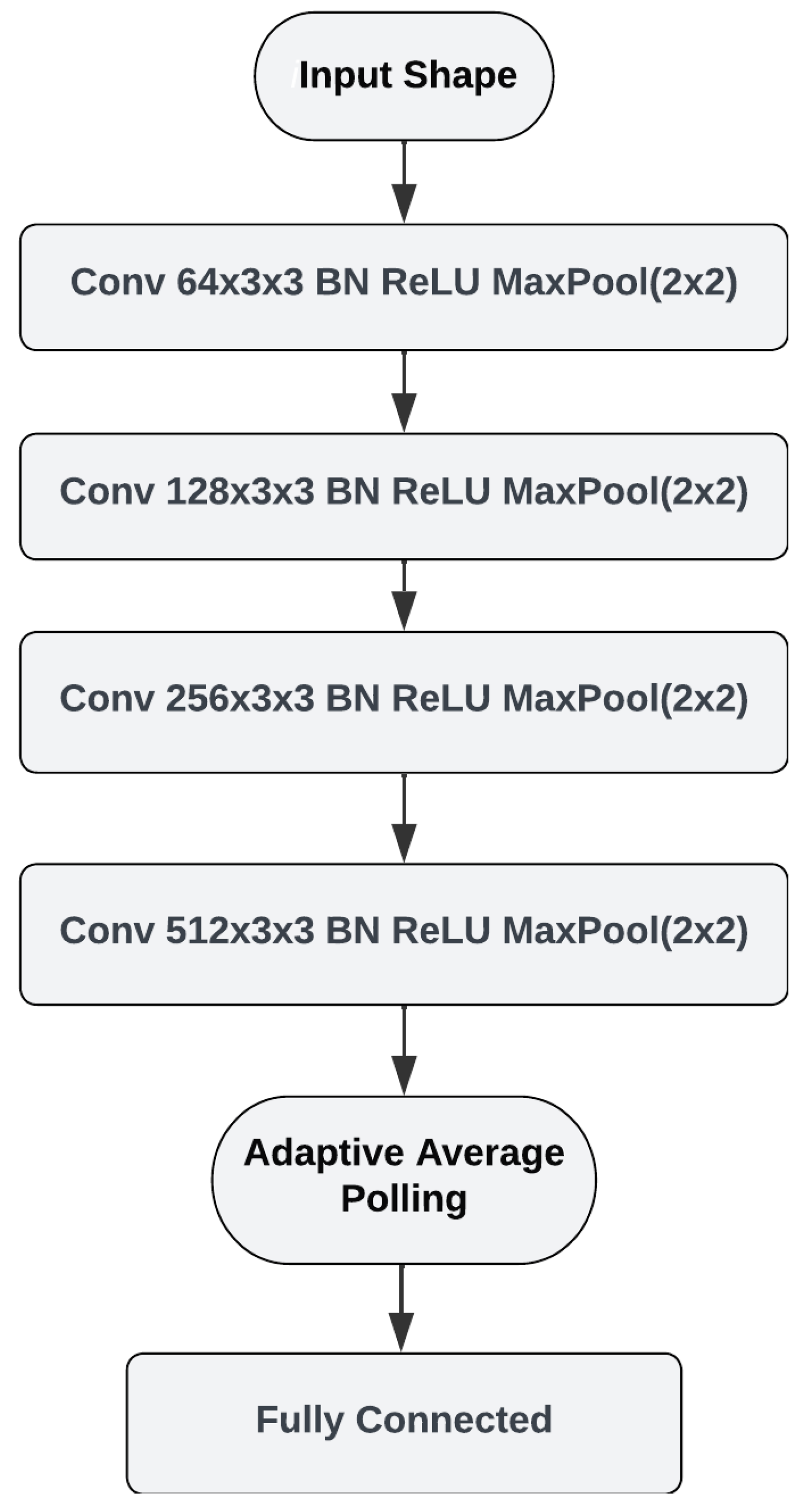
31]. To classify bioacoustic events in the FS setting, we designed a prototypical network using a four-layer CNN architecture.
Figure 4 shows the architecture of the backbone model. The input for our network is the Mel spectrogram extracted from an audio recording. The CNN architecture consists of four convolutional layers with number of filters ranging from 64 to 512. The initial learning rate was adjusted to 0.001, the scheduler gamma to 0.5, and the step size was set to update the learning rate every five epochs; a batch size = 64 was chosen because it is the right choice for the server’s GPU. The Adam optimizer with a loss function for cross-entropy and mutual information is used in the model, which is trained by varying the number of epochs between 5 and 30 for the best performance. Batch normalization is applied after each convolutional layer to normalize the output and improve training performance. MaxPooling is applied after each convolutional layer to downsample the feature maps. Average pooling is then applied to obtain the class prototypes.
Transductive Learning: The purpose of transductive learning is to create predictions for certain test cases that are already known, as opposed to making a generalization that applies to all possible test examples [
32]. Here, the model is trained on a certain subset of data points that have been labeled, and then, it is given the task of predicting labels in a subset of data points that have not been labeled. We have a support set
and a query set
for a specific FS task:
and
. We denote the feature extractor as
, and
Z expresses the set of extracted features. A base dataset,
, is given to pre-train the feature extractor. This dataset contains numerous labeled examples from various classes and provides a robust initialization for the model’s parameters, ensuring the feature extractor learns generalizable features before being fine-tuned on the few-shot learning task. The construction principle of the support set (S) is to select a small subset of labeled data points from the target class representing categories that need to be learned. It provides the labeled examples needed for the model to adapt to new, previously unseen classes. The query set (Q) contains unlabeled examples from the same target classes as the support set (S). Transductive learning aims to improve the model’s accuracy by leveraging both the labeled support set (S) and the unlabeled query set (Q). During the transductive inference process, the model iteratively refines its predictions by considering the entire distribution of the query set. This approach enhances the model’s ability to generalize from the few-shot support set to the unlabeled query set, thereby improving performance in cases with limited labeled data.
During the inference process, a technique known as transductive information maximization (TIM) increases the level of mutual information between the query instances and their projected labels [
33]. This approach is exceptional because it optimizes the mutual information between labeled and unlabeled data within a transductive framework. At this vital stage, the focus is on enhancing the correlation between data projections and labels, thereby utilizing the underlying data structure to its fullest potential. This involves not only the labeled dataset but incorporating unlabeled data. Here, transductive inference, which takes into consideration the entire data distribution, allows TIM to increase model generalizations about previously unknown data, going beyond traditional learning models. Moreover, iterative refinement entails continuously updating pseudo-labels for the unlabeled data and enhancing mutual information with each iteration until the model achieves optimal performance.
Specifically, various backbone networks underwent training using the standard supervised learning approach with the training data provided by the DCASE task organizers, and then we employed standard supervised learning. Here, cross-entropy loss is calculated between the predicted label and actual labels, and back-propagation is used to update the model parameters. A classifier is parameterized,
, and its parameters are initialized with the prototypes of each class in the support set. The backbone model is frozen and employed as a feature extractor. Regarding labels given to the features, posterior distribution is prescribed as follows:
Correspondingly, the marginal distribution across query labels is outlined:
and
are calculated:
Subsequently, the classifier is fine-tuned by employing the loss function:
Equation (
4) considers both mutual information loss and conventional cross-entropy loss on the support set:
in which
and
represent the true label of the sample within the support set and the prediction result, respectively. Thus,
is a hyper-parameter, and we set
.