Updated Toolbox for Assessing Neuronal Network Reconstruction after Cell Therapy
Abstract
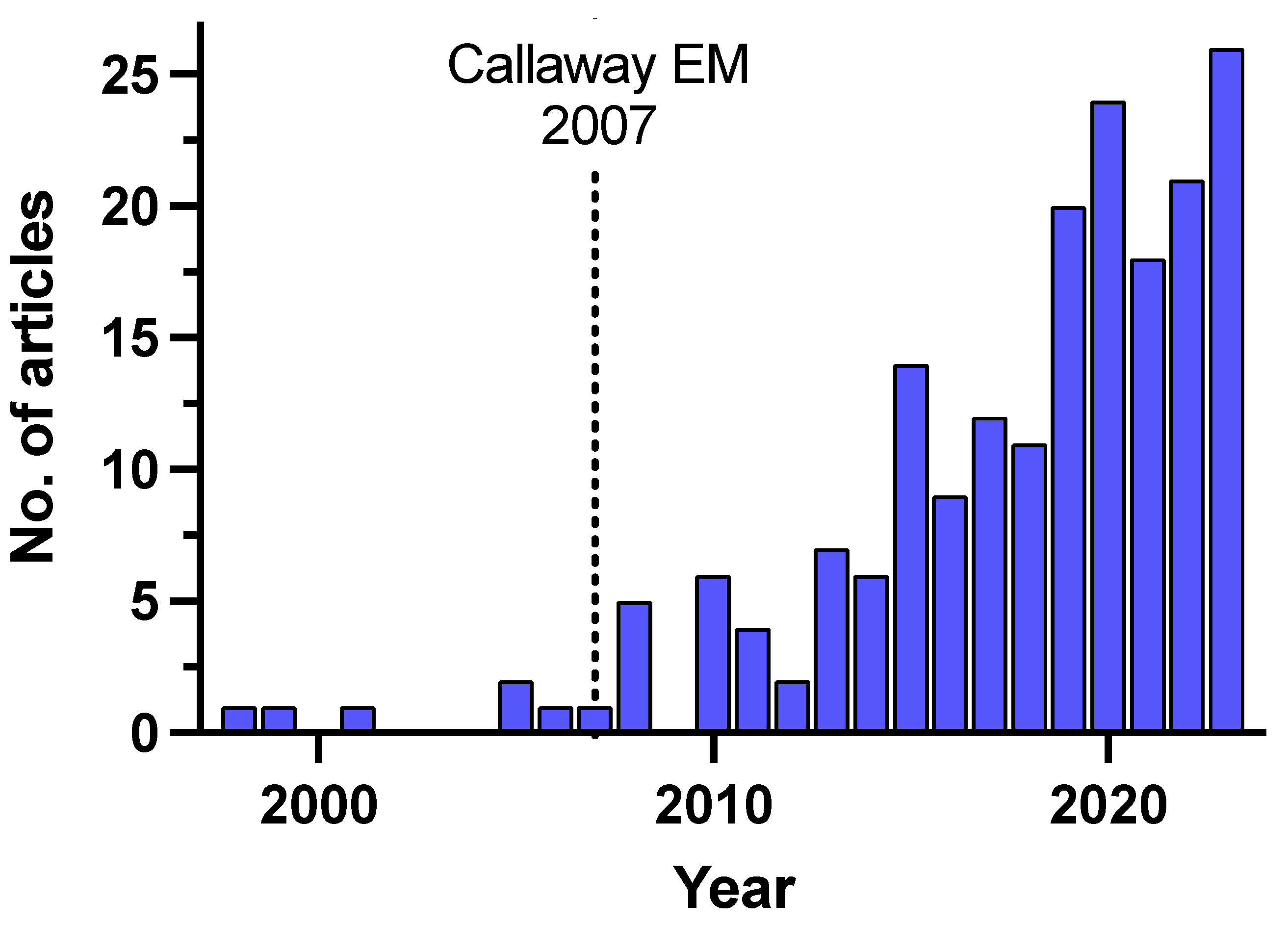
1. Introduction
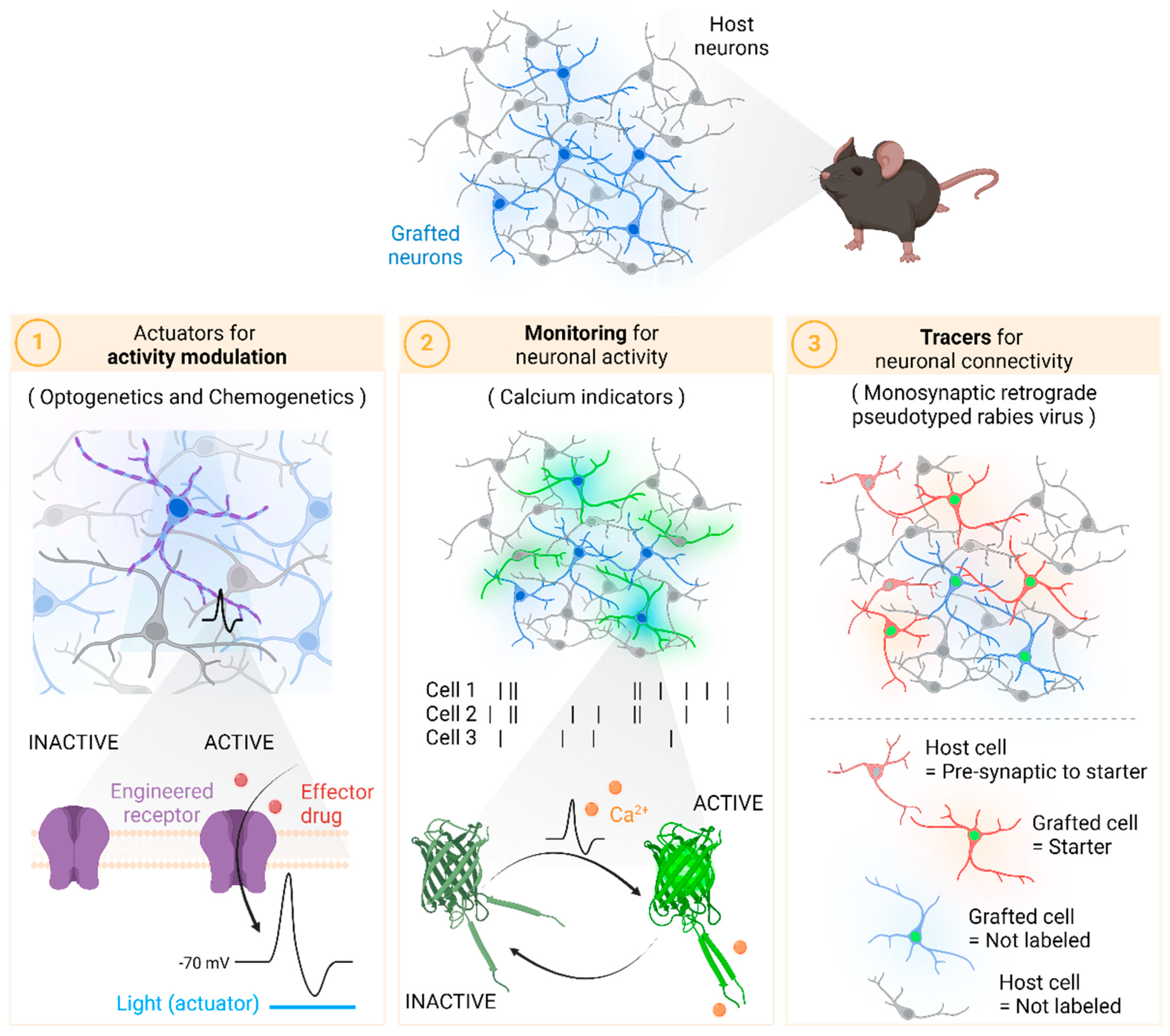
2. Actuators for Activity Modulation
2.1. Optogenetics
2.2. Chemogenetics
2.3. Advantages and Limitations
2.4. Proving Integration and Functionality
2.5. Other Applications
3. Monitoring Neuronal Activity
3.1. GECIs for In Vivo Applications
3.2. In Vivo Calcium Imaging in Head-Restrained Animals
3.3. In Vivo Calcium Imaging in Freely Moving Animals
3.4. Other Specific Indicators of Neuronal Activity
3.5. Future Perspectives in Imaging Technologies for Neuronal Connectivity
4. Tracers for Neuronal Functional Connectivity
4.1. Retrograde Tracing Using Rabies Virus
4.2. Anterograde Tracing Using Herpes Virus
4.3. Future Perspectives in Mapping Communication Pathways after Cell Transplantation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tornero, D. Neuronal Circuitry Reconstruction after Stem Cell Therapy in Damaged Brain. Neural Regen. Res. 2022, 17, 1959–1960. [Google Scholar] [CrossRef] [PubMed]
- Palma-Tortosa, S.; Coll-San Martin, B.; Kokaia, Z.; Tornero, D. Neuronal Replacement in Stem Cell Therapy for Stroke: Filling the Gap. Front. Cell Dev. Biol. 2021, 9, 662636. [Google Scholar] [CrossRef] [PubMed]
- Deisseroth, K. Optogenetics: 10 Years of Microbial Opsins in Neuroscience. Nat. Neurosci. 2015, 18, 1213–1225. [Google Scholar] [CrossRef]
- Stosiek, C.; Garaschuk, O.; Holthoff, K.; Konnerth, A. In Vivo Two-Photon Calcium Imaging of Neuronal Networks. Proc. Natl. Acad. Sci. USA 2003, 100, 7319–7324. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Jacobs, M.W.; Ito-Cole, T.; Callaway, E.M. Improved Monosynaptic Neural Circuit Tracing Using Engineered Rabies Virus Glycoproteins. Cell Rep. 2016, 15, 692–699. [Google Scholar] [CrossRef]
- Garner, A.R.; Rowland, D.C.; Hwang, S.Y.; Baumgaertel, K.; Roth, B.L.; Kentros, C.; Mayford, M. Generation of a Synthetic Memory Trace. Science 2012, 335, 1513–1516. [Google Scholar] [CrossRef] [PubMed]
- Alexander, G.M.; Rogan, S.C.; Abbas, A.I.; Armbruster, B.N.; Pei, Y.; Allen, J.A.; Nonneman, R.J.; Hartmann, J.; Moy, S.S.; Nicolelis, M.A.; et al. Remote Control of Neuronal Activity in Transgenic Mice Expressing Evolved G Protein-Coupled Receptors. Neuron 2009, 63, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Teissier, A.; Chemiakine, A.; Inbar, B.; Bagchi, S.; Ray, R.S.; Palmiter, R.D.; Dymecki, S.M.; Moore, H.; Ansorge, M.S. Activity of Raphé Serotonergic Neurons Controls Emotional Behaviors. Cell Rep. 2015, 13, 1965–1976. [Google Scholar] [CrossRef]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-Timescale, Genetically Targeted Optical Control of Neural Activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, L.-P.; Brauner, M.; Liewald, J.F.; Kay, K.; Watzke, N.; Wood, P.G.; Bamberg, E.; Nagel, G.; Gottschalk, A.; et al. Multimodal Fast Optical Interrogation of Neural Circuitry. Nature 2007, 446, 633–639. [Google Scholar] [CrossRef]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a Directly Light-Gated Cation-Selective Membrane Channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Ollig, D.; Fuhrmann, M.; Kateriya, S.; Musti, A.M.; Bamberg, E.; Hegemann, P. Channelrhodopsin-1: A Light-Gated Proton Channel in Green Algae. Science 2002, 296, 2395–2398. [Google Scholar] [CrossRef]
- Chow, B.Y.; Han, X.; Dobry, A.S.; Qian, X.; Chuong, A.S.; Li, M.; Henninger, M.A.; Belfort, G.M.; Lin, Y.; Monahan, P.E.; et al. High-Performance Genetically Targetable Optical Neural Silencing by Light-Driven Proton Pumps. Nature 2010, 463, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Chaffiol, A.; Macé, E.; Caplette, R.; Desrosiers, M.; Lampič, M.; Forster, V.; Marre, O.; Lin, J.Y.; Sahel, J.; et al. Red-shifted Channelrhodopsin Stimulation Restores Light Responses in Blind Mice, Macaque Retina, and Human Retina. EMBO Mol. Med. 2016, 8, 1248–1264. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Mendoza-Halliday, D.; Ting, J.T.; Kaiser, T.; Sun, X.; Bastos, A.M.; Wimmer, R.D.; Guo, B.; Chen, Q.; Zhou, Y.; et al. An Ultra-Sensitive Step-Function Opsin for Minimally Invasive Optogenetic Stimulation in Mice and Macaques. Neuron 2020, 107, 38–51.e8. [Google Scholar] [CrossRef] [PubMed]
- Gunaydin, L.A.; Yizhar, O.; Berndt, A.; Sohal, V.S.; Deisseroth, K.; Hegemann, P. Ultrafast Optogenetic Control. Nat. Neurosci. 2010, 13, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.K.; Flannery, J.G. Innovative Optogenetic Strategies for Vision Restoration. Front. Cell Neurosci. 2018, 12, 316. [Google Scholar] [CrossRef]
- Zhang, F.; Aravanis, A.M.; Adamantidis, A.; de Lecea, L.; Deisseroth, K. Circuit-Breakers: Optical Technologies for Probing Neural Signals and Systems. Nat. Rev. Neurosci. 2007, 8, 577–581. [Google Scholar] [CrossRef]
- Lin, J.Y. A User’s Guide to Channelrhodopsin Variants: Features, Limitations and Future Developments. Exp. Physiol. 2011, 96, 19–25. [Google Scholar] [CrossRef]
- Roth, B.L. DREADDs for Neuroscientists. Neuron 2016, 89, 683–694. [Google Scholar] [CrossRef]
- Dong, S.; Rogan, S.C.; Roth, B.L. Directed Molecular Evolution of DREADDs: A Generic Approach to Creating next-Generation RASSLs. Nat. Protoc. 2010, 5, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Strader, C.D.; Gaffney, T.; Sugg, E.E.; Candelore, M.R.; Keys, R.; Patchett, A.A.; Dixon, R.A. Allele-Specific Activation of Genetically Engineered Receptors. J. Biol. Chem. 1991, 266, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Magnus, C.J.; Lee, P.H.; Atasoy, D.; Su, H.H.; Looger, L.L.; Sternson, S.M. Chemical and Genetic Engineering of Selective Ion Channel–Ligand Interactions. Science 2011, 333, 1292–1296. [Google Scholar] [CrossRef] [PubMed]
- Arenkiel, B.R.; Klein, M.E.; Davison, I.G.; Katz, L.C.; Ehlers, M.D. Genetic Control of Neuronal Activity in Mice Conditionally Expressing TRPV1. Nat. Methods 2008, 5, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Humbert, N.; Gradinaru, J.; Ivanova, A.; Gilardoni, F.; Rusbandi, U.E.; Ward, T.R. Tailoring the Active Site of Chemzymes by Using a Chemogenetic-Optimization Procedure: Towards Substrate-Specific Artificial Hydrogenases Based on the Biotin-Avidin Technology. Angew. Chem. Int. Ed. 2005, 44, 7764–7767. [Google Scholar] [CrossRef] [PubMed]
- Collot, J.; Gradinaru, J.; Humbert, N.; Skander, M.; Zocchi, A.; Ward, T.R. Artificial Metalloenzymes for Enantioselective Catalysis Based on Biotin−Avidin. J. Am. Chem. Soc. 2003, 125, 9030–9031. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.C.; Das, T.K.; Shokat, K.M.; Cagan, R.L. Chemical Genetic Discovery of Targets and Anti-Targets for Cancer Polypharmacology. Nature 2012, 486, 80–84. [Google Scholar] [CrossRef]
- Cohen, M.S.; Zhang, C.; Shokat, K.M.; Taunton, J. Structural Bioinformatics-Based Design of Selective, Irreversible Kinase Inhibitors. Science 2005, 308, 1318–1321. [Google Scholar] [CrossRef]
- Bishop, A.C.; Shah, K.; Liu, Y.; Witucki, L.; Kung, C.; Shokat, K.M. Design of Allele-Specific Inhibitors to Probe Protein Kinase Signaling. Curr. Biol. 1998, 8, 257–266. [Google Scholar] [CrossRef]
- Conklin, B.R.; Hsiao, E.C.; Claeysen, S.; Dumuis, A.; Srinivasan, S.; Forsayeth, J.R.; Guettier, J.-M.; Chang, W.C.; Pei, Y.; McCarthy, K.D.; et al. Engineering GPCR Signaling Pathways with RASSLs. Nat. Methods 2008, 5, 673–678. [Google Scholar] [CrossRef]
- Pei, Y.; Rogan, S.C.; Yan, F.; Roth, B.L. Engineered GPCRs as Tools to Modulate Signal Transduction. Physiology 2008, 23, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Dong, S.; Roth, B.L. Generation of Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) Using Directed Molecular Evolution. Curr. Protoc. Neurosci. 2010, 50, 4–33. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, B.N.; Li, X.; Pausch, M.H.; Herlitze, S.; Roth, B.L. Evolving the Lock to Fit the Key to Create a Family of G Protein-Coupled Receptors Potently Activated by an Inert Ligand. Proc. Natl. Acad. Sci. USA 2007, 104, 5163–5168. [Google Scholar] [CrossRef] [PubMed]
- Magnus, C.J.; Lee, P.H.; Bonaventura, J.; Zemla, R.; Gomez, J.L.; Ramirez, M.H.; Hu, X.; Galvan, A.; Basu, J.; Michaelides, M.; et al. Ultrapotent Chemogenetics for Research and Potential Clinical Applications. Science 2019, 364, eaav5282. [Google Scholar] [CrossRef] [PubMed]
- Vardy, E.; Robinson, J.E.; Li, C.; Olsen, R.H.J.; DiBerto, J.F.; Giguere, P.M.; Sassano, F.M.; Huang, X.-P.; Zhu, H.; Urban, D.J.; et al. A New DREADD Facilitates the Multiplexed Chemogenetic Interrogation of Behavior. Neuron 2015, 86, 936–946. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Miyakawa, N.; Takuwa, H.; Hori, Y.; Oyama, K.; Ji, B.; Takahashi, M.; Huang, X.-P.; Slocum, S.T.; DiBerto, J.F.; et al. Deschloroclozapine, a Potent and Selective Chemogenetic Actuator Enables Rapid Neuronal and Behavioral Modulations in Mice and Monkeys. Nat. Neurosci. 2020, 23, 1157–1167. [Google Scholar] [CrossRef]
- Chen, X.; Choo, H.; Huang, X.-P.; Yang, X.; Stone, O.; Roth, B.L.; Jin, J. The First Structure–Activity Relationship Studies for Designer Receptors Exclusively Activated by Designer Drugs. ACS Chem. Neurosci. 2015, 6, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Haubensak, W.; Anthony, T.E.; Anderson, D.J. Central Amygdala PKC-Δ+ Neurons Mediate the Influence of Multiple Anorexigenic Signals. Nat. Neurosci. 2014, 17, 1240–1248. [Google Scholar] [CrossRef]
- Miao, C.; Cao, Q.; Ito, H.T.; Yamahachi, H.; Witter, M.P.; Moser, M.-B.; Moser, E.I. Hippocampal Remapping after Partial Inactivation of the Medial Entorhinal Cortex. Neuron 2015, 88, 590–603. [Google Scholar] [CrossRef]
- Betley, J.N.; Xu, S.; Cao, Z.F.H.; Gong, R.; Magnus, C.J.; Yu, Y.; Sternson, S.M. Neurons for Hunger and Thirst Transmit a Negative-Valence Teaching Signal. Nature 2015, 521, 180–185. [Google Scholar] [CrossRef]
- Portugues, R.; Severi, K.E.; Wyart, C.; Ahrens, M.B. Optogenetics in a Transparent Animal: Circuit Function in the Larval Zebrafish. Curr. Opin. Neurobiol. 2013, 23, 119–126. [Google Scholar] [CrossRef]
- Chuong, A.S.; Miri, M.L.; Busskamp, V.; Matthews, G.A.C.; Acker, L.C.; Sørensen, A.T.; Young, A.; Klapoetke, N.C.; Henninger, M.A.; Kodandaramaiah, S.B.; et al. Noninvasive Optical Inhibition with a Red-Shifted Microbial Rhodopsin. Nat. Neurosci. 2014, 17, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.; Cho, J.-H.; Leung, A.; Savvidis, G.; Ahn, S.; Moon, M.; Lee, P.K.J.; Han, J.J.; Azimi, N.; Kim, K.-S.; et al. HPSC-Derived Maturing GABAergic Interneurons Ameliorate Seizures and Abnormal Behavior in Epileptic Mice. Cell Stem Cell 2014, 15, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Waloschková, E.; Gonzalez-Ramos, A.; Mikroulis, A.; Kudláček, J.; Andersson, M.; Ledri, M.; Kokaia, M. Human Stem Cell-Derived GABAergic Interneurons Establish Efferent Synapses onto Host Neurons in Rat Epileptic Hippocampus and Inhibit Spontaneous Recurrent Seizures. Int. J. Mol. Sci. 2021, 22, 13243. [Google Scholar] [CrossRef]
- Gonzalez-Ramos, A.; Waloschková, E.; Mikroulis, A.; Kokaia, Z.; Bengzon, J.; Ledri, M.; Andersson, M.; Kokaia, M. Human Stem Cell-Derived GABAergic Neurons Functionally Integrate into Human Neuronal Networks. Sci. Rep. 2021, 11, 22050. [Google Scholar] [CrossRef] [PubMed]
- Palma-Tortosa, S.; Tornero, D.; Hansen, M.G.; Monni, E.; Hajy, M.; Kartsivadze, S.; Aktay, S.; Tsupykov, O.; Parmar, M.; Deisseroth, K.; et al. Activity in Grafted Human IPS Cell–Derived Cortical Neurons Integrated in Stroke-Injured Rat Brain Regulates Motor Behavior. Proc. Natl. Acad. Sci. USA 2020, 117, 9094–9100. [Google Scholar] [CrossRef] [PubMed]
- Steinbeck, J.A.; Choi, S.J.; Mrejeru, A.; Ganat, Y.; Deisseroth, K.; Sulzer, D.; Mosharov, E.V.; Studer, L. Optogenetics Enables Functional Analysis of Human Embryonic Stem Cell-Derived Grafts in a Parkinson’s Disease Model. Nat. Biotechnol. 2015, 33, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Upadhya, D.; Hattiangady, B.; Castro, O.W.; Shuai, B.; Kodali, M.; Attaluri, S.; Bates, A.; Dong, Y.; Zhang, S.-C.; Prockop, D.J.; et al. Human Induced Pluripotent Stem Cell-Derived MGE Cell Grafting after Status Epilepticus Attenuates Chronic Epilepsy and Comorbidities via Synaptic Integration. Proc. Natl. Acad. Sci. USA 2019, 116, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Sokolik, C.; Liu, Y.; Bauer, D.; McPherson, J.; Broeker, M.; Heimberg, G.; Qi, L.S.; Sivak, D.A.; Thomson, M. Transcription Factor Competition Allows Embryonic Stem Cells to Distinguish Authentic Signals from Noise. Cell Syst. 2015, 1, 117–129. [Google Scholar] [CrossRef]
- Giraldo, E.; Palmero-Canton, D.; Martinez-Rojas, B.; Sanchez-Martin, M.D.M.; Moreno-Manzano, V. Optogenetic Modulation of Neural Progenitor Cells Improves Neuroregenerative Potential. Int. J. Mol. Sci. 2020, 22, 365. [Google Scholar] [CrossRef]
- Inoue, M. Genetically Encoded Calcium Indicators to Probe Complex Brain Circuit Dynamics In Vivo. Neurosci. Res. 2021, 169, 2–8. [Google Scholar] [CrossRef]
- Lin, M.Z.; Schnitzer, M.J. Genetically Encoded Indicators of Neuronal Activity. Nat. Neurosci. 2016, 19, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R.Y. New Calcium Indicators and Buffers with High Selectivity against Magnesium and Protons: Design, Synthesis, and Properties of Prototype Structures. Biochemistry 1980, 19, 2396–2404. [Google Scholar] [CrossRef] [PubMed]
- Paredes, R.M.; Etzler, J.C.; Watts, L.T.; Zheng, W.; Lechleiter, J.D. Chemical Calcium Indicators. Methods 2008, 46, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, A.; Llopis, J.; Heim, R.; McCaffery, J.M.; Adams, J.A.; Ikura, M.; Tsien, R.Y. Fluorescent Indicators for Ca2+ based on Green Fluorescent Proteins and Calmodulin. Nature 1997, 388, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Pérez Koldenkova, V.; Nagai, T. Genetically Encoded Ca2+ Indicators: Properties and Evaluation. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 1787–1797. [Google Scholar] [CrossRef] [PubMed]
- Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. Circular Permutation and Receptor Insertion within Green Fluorescent Proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 11241–11246. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Sawano, A.; Park, E.S.; Miyawaki, A. Circularly Permuted Green Fluorescent Proteins Engineered to Sense Ca2+. Proc. Natl. Acad. Sci. USA 2001, 98, 3197–3202. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Nishioka, W.K.; Derecki, N.C.; Maher, M.P. High-Throughput-Compatible Assays Using a Genetically-Encoded Calcium Indicator. Sci. Rep. 2019, 9, 12692. [Google Scholar] [CrossRef]
- Nakai, J.; Ohkura, M.; Imoto, K. A High Signal-to-Noise Ca2+ Probe Composed of a Single Green Fluorescent Protein. Nat. Biotechnol. 2001, 19, 137–141. [Google Scholar] [CrossRef]
- Heinrich, R.; Hussein, W.; Berlin, S. Photo-Transformable Genetically-Encoded Optical Probes for Functional Highlighting In Vivo. J. Neurosci. Methods 2021, 355, 109129. [Google Scholar] [CrossRef]
- Hashizume, R.; Fujii, H.; Mehta, S.; Ota, K.; Qian, Y.; Zhu, W.; Drobizhev, M.; Nasu, Y.; Zhang, J.; Bito, H.; et al. A Genetically Encoded Far-red Fluorescent Calcium Ion Biosensor Derived from a Biliverdin-binding Protein. Protein Sci. 2022, 31, e4440. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive Fluorescent Proteins for Imaging Neuronal Activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef]
- Dana, H.; Mohar, B.; Sun, Y.; Narayan, S.; Gordus, A.; Hasseman, J.P.; Tsegaye, G.; Holt, G.T.; Hu, A.; Walpita, D.; et al. Sensitive Red Protein Calcium Indicators for Imaging Neural Activity. eLife 2016, 5, e12727. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, K.; Matsui, T.; Bito, H.; Ohki, K. Astrocytes in the Mouse Visual Cortex Reliably Respond to Visual Stimulation. Biochem. Biophys. Res. Commun. 2018, 505, 1216–1222. [Google Scholar] [CrossRef]
- Inoue, M.; Takeuchi, A.; Manita, S.; Horigane, S.; Sakamoto, M.; Kawakami, R.; Yamaguchi, K.; Otomo, K.; Yokoyama, H.; Kim, R.; et al. Rational Engineering of XCaMPs, a Multicolor GECI Suite for In Vivo Imaging of Complex Brain Circuit Dynamics. Cell 2019, 177, 1346–1360.e24. [Google Scholar] [CrossRef]
- Yang, W.; Yuste, R. In Vivo Imaging of Neural Activity. Nat. Methods 2017, 14, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Resta, F.; Montagni, E.; de Vito, G.; Scaglione, A.; Allegra Mascaro, A.L.; Pavone, F.S. Large-Scale All-Optical Dissection of Motor Cortex Connectivity Shows a Segregated Organization of Mouse Forelimb Representations. Cell Rep. 2022, 41, 111627. [Google Scholar] [CrossRef]
- Carrillo-Reid, L.; Han, S.; Yang, W.; Akrouh, A.; Yuste, R. Controlling Visually Guided Behavior by Holographic Recalling of Cortical Ensembles. Cell 2019, 178, 447–457.e5. [Google Scholar] [CrossRef]
- Sha, F.; Abdelfattah, A.S.; Patel, R.; Schreiter, E.R. Erasable Labeling of Neuronal Activity Using a Reversible Calcium Marker. eLife 2020, 9, e57249. [Google Scholar] [CrossRef]
- Chen, Y.; Jang, H.; Spratt, P.W.E.; Kosar, S.; Taylor, D.E.; Essner, R.A.; Bai, L.; Leib, D.E.; Kuo, T.-W.; Lin, Y.-C.; et al. Soma-Targeted Imaging of Neural Circuits by Ribosome Tethering. Neuron 2020, 107, 454–469.e6. [Google Scholar] [CrossRef] [PubMed]
- Broussard, G.J.; Liang, Y.; Fridman, M.; Unger, E.K.; Meng, G.; Xiao, X.; Ji, N.; Petreanu, L.; Tian, L. In Vivo Measurement of Afferent Activity with Axon-Specific Calcium Imaging. Nat. Neurosci. 2018, 21, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, L.; Sun, L.; Yu, J.; Hu, Z.; Lian, K.; Cao, G.; Dai, J. In Vivo Two-Photon Calcium Imaging in Dendrites of Rabies Virus-Labeled V1 Corticothalamic Neurons. Neurosci. Bull. 2020, 36, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Wu, R.; Li, M.; Hu, Y.; Li, Y.; Li, J.; Rong, H.; Wu, H.; Xu, Y.; Lu, Y.; et al. Fast High-Resolution Miniature Two-Photon Microscopy for Brain Imaging in Freely Behaving Mice. Nat. Methods 2017, 14, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Chen, T.-W.; Hu, A.; Shields, B.C.; Guo, C.; Looger, L.L.; Kim, D.S.; Svoboda, K. Thy1-GCaMP6 Transgenic Mice for Neuronal Population Imaging In Vivo. PLoS ONE 2014, 9, e108697. [Google Scholar] [CrossRef] [PubMed]
- Rynes, M.L.; Surinach, D.A.; Linn, S.; Laroque, M.; Rajendran, V.; Dominguez, J.; Hadjistamoulou, O.; Navabi, Z.S.; Ghanbari, L.; Johnson, G.W.; et al. Miniaturized Head-Mounted Microscope for Whole-Cortex Mesoscale Imaging in Freely Behaving Mice. Nat. Methods 2021, 18, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Cramer, J.V.; Gesierich, B.; Roth, S.; Dichgans, M.; Düring, M.; Liesz, A. In Vivo Widefield Calcium Imaging of the Mouse Cortex for Analysis of Network Connectivity in Health and Brain Disease. Neuroimage 2019, 199, 570–584. [Google Scholar] [CrossRef]
- Scott, B.B.; Thiberge, S.Y.; Guo, C.; Tervo, D.G.R.; Brody, C.D.; Karpova, A.Y.; Tank, D.W. Imaging Cortical Dynamics in GCaMP Transgenic Rats with a Head-Mounted Widefield Macroscope. Neuron 2018, 100, 1045–1058.e5. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Li, M.; Zhou, Y.; Ma, L.; Qiao, Q.; Hu, W.; Li, W.; Wills, Z.P.; Gan, W.-B. Abnormal Dendritic Calcium Activity and Synaptic Depotentiation Occur Early in a Mouse Model of Alzheimer’s Disease. Mol. Neurodegener. 2017, 12, 86. [Google Scholar] [CrossRef]
- Calvo-Rodriguez, M.; Kharitonova, E.K.; Bacskai, B.J. In Vivo Brain Imaging of Mitochondrial Ca2+ in Neurodegenerative Diseases with Multiphoton Microscopy. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118998. [Google Scholar] [CrossRef]
- Linaro, D.; Vermaercke, B.; Iwata, R.; Ramaswamy, A.; Libé-Philippot, B.; Boubakar, L.; Davis, B.A.; Wierda, K.; Davie, K.; Poovathingal, S.; et al. Xenotransplanted Human Cortical Neurons Reveal Species-Specific Development and Functional Integration into Mouse Visual Circuits. Neuron 2019, 104, 972–986.e6. [Google Scholar] [CrossRef] [PubMed]
- Falkner, S.; Grade, S.; Dimou, L.; Conzelmann, K.K.; Bonhoeffer, T.; Götz, M.; Hübener, M. Transplanted Embryonic Neurons Integrate into Adult Neocortical Circuits. Nature 2016, 539, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Real, R.; Peter, M.; Trabalza, A.; Khan, S.; Smith, M.A.; Dopp, J.; Barnes, S.J.; Momoh, A.; Strano, A.; Volpi, E.; et al. In Vivo Modeling of Human Neuron Dynamics and Down Syndrome. Science 2018, 362, eaau1810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chen, C.; Yang, Z.; He, W.; Liao, X.; Ma, Q.; Deng, P.; Lu, J.; Li, J.; Wang, M.; et al. Sensory Response of Transplanted Astrocytes in Adult Mammalian Cortex In Vivo. Cereb. Cortex 2016, 26, 3690–3704. [Google Scholar] [CrossRef] [PubMed]
- Cichon, J.; Gan, W.-B. Branch-Specific Dendritic Ca2+ Spikes Cause Persistent Synaptic Plasticity. Nature 2015, 520, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Dombeck, D.A.; Harvey, C.D.; Tian, L.; Looger, L.L.; Tank, D.W. Functional Imaging of Hippocampal Place Cells at Cellular Resolution during Virtual Navigation. Nat. Neurosci. 2010, 13, 1433–1440. [Google Scholar] [CrossRef] [PubMed]
- Morcos, A.S.; Harvey, C.D. History-Dependent Variability in Population Dynamics during Evidence Accumulation in Cortex. Nat. Neurosci. 2016, 19, 1672–1681. [Google Scholar] [CrossRef]
- Umpierre, A.D.; Bystrom, L.L.; Ying, Y.; Liu, Y.U.; Worrell, G.; Wu, L.-J. Microglial Calcium Signaling Is Attuned to Neuronal Activity in Awake Mice. eLife 2020, 9, e56502. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Hill, R.A.; Damisah, E.C.; Murray, K.N.; Yuan, P.; Bordey, A.; Grutzendler, J. Imaging and Optogenetic Modulation of Vascular Mural Cells in the Live Brain. Nat. Protoc. 2021, 16, 472–496. [Google Scholar] [CrossRef]
- Werner, C.T.; Williams, C.J.; Fermelia, M.R.; Lin, D.-T.; Li, Y. Circuit Mechanisms of Neurodegenerative Diseases: A New Frontier With Miniature Fluorescence Microscopy. Front. Neurosci. 2019, 13, 494308. [Google Scholar] [CrossRef]
- Helmchen, F.; Fee, M.S.; Tank, D.W.; Denk, W. A Miniature Head-Mounted Two-Photon Microscope. Neuron 2001, 31, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Myaing, M.T.; MacDonald, D.J.; Li, X. Fiber-Optic Scanning Two-Photon Fluorescence Endoscope. Opt. Lett. 2006, 31, 1076. [Google Scholar] [CrossRef]
- Ghosh, K.K.; Burns, L.D.; Cocker, E.D.; Nimmerjahn, A.; Ziv, Y.; El Gamal, A.; Schnitzer, M.J. Miniaturized Integration of a Fluorescence Microscope. Nat. Methods 2011, 8, 871–878. [Google Scholar] [CrossRef]
- Ziv, Y.; Ghosh, K.K. Miniature Microscopes for Large-Scale Imaging of Neuronal Activity in Freely Behaving Rodents. Curr. Opin. Neurobiol. 2015, 32, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Cameron, C.M.; Murugan, M.; Choi, J.Y.; Engel, E.A.; Witten, I.B. Increased Cocaine Motivation Is Associated with Degraded Spatial and Temporal Representations in IL-NAc Neurons. Neuron 2019, 103, 80–91.e7. [Google Scholar] [CrossRef] [PubMed]
- Szabo, V.; Ventalon, C.; De Sars, V.; Bradley, J.; Emiliani, V. Spatially Selective Holographic Photoactivation and Functional Fluorescence Imaging in Freely Behaving Mice with a Fiberscope. Neuron 2014, 84, 1157–1169. [Google Scholar] [CrossRef]
- Accanto, N.; Blot, F.G.C.; Lorca-Cámara, A.; Zampini, V.; Bui, F.; Tourain, C.; Badt, N.; Katz, O.; Emiliani, V. A Flexible Two-Photon Fiberscope for Fast Activity Imaging and Precise Optogenetic Photostimulation of Neurons in Freely Moving Mice. Neuron 2023, 111, 176–189.e6. [Google Scholar] [CrossRef]
- Zhang, J.; Hughes, R.N.; Kim, N.; Fallon, I.P.; Bakhurin, K.; Kim, J.; Severino, F.P.U.; Yin, H.H. A One-Photon Endoscope for Simultaneous Patterned Optogenetic Stimulation and Calcium Imaging in Freely Behaving Mice. Nat. Biomed. Eng. 2022, 7, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Gunaydin, L.A.; Grosenick, L.; Finkelstein, J.C.; Kauvar, I.V.; Fenno, L.E.; Adhikari, A.; Lammel, S.; Mirzabekov, J.J.; Airan, R.D.; Zalocusky, K.A.; et al. Natural Neural Projection Dynamics Underlying Social Behavior. Cell 2014, 157, 1535–1551. [Google Scholar] [CrossRef]
- Kim, C.K.; Yang, S.J.; Pichamoorthy, N.; Young, N.P.; Kauvar, I.; Jennings, J.H.; Lerner, T.N.; Berndt, A.; Lee, S.Y.; Ramakrishnan, C.; et al. Simultaneous Fast Measurement of Circuit Dynamics at Multiple Sites across the Mammalian Brain. Nat. Methods 2016, 13, 325–328. [Google Scholar] [CrossRef]
- Nieh, E.H.; Vander Weele, C.M.; Matthews, G.A.; Presbrey, K.N.; Wichmann, R.; Leppla, C.A.; Izadmehr, E.M.; Tye, K.M. Inhibitory Input from the Lateral Hypothalamus to the Ventral Tegmental Area Disinhibits Dopamine Neurons and Promotes Behavioral Activation. Neuron 2016, 90, 1286–1298. [Google Scholar] [CrossRef] [PubMed]
- Legaria, A.A.; Matikainen-Ankney, B.A.; Yang, B.; Ahanonu, B.; Licholai, J.A.; Parker, J.G.; Kravitz, A.V. Fiber Photometry in Striatum Reflects Primarily Nonsomatic Changes in Calcium. Nat. Neurosci. 2022, 25, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Sawinski, J.; Wallace, D.J.; Greenberg, D.S.; Grossmann, S.; Denk, W.; Kerr, J.N.D. Visually Evoked Activity in Cortical Cells Imaged in Freely Moving Animals. Proc. Natl. Acad. Sci. USA 2009, 106, 19557–19562. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Li, D.; Park, H.; Li, A.; Yue, Y.; Gau, Y.-T.A.; Li, M.-J.; Bergles, D.E.; Lu, H.; Li, X. Deep-Learning Two-Photon Fiberscopy for Video-Rate Brain Imaging in Freely-Behaving Mice. Nat. Commun. 2022, 13, 1534. [Google Scholar] [CrossRef] [PubMed]
- Flusberg, B.A.; Nimmerjahn, A.; Cocker, E.D.; Mukamel, E.A.; Barretto, R.P.J.; Ko, T.H.; Burns, L.D.; Jung, J.C.; Schnitzer, M.J. High-Speed, Miniaturized Fluorescence Microscopy in Freely Moving Mice. Nat. Methods 2008, 5, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Guan, H.; Park, H.-C.; Yue, Y.; Chen, D.; Liang, W.; Li, M.-J.; Lu, H.; Li, X. Twist-Free Ultralight Two-Photon Fiberscope Enabling Neuroimaging on Freely Rotating/Walking Mice. Optica 2021, 8, 870. [Google Scholar] [CrossRef]
- Gengatharan, A.; Malvaut, S.; Marymonchyk, A.; Ghareghani, M.; Snapyan, M.; Fischer-Sternjak, J.; Ninkovic, J.; Götz, M.; Saghatelyan, A. Adult Neural Stem Cell Activation in Mice Is Regulated by the Day/Night Cycle and Intracellular Calcium Dynamics. Cell 2021, 184, 709–722.e13. [Google Scholar] [CrossRef]
- Barbera, G.; Liang, B.; Zhang, L.; Li, Y.; Lin, D.-T. A Wireless MiniScope for Deep Brain Imaging in Freely Moving Mice. J. Neurosci. Methods 2019, 323, 56–60. [Google Scholar] [CrossRef]
- Skocek, O.; Nöbauer, T.; Weilguny, L.; Martínez Traub, F.; Xia, C.N.; Molodtsov, M.I.; Grama, A.; Yamagata, M.; Aharoni, D.; Cox, D.D.; et al. High-Speed Volumetric Imaging of Neuronal Activity in Freely Moving Rodents. Nat. Methods 2018, 15, 429–432. [Google Scholar] [CrossRef]
- Zong, W.; Wu, R.; Chen, S.; Wu, J.; Wang, H.; Zhao, Z.; Chen, G.; Tu, R.; Wu, D.; Hu, Y.; et al. Miniature Two-Photon Microscopy for Enlarged Field-of-View, Multi-Plane and Long-Term Brain Imaging. Nat. Methods 2021, 18, 46–49. [Google Scholar] [CrossRef]
- Supekar, O.D.; Sias, A.; Hansen, S.R.; Martinez, G.; Peet, G.C.; Peng, X.; Bright, V.M.; Hughes, E.G.; Restrepo, D.; Shepherd, D.P.; et al. Miniature Structured Illumination Microscope for in Vivo 3D Imaging of Brain Structures with Optical Sectioning. Biomed. Opt. Express 2022, 13, 2530. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Matlashov, M.E.; Shcherbakova, D.M.; Antic, S.D.; Verkhusha, V.V.; Knöpfel, T. Characterization of Two Near-Infrared Genetically Encoded Voltage Indicators. Neurophotonics 2023, 11, 024201. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, J.; Li, Y.; Chen, Y.; Ren, H.; Ding, R.; Qian, W.; Ren, K.; Xie, B.; Deng, M.; et al. Bright and Sensitive Red Voltage Indicators for Imaging Action Potentials in Brain Slices and Pancreatic Islets. Sci. Adv. 2023, 9, eadi4208. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.; Vasan, G.; Haziza, S.; Huang, C.; Chrapkiewicz, R.; Luo, J.; Cardin, J.A.; Schnitzer, M.J.; Pieribone, V.A. Dual-Polarity Voltage Imaging of the Concurrent Dynamics of Multiple Neuron Types. Science 2022, 378, eabm8797. [Google Scholar] [CrossRef]
- Davis, H.C.; III, F.P.B.; Wong-Campos, J.D.; Cohen, A.E. Optical Constraints on Two-Photon Voltage Imaging. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Leopold, A.V.; Shcherbakova, D.M.; Verkhusha, V.V. Fluorescent Biosensors for Neurotransmission and Neuromodulation: Engineering and Applications. Front. Cell Neurosci. 2019, 13, 474. [Google Scholar] [CrossRef] [PubMed]
- Marvin, J.S.; Scholl, B.; Wilson, D.E.; Podgorski, K.; Kazemipour, A.; Müller, J.A.; Schoch, S.; Quiroz, F.J.U.; Rebola, N.; Bao, H.; et al. Stability, Affinity, and Chromatic Variants of the Glutamate Sensor IGluSnFR. Nat. Methods 2018, 15, 936–939. [Google Scholar] [CrossRef] [PubMed]
- Marvin, J.S.; Shimoda, Y.; Magloire, V.; Leite, M.; Kawashima, T.; Jensen, T.P.; Kolb, I.; Knott, E.L.; Novak, O.; Podgorski, K.; et al. A Genetically Encoded Fluorescent Sensor for in Vivo Imaging of GABA. Nat. Methods 2019, 16, 763–770. [Google Scholar] [CrossRef]
- Revah, O.; Gore, F.; Kelley, K.W.; Andersen, J.; Sakai, N.; Chen, X.; Li, M.-Y.; Birey, F.; Yang, X.; Saw, N.L.; et al. Maturation and Circuit Integration of Transplanted Human Cortical Organoids. Nature 2022, 610, 319–326. [Google Scholar] [CrossRef]
- Card, J.; Rinaman, L.; Lynn, R.; Lee, B.; Meade, R.; Miselis, R.; Enquist, L. Pseudorabies Virus Infection of the Rat Central Nervous System: Ultrastructural Characterization of Viral Replication, Transport, and Pathogenesis. J. Neurosci. 1993, 13, 2515–2539. [Google Scholar] [CrossRef]
- Lee, H.S.; Lee, I.S.; Kang, T.-C.; Seo, J.; Lee, B.H. The Distribution of Calcitonin Gene-Related Peptide in Gastric Vagal Circuit of Rats. Anat. Histol. Embryol. 1998, 27, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Wickersham, I.R.; Lyon, D.C.; Barnard, R.J.O.; Mori, T.; Finke, S.; Conzelmann, K.K.; Young, J.A.T.; Callaway, E.M. Monosynaptic Restriction of Transsynaptic Tracing from Single, Genetically Targeted Neurons. Neuron 2007, 53, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Tornero, D.; Tsupykov, O.; Granmo, M.; Rodriguez, C.; Grønning-Hansen, M.; Thelin, J.; Smozhanik, E.; Laterza, C.; Wattananit, S.; Ge, R.; et al. Synaptic Inputs from Stroke-Injured Brain to Grafted Human Stem Cell-Derived Neurons Activated by Sensory Stimuli. Brain 2017, 140, 692–706. [Google Scholar] [CrossRef] [PubMed]
- Masaki, Y.; Yamaguchi, M.; Takeuchi, R.F.; Osakada, F. Monosynaptic Rabies Virus Tracing from Projection-Targeted Single Neurons. Neurosci. Res. 2022, 178, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-Z.; Li, L.; Ma, W.-Y.; Yang, X.; Han, Z.-P.; Luo, N.-S.; Wang, J.; Xu, F.-Q. A Rabies Virus–Based Toolkit for Efficient Retrograde Labeling and Monosynaptic Tracing. Neural Regen. Res. 2022, 18, 1827–1833. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E.M.; Luo, L. Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses. J. Neurosci. 2015, 35, 8979–8985. [Google Scholar] [CrossRef] [PubMed]
- Grealish, S.; Heuer, A.; Cardoso, T.; Kirkeby, A.; Jönsson, M.; Johansson, J.; Björklund, A.; Jakobsson, J.; Parmar, M. Monosynaptic Tracing Using Modified Rabies Virus Reveals Early and Extensive Circuit Integration of Human Embryonic Stem Cell-Derived Neurons. Stem Cell Rep. 2015, 4, 975–983. [Google Scholar] [CrossRef]
- Grade, S.; Thomas, J.; Zarb, Y.; Thorwirth, M.; Conzelmann, K.-K.; Hauck, S.M.; Götz, M. Brain Injury Environment Critically Influences the Connectivity of Transplanted Neurons. Sci. Adv. 2022, 8, eabg9445. [Google Scholar] [CrossRef]
- Thomas, J.; Fernanda Martinez-Reza, M.; Thorwirth, M.; Zarb, Y.; Conzelmann, K.-K.; Hauck, S.M.; Grade, S.; Götz, M. Excessive Local Host-Graft Connectivity in Aging and Amyloid-Loaded Brain. Sci. Adv. 2022, 8, eabg9287. [Google Scholar] [CrossRef]
- Grønning Hansen, M.; Laterza, C.; Palma-Tortosa, S.; Kvist, G.; Monni, E.; Tsupykov, O.; Tornero, D.; Uoshima, N.; Soriano, J.; Bengzon, J.; et al. Grafted Human Pluripotent Stem Cell-Derived Cortical Neurons Integrate into Adult Human Cortical Neural Circuitry. Stem Cells Transl. Med. 2020, 9, 1365–1377. [Google Scholar] [CrossRef]
- Yang, H.; Xiong, F.; Qin, H.B.; Yu, Q.T.; Sun, J.Y.; Zhao, H.W.; Li, D.; Zhou, Y.; Zhang, F.K.; Zhu, X.W.; et al. A Novel H129-Based Anterograde Monosynaptic Tracer Exhibits Features of Strong Labeling Intensity, High Tracing Efficiency, and Reduced Retrograde Labeling. Mol. Neurodegener. 2022, 17, 6. [Google Scholar] [CrossRef]
- Fischer, K.B.; Collins, H.K.; Pang, Y.; Roy, D.S.; Zhang, Y.; Feng, G.; Li, S.; Kepecs, A.; Callaway, E.M. Monosynaptic Restriction of the Anterograde Herpes Simplex Virus Strain H129 for Neural Circuit Tracing. J. Comp. Neurol. 2023, 531, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Yang, H.; Song, Y.G.; Qin, H.B.; Zhang, Q.Y.; Huang, X.; Jing, W.; Deng, M.; Liu, Y.; Liu, Z.; et al. An HSV-1-H129 Amplicon Tracer System for Rapid and Efficient Monosynaptic Anterograde Neural Circuit Tracing. Nat. Commun. 2022, 13, 7645. [Google Scholar] [CrossRef]
- Vivar, C.; Potter, M.C.; Choi, J.; Lee, J.; Stringer, T.P.; Callaway, E.M.; Gage, F.H.; Suh, H.; van Praag, H. Monosynaptic Inputs to New Neurons in the Dentate Gyrus. Nat. Commun. 2012, 3, 1107. [Google Scholar] [CrossRef]
- Kato, S.; Kobayashi, K. Pseudotyped Lentiviral Vectors for Tract-Targeting and Application for the Functional Control of Selective Neural Circuits. J. Neurosci. Methods 2020, 344, 108854. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Urgellés, E.; Casas-Torremocha, D.; Sancho-Balsells, A.; Ballasch, I.; García-García, E.; Miquel-Rio, L.; Manasanch, A.; del Castillo, I.; Chen, W.; Pupak, A.; et al. Thalamic Foxp2 Regulates Output Connectivity and Sensory-Motor Impairments in a Model of Huntington’s Disease. Cell. Mol. Life Sci. 2023, 80, 367. [Google Scholar] [CrossRef]
- Miyakawa, N.; Nagai, Y.; Hori, Y.; Mimura, K.; Orihara, A.; Oyama, K.; Matsuo, T.; Inoue, K.; Suzuki, T.; Hirabayashi, T.; et al. Chemogenetic Attenuation of Cortical Seizures in Nonhuman Primates. Nat. Commun. 2023, 14, 971. [Google Scholar] [CrossRef]
- Sahel, J.-A.; Boulanger-Scemama, E.; Pagot, C.; Arleo, A.; Galluppi, F.; Martel, J.N.; Esposti, S.D.; Delaux, A.; de Saint Aubert, J.-B.; de Montleau, C.; et al. Partial Recovery of Visual Function in a Blind Patient after Optogenetic Therapy. Nat. Med. 2021, 27, 1223–1229. [Google Scholar] [CrossRef]
- Bryson, J.B.; Kourgiantaki, A.; Jiang, D.; Demosthenous, A.; Greensmith, L. An Optogenetic Cell Therapy to Restore Control of Target Muscles in an Aggressive Mouse Model of Amyotrophic Lateral Sclerosis. eLife 2024, 12, RP88250. [Google Scholar] [CrossRef]

| Optogenetics | Chemogenetics | |
|---|---|---|
| Timing of the response | Fast (milliseconds) | Prolonged, rely on pharmacokinetics |
| Control of stimulation | Exogenous | Endogenous |
| Target area | Restricted to the illuminated area | Engineered cells in all the body |
| Invasiveness | Invasive | Noninvasive |
| Examples | ||
| Excitation | ChR2, ChETA, ChrimsonR | hM3Dq, PSAM4-5HT3 |
| Inhibition | Halo, eNpHR3.0, ArchT | hM4Di, KORD, PSAM4-GlyR |
| Calcium Sensor | Category | Indicators | Description | Ref. |
|---|---|---|---|---|
| Chemical calcium indicators | Synthetic ratiometric | Fura-2 | High-affinity Ca2+ indicator that shifts its excitation wavelength depending on whether Ca2+ is bound. | [53,54] |
| Single-wavelength | Fluo-4 | High-affinity Ca2+ indicator that boosts fluorescence upon Ca2+ binding. | ||
| Genetically encoded calcium indicators (GECIs) | Fluorescence resonance energy transfer (FRET)-based | Cameleon | ECFP and EYFP are fused to calmodulin (CaM) and calmodulin-target peptide (M13), resulting in an increase in the FRET signal upon Ca2+ binding. | [55,56] |
| Single fluorescent protein (FP)-based | Camgaroo-1 | CaM is fused to YFP, thus allowing Ca2+ to induce an increase in fluorescence by causing a conformational change in CaM. | [52,57] | |
| Pericams | Circularly permuted YFP (cpYFP) is fused to CaM and M13. The binding of calcium allows CaM/M13 to fold tightly against YFP, greatly increasing fluorescence. | [58,59] | ||
| GCaMP family | Circularly permuted GFP is fused to CaM and M13. Many different GCaMP variants have been designed (from GCaMP1 to GCaMP8). | [60,61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Ramos, A.; Puigsasllosas-Pastor, C.; Arcas-Marquez, A.; Tornero, D. Updated Toolbox for Assessing Neuronal Network Reconstruction after Cell Therapy. Bioengineering 2024, 11, 487. https://doi.org/10.3390/bioengineering11050487
Gonzalez-Ramos A, Puigsasllosas-Pastor C, Arcas-Marquez A, Tornero D. Updated Toolbox for Assessing Neuronal Network Reconstruction after Cell Therapy. Bioengineering. 2024; 11(5):487. https://doi.org/10.3390/bioengineering11050487
Chicago/Turabian StyleGonzalez-Ramos, Ana, Claudia Puigsasllosas-Pastor, Ainhoa Arcas-Marquez, and Daniel Tornero. 2024. "Updated Toolbox for Assessing Neuronal Network Reconstruction after Cell Therapy" Bioengineering 11, no. 5: 487. https://doi.org/10.3390/bioengineering11050487
APA StyleGonzalez-Ramos, A., Puigsasllosas-Pastor, C., Arcas-Marquez, A., & Tornero, D. (2024). Updated Toolbox for Assessing Neuronal Network Reconstruction after Cell Therapy. Bioengineering, 11(5), 487. https://doi.org/10.3390/bioengineering11050487








