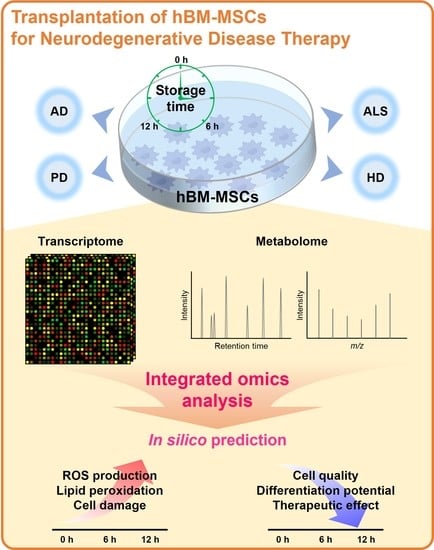
Human Bone Marrow-Derived Mesenchymal Stem Cell Applications in Neurodegenerative Disease Treatment and Integrated Omics Analysis for Successful Stem Cell Therapy
Abstract
1. Introduction
2. Application of hBM-MSCs for the Treatment of Various Neurodegenerative Diseases
2.1. AD
2.2. PD
2.3. ALS
2.4. HD
3. Analysis of the Quality and Differentiation Ability of hBM-MSCs
3.1. Optimizing hBM-MSCs While Preserving Cell Quality
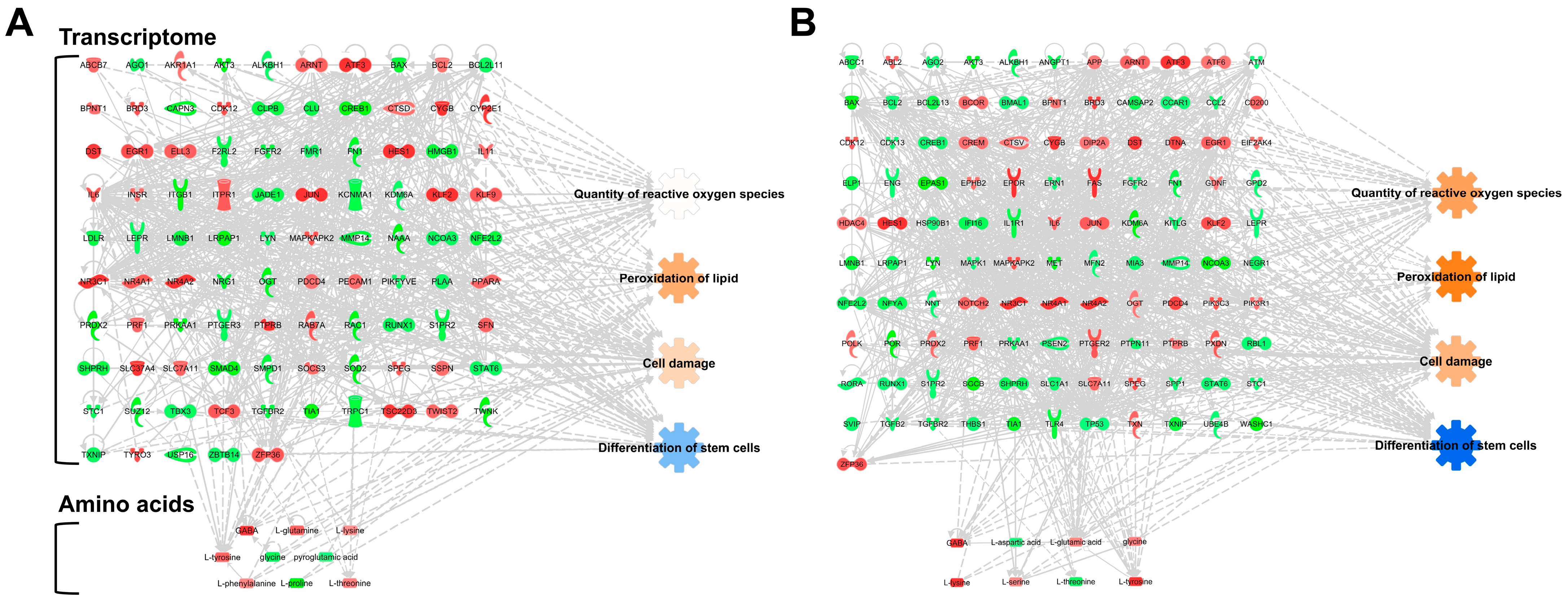
3.2. PBS Storage Time Is a Critical Factor for the Differentiation of hBM-MSCs in Gene Expression and Amino Acid Levels
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ayeni, E.A.; Aldossary, A.M.; Ayejoto, D.A.; Gbadegesin, L.A.; Alshehri, A.A.; Alfassam, H.A.; Afewerky, H.K.; Almughem, F.A.; Bello, S.M.; Tawfik, E.A. Neurodegenerative Diseases: Implications of Environmental and Climatic Influences on Neurotransmitters and Neuronal Hormones Activities. Int. J. Environ. Res. Public Health 2022, 19, 12495. [Google Scholar] [CrossRef] [PubMed]
- Parsippany, N. Understanding Neuromuscular Disease Care. Available online: https://www.iqvia.com/-/media/iqvia/pdfs/institute-reports/understanding-neuromuscular-disease-care.pdf (accessed on 6 November 2022).
- Dugger, B.N.; Dickson, D.W. Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 2017, 9, a028035. [Google Scholar] [CrossRef] [PubMed]
- Ahani-Nahayati, M.; Shariati, A.; Mahmoodi, M.; Olegovna Zekiy, A.; Javidi, K.; Shamlou, S.; Mousakhani, A.; Zamani, M.; Hassanzadeh, A. Stem cell in neurodegenerative disorders; an emerging strategy. Int. J. Dev. Neurosci. 2021, 81, 291–311. [Google Scholar] [CrossRef] [PubMed]
- Basith, S.; Manavalan, B.; Lee, G. Amyotrophic lateral sclerosis disease-related mutations disrupt the dimerization of superoxide dismutase 1—A comparative molecular dynamics simulation study. Comput. Biol. Med. 2022, 151, 106319. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.R. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 364, 362. [Google Scholar] [CrossRef]
- Mendizabal, A.; Diaz, J.M.; Bustamante, A.V.; Bordelon, Y. Health Services in Huntington Disease: A Systematic Literature Review. Neurol. Clin. Pract. 2023, 13, e200108. [Google Scholar] [CrossRef]
- Feigin, V.L.; Vos, T.; Nichols, E.; Owolabi, M.O.; Carroll, W.M.; Dichgans, M.; Deuschl, G.; Parmar, P.; Brainin, M.; Murray, C. The global burden of neurological disorders: Translating evidence into policy. Lancet Neurol. 2020, 19, 255–265. [Google Scholar] [CrossRef]
- Baraniak, P.R.; McDevitt, T.C. Stem cell paracrine actions and tissue regeneration. Regen Med. 2010, 5, 121–143. [Google Scholar] [CrossRef]
- Chang, C.; Yan, J.; Yao, Z.; Zhang, C.; Li, X.; Mao, H.Q. Effects of Mesenchymal Stem Cell-Derived Paracrine Signals and Their Delivery Strategies. Adv. Healthc. Mater. 2021, 10, e2001689. [Google Scholar] [CrossRef]
- Madigan, M.; Atoui, R. Therapeutic Use of Stem Cells for Myocardial Infarction. Bioengineering 2018, 5, 28. [Google Scholar] [CrossRef]
- Bang, O.Y.; Lee, J.S.; Lee, P.H.; Lee, G. Autologous mesenchymal stem cell transplantation in stroke patients. Ann. Neurol. 2005, 57, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Wang, M.; Zhang, B.; Wang, X.; Wanyan, P. Clinical translation of stem cell therapy for spinal cord injury still premature: Results from a single-arm meta-analysis based on 62 clinical trials. BMC Med. 2022, 20, 284. [Google Scholar] [CrossRef] [PubMed]
- Venkataramana, N.K.; Kumar, S.K.; Balaraju, S.; Radhakrishnan, R.C.; Bansal, A.; Dixit, A.; Rao, D.K.; Das, M.; Jan, M.; Gupta, P.K.; et al. Open-labeled study of unilateral autologous bone-marrow-derived mesenchymal stem cell transplantation in Parkinson’s disease. Transl. Res. 2010, 155, 62–70. [Google Scholar] [CrossRef]
- Lee, P.H.; Kim, J.W.; Bang, O.Y.; Ahn, Y.H.; Joo, I.S.; Huh, K. Autologous mesenchymal stem cell therapy delays the progression of neurological deficits in patients with multiple system atrophy. Clin. Pharmacol. Ther. 2008, 83, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Torrest, A.; Pollock, K.; Dahlenburg, H.; Annett, G.; Nolta, J.A.; Fink, K.D. Clinical trial perspective for adult and juvenile Huntington’s disease using genetically-engineered mesenchymal stem cells. Neural. Regen. Res. 2016, 11, 702–705. [Google Scholar]
- Lee, H.J.; Kim, Y.H.; Choi, D.W.; Cho, K.A.; Park, J.W.; Shin, S.J.; Jo, I.; Woo, S.Y.; Ryu, K.H. Tonsil-derived mesenchymal stem cells enhance allogeneic bone marrow engraftment via collagen IV degradation. Stem. Cell Res. Ther. 2021, 12, 329. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Martinez, E.; Mendoza-Nunez, V.M.; Santiago-Osorio, E. Mesenchymal Stem Cells Derived from Dental Pulp: A Review. Stem. Cells Int. 2016, 2016, 4709572. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Chailakhjan, R.K.; Lalykina, K.S. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970, 3, 393–403. [Google Scholar] [CrossRef]
- Liu, Y.; Holmes, C. Tissue Regeneration Capacity of Extracellular Vesicles Isolated From Bone Marrow-Derived and Adipose-Derived Mesenchymal Stromal/Stem Cells. Front. Cell Dev. Biol. 2021, 9, 648098. [Google Scholar] [CrossRef]
- Bhat, S.; Viswanathan, P.; Chandanala, S.; Prasanna, S.J.; Seetharam, R.N. Expansion and characterization of bone marrow derived human mesenchymal stromal cells in serum-free conditions. Sci. Rep. 2021, 11, 3403. [Google Scholar] [CrossRef]
- Huo, J.; Sun, S.; Geng, Z.; Sheng, W.; Chen, R.; Ma, K.; Sun, X.; Fu, X. Bone Marrow-Derived Mesenchymal Stem Cells Promoted Cutaneous Wound Healing by Regulating Keratinocyte Migration via beta(2)-Adrenergic Receptor Signaling. Mol. Pharm. 2018, 15, 2513–2527. [Google Scholar] [CrossRef]
- Musial-Wysocka, A.; Kot, M.; Majka, M. The Pros and Cons of Mesenchymal Stem Cell-Based Therapies. Cell Transplant. 2019, 28, 801–812. [Google Scholar] [CrossRef]
- Klimczak, A.; Kozlowska, U.; Kurpisz, M. Muscle Stem/Progenitor Cells and Mesenchymal Stem Cells of Bone Marrow Origin for Skeletal Muscle Regeneration in Muscular Dystrophies. Arch. Immunol. Ther. Exp. 2018, 66, 341–354. [Google Scholar] [CrossRef]
- Wang, Y.H.; Wang, D.R.; Guo, Y.C.; Liu, J.Y.; Pan, J. The application of bone marrow mesenchymal stem cells and biomaterials in skeletal muscle regeneration. Regen. Ther. 2020, 15, 285–294. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, M.; Shi, L.; Yang, X.; Chen, L.; Cao, N.; Lei, A.; Cao, Y. Neural stemness contributes to cell tumorigenicity. Cell Biosci. 2021, 11, 21. [Google Scholar] [CrossRef]
- Paik, M.J.; Li, W.Y.; Ahn, Y.H.; Lee, P.H.; Choi, S.; Kim, K.R.; Kim, Y.M.; Bang, O.Y.; Lee, G. The free fatty acid metabolome in cerebral ischemia following human mesenchymal stem cell transplantation in rats. Clin. Chim. Acta 2009, 402, 25–30. [Google Scholar] [CrossRef]
- Rustad, K.C.; Gurtner, G.C. Mesenchymal Stem Cells Home to Sites of Injury and Inflammation. Adv. Wound Care 2012, 1, 147–152. [Google Scholar] [CrossRef]
- Xie, C.; Yang, Z.; Suo, Y.; Chen, Q.; Wei, D.; Weng, X.; Gu, Z.; Wei, X. Systemically Infused Mesenchymal Stem Cells Show Different Homing Profiles in Healthy and Tumor Mouse Models. Stem. Cells Transl. Med. 2017, 6, 1120–1131. [Google Scholar] [CrossRef]
- Shin, T.H.; Phukan, G.; Shim, J.S.; Nguyen, D.T.; Kim, Y.; Oh-Lee, J.D.; Lee, H.S.; Paik, M.J.; Lee, G. Restoration of Polyamine Metabolic Patterns in In Vivo and In Vitro Model of Ischemic Stroke following Human Mesenchymal Stem Cell Treatment. Stem. Cells Int. 2016, 2016, 4612531. [Google Scholar] [CrossRef]
- Zhang, T.; Lee, Y.W.; Rui, Y.F.; Cheng, T.Y.; Jiang, X.H.; Li, G. Bone marrow-derived mesenchymal stem cells promote growth and angiogenesis of breast and prostate tumors. Stem. Cell Res. Ther. 2013, 4, 70. [Google Scholar] [CrossRef]
- Saccu, G.; Menchise, V.; Gai, C.; Bertolin, M.; Ferrari, S.; Giordano, C.; Manco, M.; Dastru, W.; Tolosano, E.; Bussolati, B.; et al. Bone Marrow Mesenchymal Stromal/Stem Cell-Derived Extracellular Vesicles Promote Corneal Wound Repair by Regulating Inflammation and Angiogenesis. Cells 2022, 11, 3892. [Google Scholar] [CrossRef]
- Almeida, A.; Lira, R.; Oliveira, M.; Martins, M.; Azevedo, Y.; Silva, K.R.; Carvalho, S.; Cortez, E.; Stumbo, A.C.; Carvalho, L.; et al. Bone marrow-derived mesenchymal stem cells transplantation ameliorates renal injury through anti-fibrotic and anti-inflammatory effects in chronic experimental renovascular disease. Biomed. J. 2022, 45, 629–641. [Google Scholar] [CrossRef]
- Kim, Y.J.; Park, H.J.; Lee, G.; Bang, O.Y.; Ahn, Y.H.; Joe, E.; Kim, H.O.; Lee, P.H. Neuroprotective effects of human mesenchymal stem cells on dopaminergic neurons through anti-inflammatory action. Glia 2009, 57, 13–23. [Google Scholar] [CrossRef]
- Dai, W.; Hale, S.L.; Martin, B.J.; Kuang, J.Q.; Dow, J.S.; Wold, L.E.; Kloner, R.A. Allogeneic mesenchymal stem cell transplantation in postinfarcted rat myocardium: Short- and long-term effects. Circulation 2005, 112, 214–223. [Google Scholar] [CrossRef]
- Fan, L.; Yang, K.; Yu, R.; Hui, H.; Wu, W. circ-Iqsec1 induces bone marrow-derived mesenchymal stem cell (BMSC) osteogenic differentiation through the miR-187-3p/Satb2 signaling pathway. Arthritis. Res. Ther. 2022, 24, 273. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, Y.; Wang, Y.; Zhao, Z.; Li, T. LINC00473 rescues human bone marrow mesenchymal stem cells from apoptosis induced by dexamethasone through the PEBP1-mediated Akt/Bad/Bcl-2 signaling pathway. Int. J. Mol. Med. 2021, 47, 171–182. [Google Scholar] [CrossRef]
- Sharifi, A.M.; Ghazanfari, R.; Tekiyehmaroof, N.; Sharifi, M.A. Investigating the effect of lead acetate on rat bone marrow-derived mesenchymal stem cells toxicity: Role of apoptosis. Toxicol. Mech. Methods 2011, 21, 225–230. [Google Scholar] [CrossRef]
- Jones, J.; Jaramillo-Merchan, J.; Bueno, C.; Pastor, D.; Viso-Leon, M.; Martinez, S. Mesenchymal stem cells rescue Purkinje cells and improve motor functions in a mouse model of cerebellar ataxia. Neurobiol. Dis. 2010, 40, 415–423. [Google Scholar] [CrossRef]
- Shim, E.K.; Lee, J.S.; Kim, D.E.; Kim, S.K.; Jung, B.J.; Choi, E.Y.; Kim, C.S. Autogenous Mesenchymal Stem Cells from the Vertebral Body Enhance Intervertebral Disc Regeneration via Paracrine Interaction: An in Vitro Pilot Study. Cell Transplant. 2016, 25, 1819–1832. [Google Scholar] [CrossRef]
- Li, W.Y.; Choi, Y.J.; Lee, P.H.; Huh, K.; Kang, Y.M.; Kim, H.S.; Ahn, Y.H.; Lee, G.; Bang, O.Y. Mesenchymal stem cells for ischemic stroke: Changes in effects after ex vivo culturing. Cell Transplant. 2008, 17, 1045–1059. [Google Scholar] [CrossRef]
- Shi, H.; Li, X.; Yang, J.; Zhao, Y.; Xue, C.; Wang, Y.; He, Q.; Shen, M.; Zhang, Q.; Yang, Y.; et al. Bone marrow-derived neural crest precursors improve nerve defect repair partially through secreted trophic factors. Stem. Cell Res. Ther. 2019, 10, 397. [Google Scholar] [CrossRef]
- Scheper, V.; Schwieger, J.; Hamm, A.; Lenarz, T.; Hoffmann, A. BDNF-overexpressing human mesenchymal stem cells mediate increased neuronal protection in vitro. J. Neurosci. Res. 2019, 97, 1414–1429. [Google Scholar] [CrossRef]
- Park, K.W.; Eglitis, M.A.; Mouradian, M.M. Protection of nigral neurons by GDNF-engineered marrow cell transplantation. Neurosci. Res. 2001, 40, 315–323. [Google Scholar] [CrossRef]
- Hoban, D.B.; Howard, L.; Dowd, E. GDNF-secreting mesenchymal stem cells provide localized neuroprotection in an inflammation-driven rat model of Parkinson’s disease. Neuroscience 2015, 303, 402–411. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, P.; Chen, F.; Zhao, Y.; Li, Y.; He, X.; Huselstein, C.; Ye, Q.; Tong, Z.; Chen, Y. Brain Derived Neurotrophic Factor and Glial Cell Line-Derived Neurotrophic Factor-Transfected Bone Mesenchymal Stem Cells for the Repair of Periphery Nerve Injury. Front. Bioeng. Biotechnol. 2020, 8, 874. [Google Scholar] [CrossRef]
- Mortazavi, Y.; Sheikhsaran, F.; Khamisipour, G.K.; Soleimani, M.; Teimuri, A.; Shokri, S. The Evaluation of Nerve Growth Factor Over Expression on Neural Lineage Specific Genes in Human Mesenchymal Stem Cells. Cell J. 2016, 18, 189–196. [Google Scholar]
- Li, S.; Liao, X.; He, Y.; Chen, R.; Zheng, W.V.; Tang, M.; Guo, X.; Chen, J.; Hu, S.; Sun, J. Exosomes derived from NGF-overexpressing bone marrow mesenchymal stem cell sheet promote spinal cord injury repair in a mouse model. Neurochem. Int. 2022, 157, 105339. [Google Scholar] [CrossRef]
- Lindvall, O.; Kokaia, Z. Stem cells in human neurodegenerative disorders--time for clinical translation? J. Clin. Investig. 2010, 120, 29–40. [Google Scholar] [CrossRef]
- Andrzejewska, A.; Dabrowska, S.; Lukomska, B.; Janowski, M. Mesenchymal Stem Cells for Neurological Disorders. Adv. Sci. 2021, 8, 2002944. [Google Scholar] [CrossRef]
- Vacanti, J.P.; Langer, R. Tissue engineering: The design and fabrication of living replacement devices for surgical reconstruction and transplantation. Lancet 1999, 354, S32–S34. [Google Scholar] [CrossRef]
- Kassem, M.; Abdallah, B.M. Human bone-marrow-derived mesenchymal stem cells: Biological characteristics and potential role in therapy of degenerative diseases. Cell Tissue Res. 2008, 331, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Y.; Zhang, Y.; Yao, B.; Enhejirigala; Li, Z.; Song, W.; Wang, Y.; Duan, X.; Yuan, X.; et al. Biophysical and Biochemical Cues of Biomaterials Guide Mesenchymal Stem Cell Behaviors. Front. Cell Dev. Biol. 2021, 9, 640388. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Serra, R.; Gallego, R.; Lozano, P.; González-Nieto, D. Hydrogels for neuroprotection and functional rewiring: A new era for brain engineering. Neural. Regen. Res. 2020, 15, 783. [Google Scholar] [PubMed]
- Yan, F.; Li, M.; Zhang, H.-Q.; Li, G.-L.; Hua, Y.; Shen, Y.; Ji, X.-M.; Wu, C.-J.; An, H.; Ren, M. Collagen-chitosan scaffold impregnated with bone marrow mesenchymal stem cells for treatment of traumatic brain injury. Neural. Regen. Res. 2019, 14, 1780. [Google Scholar] [PubMed]
- Lee, D.Y.; Lee, S.E.; Kwon, D.H.; Nithiyanandam, S.; Lee, M.H.; Hwang, J.S.; Basith, S.; Ahn, J.H.; Shin, T.H.; Lee, G. Strategies to Improve the Quality and Freshness of Human Bone Marrow-Derived Mesenchymal Stem Cells for Neurological Diseases. Stem. Cells Int. 2021, 2021, 8444599. [Google Scholar] [CrossRef] [PubMed]
- Shin, T.H.; Lee, S.; Choi, K.R.; Lee, D.Y.; Kim, Y.; Paik, M.J.; Seo, C.; Kang, S.; Jin, M.S.; Yoo, T.H.; et al. Quality and freshness of human bone marrow-derived mesenchymal stem cells decrease over time after trypsinization and storage in phosphate-buffered saline. Sci. Rep. 2017, 7, 1106. [Google Scholar] [CrossRef]
- Lee, K.A.; Shim, W.; Paik, M.J.; Lee, S.C.; Shin, J.Y.; Ahn, Y.H.; Park, K.; Kim, J.H.; Choi, S.; Lee, G. Analysis of changes in the viability and gene expression profiles of human mesenchymal stromal cells over time. Cytotherapy 2009, 11, 688–697. [Google Scholar] [CrossRef]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef]
- Milward, E.A.; Shahandeh, A.; Heidari, M.; Johnstone, D.M.; Daneshi, N.; Hondermarck, H. Transcriptomics. Encycl. Cell Biol. 2016, 4, 160–165. [Google Scholar]
- Kumar, K. Integrated benchmarking standard and decision support system for structured, semi structured, unstructured retail data. Wirel. Netw. 2021, 1–11. [Google Scholar] [CrossRef]
- Cosgriff, C.V.; Stone, D.J.; Weissman, G.; Pirracchio, R.; Celi, L.A. The clinical artificial intelligence department: A prerequisite for success. BMJ Health Care Inform. 2020, 27, e100183. [Google Scholar] [CrossRef] [PubMed]
- Basith, S.; Manavalan, B.; Hwan Shin, T.; Lee, G. Machine intelligence in peptide therapeutics: A next-generation tool for rapid disease screening. Med. Res. Rev. 2020, 40, 1276–1314. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, L.; Xu, Y.; Yang, J. Machine learning meets omics: Applications and perspectives. Brief. Bioinform. 2021, 23, bbab460. [Google Scholar] [CrossRef] [PubMed]
- Shin, T.H.; Manavalan, B.; Lee, D.Y.; Basith, S.; Seo, C.; Paik, M.J.; Kim, S.W.; Seo, H.; Lee, J.Y.; Kim, J.Y.; et al. Silica-coated magnetic-nanoparticle-induced cytotoxicity is reduced in microglia by glutathione and citrate identified using integrated omics. Part Fibre. Toxicol. 2021, 18, 42. [Google Scholar] [CrossRef]
- Shin, T.H.; Lee, D.Y.; Jang, Y.E.; Kwon, D.H.; Hwang, J.S.; Kim, S.G.; Seo, C.; Paik, M.J.; Lee, J.Y.; Kim, J.Y.; et al. Reduction in the Migration Activity of Microglia Treated with Silica-Coated Magnetic Nanoparticles and their Recovery Using Citrate. Cells 2022, 11, 2393. [Google Scholar] [CrossRef]
- Picard, M.; Scott-Boyer, M.-P.; Bodein, A.; Périn, O.; Droit, A. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef]
- Lamptey, R.N.L.; Chaulagain, B.; Trivedi, R.; Gothwal, A.; Layek, B.; Singh, J. A Review of the Common Neurodegenerative Disorders: Current Therapeutic Approaches and the Potential Role of Nanotherapeutics. Int. J. Mol. Sci. 2022, 23, 1851. [Google Scholar] [CrossRef]
- Ramanathan, S.; Archunan, G.; Sivakumar, M.; Tamil Selvan, S.; Fred, A.L.; Kumar, S.; Gulyás, B.; Padmanabhan, P. Theranostic applications of nanoparticles in neurodegenerative disorders. Int. J. Nanomed. 2018, 13, 5561–5576. [Google Scholar] [CrossRef]
- Scopetti, M.; Santurro, A.; Gatto, V.; La Russa, R.; Manetti, F.; D′Errico, S.; Frati, P.; Fineschi, V. Mesenchymal stem cells in neurodegenerative diseases: Opinion review on ethical dilemmas. World J. Stem. Cells 2020, 12, 168–177. [Google Scholar] [CrossRef]
- Qin, C.; Wang, K.; Zhang, L.; Bai, L. Stem cell therapy for Alzheimer’s disease: An overview of experimental models and reality. Anim. Model Exp. Med. 2022, 5, 15–26. [Google Scholar] [CrossRef]
- Hernández, A.E.; García, E. Mesenchymal Stem Cell Therapy for Alzheimer’s Disease. Stem. Cells Int. 2021, 2021, 7834421. [Google Scholar] [CrossRef]
- Lee, J.K.; Jin, H.K.; Bae, J.S. Bone marrow-derived mesenchymal stem cells attenuate amyloid β-induced memory impairment and apoptosis by inhibiting neuronal cell death. Curr. Alzheimer Res. 2010, 7, 540–548. [Google Scholar] [CrossRef]
- Cone, A.S.; Yuan, X.; Sun, L.; Duke, L.C.; Vreones, M.P.; Carrier, A.N.; Kenyon, S.M.; Carver, S.R.; Benthem, S.D.; Stimmell, A.C.; et al. Mesenchymal stem cell-derived extracellular vesicles ameliorate Alzheimer’s disease-like phenotypes in a preclinical mouse model. Theranostics 2021, 11, 8129–8142. [Google Scholar] [CrossRef]
- Nakano, M.; Kubota, K.; Kobayashi, E.; Chikenji, T.S.; Saito, Y.; Konari, N.; Fujimiya, M. Bone marrow-derived mesenchymal stem cells improve cognitive impairment in an Alzheimer’s disease model by increasing the expression of microRNA-146a in hippocampus. Sci. Rep. 2020, 10, 10772. [Google Scholar] [CrossRef]
- Fields, C.R.; Bengoa-Vergniory, N.; Wade-Martins, R. Targeting alpha-synuclein as a therapy for Parkinson’s disease. Front. Mol. Neurosci. 2019, 12, 299. [Google Scholar] [CrossRef]
- Venkatesh, K.; Sen, D. Mesenchymal Stem Cells as a Source of Dopaminergic Neurons: A Potential Cell Based Therapy for Parkinson’s Disease. Curr. Stem. Cell Res. Ther. 2017, 12, 326–347. [Google Scholar] [CrossRef]
- Blandini, F.; Cova, L.; Armentero, M.-T.; Zennaro, E.; Levandis, G.; Bossolasco, P.; Calzarossa, C.; Mellone, M.; Giuseppe, B.; Deliliers, G.L.; et al. Transplantation of Undifferentiated Human Mesenchymal Stem Cells Protects against 6-Hydroxydopamine Neurotoxicity in the Rat. Cell Transplant. 2010, 19, 203–218. [Google Scholar] [CrossRef]
- Heris, R.M.; Shirvaliloo, M.; Abbaspour-Aghdam, S.; Hazrati, A.; Shariati, A.; Youshanlouei, H.R.; Niaragh, F.J.; Valizadeh, H.; Ahmadi, M. The potential use of mesenchymal stem cells and their exosomes in Parkinson’s disease treatment. Stem. Cell Res. Ther. 2022, 13, 371. [Google Scholar] [CrossRef]
- Ramalingam, M.; Jang, S.; Jeong, H.S. Therapeutic Effects of Conditioned Medium of Neural Differentiated Human Bone Marrow-Derived Stem Cells on Rotenone-Induced Alpha-Synuclein Aggregation and Apoptosis. Stem. Cells Int. 2021, 2021, 6658271. [Google Scholar] [CrossRef]
- Schiess, M.; Suescun, J.; Doursout, M.-F.; Adams, C.; Green, C.; Saltarrelli, J.G.; Savitz, S.; Ellmore, T.M. Allogeneic Bone Marrow–Derived Mesenchymal Stem Cell Safety in Idiopathic Parkinson’s Disease. Mov. Dis. 2021, 36, 1825–1834. [Google Scholar] [CrossRef]
- Staff, N.P.; Jones, D.T.; Singer, W. Mesenchymal Stromal Cell Therapies for Neurodegenerative Diseases. Mayo. Clin. Proc. 2019, 94, 892–905. [Google Scholar] [CrossRef]
- Chiò, A.; Mazzini, L.; Mora, G. Disease-modifying therapies in amyotrophic lateral sclerosis. Neuropharmacology 2020, 167, 107986. [Google Scholar] [CrossRef]
- Vercelli, A.; Mereuta, O.M.; Garbossa, D.; Muraca, G.; Mareschi, K.; Rustichelli, D.; Ferrero, I.; Mazzini, L.; Madon, E.; Fagioli, F. Human mesenchymal stem cell transplantation extends survival, improves motor performance and decreases neuroinflammation in mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2008, 31, 395–405. [Google Scholar] [CrossRef]
- Stavely, R.; Nurgali, K. The emerging antioxidant paradigm of mesenchymal stem cell therapy. Stem. Cells Transl. Med. 2020, 9, 985–1006. [Google Scholar] [CrossRef]
- Morata-Tarifa, C.; Azkona, G.; Glass, J.; Mazzini, L.; Sanchez-Pernaute, R. Looking backward to move forward: A meta-analysis of stem cell therapy in amyotrophic lateral sclerosis. NPJ Regen. Med. 2021, 6, 20. [Google Scholar] [CrossRef]
- Oh, K.-W.; Moon, C.; Kim, H.Y.; Oh, S.-i.; Park, J.; Lee, J.H.; Chang, I.Y.; Kim, K.S.; Kim, S.H. Phase I Trial of Repeated Intrathecal Autologous Bone Marrow-Derived Mesenchymal Stromal Cells in Amyotrophic Lateral Sclerosis. Stem. Cells Transl. Med. 2015, 4, 590–597. [Google Scholar] [CrossRef]
- Tavakol-Afshari, J.; Boroumand, A.R.; Farkhad, N.K.; Adhami Moghadam, A.; Sahab-Negah, S.; Gorji, A. Safety and efficacy of bone marrow derived-mesenchymal stem cells transplantation in patients with amyotrophic lateral sclerosis. Regen. Ther. 2021, 18, 268–274. [Google Scholar] [CrossRef]
- Lewis, C.M.; Suzuki, M. Therapeutic applications of mesenchymal stem cells for amyotrophic lateral sclerosis. Stem. Cell Res. Ther. 2014, 5, 32. [Google Scholar] [CrossRef]
- Choi, K.-A.; Choi, Y.; Hong, S. Stem cell transplantation for Huntington’s diseases. Methods 2018, 133, 104–112. [Google Scholar] [CrossRef]
- Alberch, J.; Pérez-Navarro, E.; Canals, J.M. Neurotrophic factors in Huntington’s disease. Prog. Brain. Res. 2004, 146, 197–229. [Google Scholar]
- Lin, Y.-T.; Chern, Y.; Shen, C.-K.J.; Wen, H.-L.; Chang, Y.-C.; Li, H.; Cheng, T.-H.; Hsieh-Li, H.M. Human Mesenchymal Stem Cells Prolong Survival and Ameliorate Motor Deficit through Trophic Support in Huntington’s Disease Mouse Models. PLoS ONE 2011, 6, e22924. [Google Scholar] [CrossRef]
- Pollock, K.; Dahlenburg, H.; Nelson, H.; Fink, K.D.; Cary, W.; Hendrix, K.; Annett, G.; Torrest, A.; Deng, P.; Gutierrez, J.; et al. Human Mesenchymal Stem Cells Genetically Engineered to Overexpress Brain-derived Neurotrophic Factor Improve Outcomes in Huntington’s Disease Mouse Models. Mol. Ther. 2016, 24, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Sadan, O.; Shemesh, N.; Barzilay, R.; Dadon-Nahum, M.; Blumenfeld-Katzir, T.; Assaf, Y.; Yeshurun, M.; Djaldetti, R.; Cohen, Y.; Melamed, E.; et al. Mesenchymal stem cells induced to secrete neurotrophic factors attenuate quinolinic acid toxicity: A potential therapy for Huntington’s disease. Exp. Neurol. 2012, 234, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Chang, H.-Y.; Sang, T.-K. Neuronal cell death mechanisms in major neurodegenerative diseases. Int. J. Mol. Sci. 2018, 19, 3082. [Google Scholar] [CrossRef] [PubMed]
- Marde, V.S.; Tiwari, P.L.; Wankhede, N.L.; Taksande, B.G.; Upaganlawar, A.B.; Umekar, M.J.; Kale, M.B. Neurodegenerative disorders associated with genes of mitochondria. Future J. Pharm. Sci. 2021, 7, 1–8. [Google Scholar] [CrossRef]
- Wilson, D.M.; Cookson, M.R.; Van Den Bosch, L.; Zetterberg, H.; Holtzman, D.M.; Dewachter, I. Hallmarks of neurodegenerative diseases. Cell 2023, 186, 693–714. [Google Scholar] [CrossRef]
- Mehta, S.R.; Tom, C.M.; Wang, Y.; Bresee, C.; Rushton, D.; Mathkar, P.P.; Tang, J.; Mattis, V.B. Human Huntington’s disease iPSC-derived cortical neurons display altered transcriptomics, morphology, and maturation. Cell Rep. 2018, 25, 1081–1096.e6. [Google Scholar] [CrossRef]
- Franco-Bocanegra, D.K.; Gourari, Y.; McAuley, C.; Chatelet, D.S.; Johnston, D.A.; Nicoll, J.A.; Boche, D. Microglial morphology in Alzheimer’s disease and after Aβ immunotherapy. Sci. Rep. 2021, 11, 15955. [Google Scholar] [CrossRef]
- Peng, J.; Peng, L.; Stevenson, F.F.; Doctrow, S.R.; Andersen, J.K. Iron and paraquat as synergistic environmental risk factors in sporadic Parkinson’s disease accelerate age-related neurodegeneration. J. Neurosci. 2007, 27, 6914–6922. [Google Scholar] [CrossRef]
- Sharifi-Rad, M.; Anil Kumar, N.V.; Zucca, P.; Varoni, E.M.; Dini, L.; Panzarini, E.; Rajkovic, J.; Tsouh Fokou, P.V.; Azzini, E.; Peluso, I. Lifestyle, oxidative stress, and antioxidants: Back and forth in the pathophysiology of chronic diseases. Front. Physiol. 2020, 11, 694. [Google Scholar] [CrossRef]
- Ajoolabady, A.; Lindholm, D.; Ren, J.; Pratico, D. ER stress and UPR in Alzheimer’s disease: Mechanisms, pathogenesis, treatments. Cell Death Dis. 2022, 13, 706. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, W.; Dobrzyński, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem. Cell Res. Ther. 2019, 10, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Stolzing, A.; Jones, E.; Mcgonagle, D.; Scutt, A. Age-related changes in human bone marrow-derived mesenchymal stem cells: Consequences for cell therapies. Mech. Ageing Dev. 2008, 129, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Geissler, S.; Textor, M.; Kühnisch, J.; Könnig, D.; Klein, O.; Ode, A.; Pfitzner, T.; Adjaye, J.; Kasper, G.; Duda, G.N. Functional comparison of chronological and in vitro aging: Differential role of the cytoskeleton and mitochondria in mesenchymal stromal cells. PLoS ONE 2012, 7, e52700. [Google Scholar] [CrossRef]
- Sammour, I.; Somashekar, S.; Huang, J.; Batlahally, S.; Breton, M.; Valasaki, K.; Khan, A.; Wu, S.; Young, K.C. The Effect of Gender on Mesenchymal Stem Cell (MSC) Efficacy in Neonatal Hyperoxia-Induced Lung Injury. PLoS ONE 2016, 11, e0164269. [Google Scholar] [CrossRef]
- Crisostomo, P.R.; Wang, M.; Herring, C.M.; Markel, T.A.; Meldrum, K.K.; Lillemoe, K.D.; Meldrum, D.R. Gender differences in injury induced mesenchymal stem cell apoptosis and VEGF, TNF, IL-6 expression: Role of the 55 kDa TNF receptor (TNFR1). J. Mol. Cell Cardiol. 2007, 42, 142–149. [Google Scholar] [CrossRef]
- Siegel, G.; Kluba, T.; Hermanutz-Klein, U.; Bieback, K.; Northoff, H.; Schafer, R. Phenotype, donor age and gender affect function of human bone marrow-derived mesenchymal stromal cells. BMC Med. 2013, 11, 146. [Google Scholar] [CrossRef]
- Wang, M.; Yuan, Q.; Xie, L. Mesenchymal stem cell-based immunomodulation: Properties and clinical application. Stem. Cells Int. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
- Salari, V.; Mengoni, F.; Del Gallo, F.; Bertini, G.; Fabene, P.F. The anti-inflammatory properties of mesenchymal stem cells in epilepsy: Possible treatments and future perspectives. Int. J. Mol. Sci. 2020, 21, 9683. [Google Scholar] [CrossRef]
- Crippa, S.; Santi, L.; Berti, M.; De Ponti, G.; Bernardo, M.E. Role of ex vivo expanded mesenchymal stromal cells in determining hematopoietic stem cell transplantation outcome. Front. Cell Dev. Biol. 2021, 9, 663316. [Google Scholar] [CrossRef]
- Binato, R.; de Souza Fernandez, T.; Lazzarotto-Silva, C.; Du Rocher, B.; Mencalha, A.; Pizzatti, L.; Bouzas, L.; Abdelhay, E. Stability of human mesenchymal stem cells during in vitro culture: Considerations for cell therapy. Cell Prolif. 2013, 46, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Shakouri-Motlagh, A.; O′Connor, A.J.; Kalionis, B.; Heath, D.E. Improved ex vivo expansion of mesenchymal stem cells on solubilized acellular fetal membranes. J. Biomed. Mater Res. A 2019, 107, 232–242. [Google Scholar] [CrossRef]
- Li, X.-Y.; Ding, J.; Zheng, Z.-H.; Li, X.-Y.; Wu, Z.-B.; Zhu, P. Long-term culture in vitro impairs the immunosuppressive activity of mesenchymal stem cells on T cells. Mol. Med. Rep. 2012, 6, 1183–1189. [Google Scholar] [CrossRef]
- Bonab, M.M.; Alimoghaddam, K.; Talebian, F.; Ghaffari, S.H.; Ghavamzadeh, A.; Nikbin, B. Aging of mesenchymal stem cell in vitro. BMC Cell Biol. 2006, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kretlow, J.D.; Jin, Y.-Q.; Liu, W.; Zhang, W.J.; Hong, T.-H.; Zhou, G.; Baggett, L.S.; Mikos, A.G.; Cao, Y. Donor age and cell passage affects differentiation potential of murine bone marrow-derived stem cells. BMC Cell Biol. 2008, 9, 1–13. [Google Scholar] [CrossRef]
- Stenderup, K.; Justesen, J.; Clausen, C.; Kassem, M. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone 2003, 33, 919–926. [Google Scholar] [CrossRef]
- Zaim, M.; Karaman, S.; Cetin, G.; Isik, S. Donor age and long-term culture affect differentiation and proliferation of human bone marrow mesenchymal stem cells. Ann. Hematol. 2012, 91, 1175–1186. [Google Scholar] [CrossRef]
- Baxter, M.A.; Wynn, R.F.; Jowitt, S.N.; Wraith, J.E.; Fairbairn, L.J.; Bellantuono, I. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem. Cells 2004, 22, 675–682. [Google Scholar] [CrossRef]
- Watanabe, J.; Yamada, M.; Niibe, K.; Zhang, M.; Kondo, T.; Ishibashi, M.; Egusa, H. Preconditioning of bone marrow-derived mesenchymal stem cells with N-acetyl-L-cysteine enhances bone regeneration via reinforced resistance to oxidative stress. Biomaterials 2018, 185, 25–38. [Google Scholar] [CrossRef]
- Nandy, S.B.; Mohanty, S.; Singh, M.; Behari, M.; Airan, B. Fibroblast Growth Factor-2 alone as an efficient inducer for differentiation of human bone marrow mesenchymal stem cells into dopaminergic neurons. J. Biomed. Sci. 2014, 21, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Longobardi, L.; O’Rear, L.; Aakula, S.; Johnstone, B.; Shimer, K.; Chytil, A.; Horton, W.A.; Moses, H.L.; Spagnoli, A. Effect of IGF-I in the chondrogenesis of bone marrow mesenchymal stem cells in the presence or absence of TGF-β signaling. J. Bone. Miner. Res. 2006, 21, 626–636. [Google Scholar] [CrossRef]
- Xian, L.; Wu, X.; Pang, L.; Lou, M.; Rosen, C.J.; Qiu, T.; Crane, J.; Frassica, F.; Zhang, L.; Rodriguez, J.P. Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat. Med. 2012, 18, 1095–1101. [Google Scholar] [CrossRef]
- Heng, B.C.; Cowan, C.M.; Basu, S. Temperature and calcium ions affect aggregation of mesenchymal stem cells in phosphate buffered saline. Cytotechnology 2008, 58, 69–75. [Google Scholar] [CrossRef]
- Karahalil, B. Overview of Systems Biology and Omics Technologies. Curr. Med. Chem. 2016, 23, 4221–4230. [Google Scholar] [CrossRef]
- Johnson, C.H.; Gonzalez, F.J. Challenges and opportunities of metabolomics. J. Cell Physiol. 2012, 227, 2975–2981. [Google Scholar] [CrossRef]
- Shin, T.H.; Nithiyanandam, S.; Lee, D.Y.; Kwon, D.H.; Hwang, J.S.; Kim, S.G.; Jang, Y.E.; Basith, S.; Park, S.; Mo, J.S.; et al. Analysis of Nanotoxicity with Integrated Omics and Mechanobiology. Nanomaterials 2021, 11, 2385. [Google Scholar] [CrossRef]
- Shin, T.H.; Kim, S.G.; Ji, M.; Kwon, D.H.; Hwang, J.S.; George, N.P.; Ergando, D.S.; Park, C.B.; Paik, M.J.; Lee, G. Diesel-derived PM(2.5) induces impairment of cardiac movement followed by mitochondria dysfunction in cardiomyocytes. Front. Endocrinol. 2022, 13, 999475. [Google Scholar] [CrossRef]
- Shim, W.; Paik, M.J.; Nguyen, D.T.; Lee, J.K.; Lee, Y.; Kim, J.H.; Shin, E.H.; Kang, J.S.; Jung, H.S.; Choi, S.; et al. Analysis of changes in gene expression and metabolic profiles induced by silica-coated magnetic nanoparticles. ACS Nano 2012, 6, 7665–7680. [Google Scholar] [CrossRef]
- Khan, S.; Ince-Dunn, G.; Suomalainen, A.; Elo, L.L. Integrative omics approaches provide biological and clinical insights: Examples from mitochondrial diseases. J. Clin. Investig. 2020, 130, 20–28. [Google Scholar] [CrossRef]
- Paik, M.J.; Cho, I.S.; Mook-Jung, I.; Lee, G.; Kim, K.R. Altered free amino acid levels in brain cortex tissues of mice with Alzheimer’s disease as their N(O,S)-ethoxycarbonyl/tert-butyldimethylsilyl derivatives. BMB Rep. 2008, 41, 23–28. [Google Scholar] [CrossRef]


| Substances | Differentiation Direction | Functions and Effects | References |
|---|---|---|---|
| N-acetyl-L-cysteine (NAC) | Osteoblast | Decreased apoptosis Increased survival Increased GSH level Enhanced bone regeneration | [120] |
| Fibroblast growth factor-2 (FGF-2) | Dopaminergic neurons | Decreased cell death Increased upregulation of tyrosine hydroxylase Increased dopamine release | [121] |
| Transforming growth factor-beta (TGF-β) | Chondrocyte | Decreased apoptosis Increased cell proliferation Increased chondrogenic condensation and markers | [122] |
| Insulin-like growth factor-1 (IGF-1) | Chondrocyte | Decreased apoptosis Increased cell proliferation Increased chondrogenic condensation and markers | [122] |
| Osteoblast | Induction of osteoblastic differentiation | [123] |
| Entrez Gene Name | Symbol | Entrez Gene ID a | Location | Fold Change b |
|---|---|---|---|---|
| ATP binding cassette subfamily B member 7 | ABCB7 | 22 | Cytoplasm | 1.637 |
| argonaute RISC component 1 | AGO1 | 26,523 | Cytoplasm | −1.542 |
| aldo-keto reductase family 1 member A1 | AKR1A1 | 10,327 | Cytoplasm | 1.512 |
| AKT serine/threonine kinase 3 | AKT3 | 10,000 | Cytoplasm | −2.379 |
| alkB homolog 1, histone H2A dioxygenase | ALKBH1 | 8846 | Cytoplasm | −1.503 |
| aryl hydrocarbon receptor nuclear translocator | ARNT | 405 | Nucleus | 1.715 |
| activating transcription factor 3 | ATF3 | 467 | Nucleus | 2.88 |
| BCL2 associated X, apoptosis regulator | BAX | 581 | Cytoplasm | −1.946 |
| BCL2 apoptosis regulator | BCL2 | 596 | Cytoplasm | 1.534 |
| BCL2 like 11 | BCL2L11 | 10,018 | Cytoplasm | −1.579 |
| 3′(2′), 5′-bisphosphate nucleotidase 1 | BPNT1 | 10,380 | Nucleus | 1.564 |
| bromodomain containing 3 | BRD3 | 8019 | Nucleus | 1.855 |
| calpain 3 | CAPN3 | 825 | Cytoplasm | −1.974 |
| cyclin dependent kinase 12 | CDK12 | 51,755 | Nucleus | 1.925 |
| caseinolytic mitochondrial matrix peptidase chaperone subunit B | CLPB | 81,570 | Nucleus | −1.785 |
| Clusterin | CLU | 1191 | Cytoplasm | −1.768 |
| cAMP responsive element binding protein 1 | CREB1 | 1385 | Nucleus | −2.252 |
| cathepsin D | CTSD | 1509 | Cytoplasm | 1.527 |
| Cytoglobin | CYGB | 114,757 | Cytoplasm | 2.368 |
| cytochrome P450 family 2 subfamily E member 1 | CYP2E1 | 1571 | Cytoplasm | 2.095 |
| Dystonin | DST | 667 | Plasma Membrane | 2.126 |
| early growth response 1 | EGR1 | 1958 | Nucleus | 1.915 |
| elongation factor for RNA polymerase II 3 | ELL3 | 80,237 | Nucleus | 1.749 |
| coagulation factor II thrombin receptor like 2 | F2RL2 | 2151 | Plasma Membrane | −1.714 |
| fibroblast growth factor receptor 2 | FGFR2 | 2263 | Plasma Membrane | −1.859 |
| fragile X messenger ribonucleoprotein 1 | FMR1 | 2332 | Cytoplasm | −1.569 |
| fibronectin 1 | FN1 | 2335 | Extracellular Space | −1.951 |
| hes family bHLH transcription factor 1 | HES1 | 3280 | Nucleus | 8.134 |
| high mobility group box 1 | HMGB1 | 3146 | Nucleus | −1.903 |
| interleukin 11 | IL11 | 3589 | Extracellular Space | 1.596 |
| interleukin 6 | IL6 | 3569 | Extracellular Space | 1.784 |
| insulin receptor | INSR | 3643 | Plasma Membrane | 1.561 |
| integrin subunit beta 1 | ITGB1 | 3688 | Plasma Membrane | −2.31 |
| inositol 1,4,5-trisphosphate receptor type 1 | ITPR1 | 3708 | Cytoplasm | 1.775 |
| jade family PHD finger 1 | JADE1 | 79,960 | Nucleus | −1.76 |
| Jun proto-oncogene, AP-1 transcription factor subunit | JUN | 3725 | Nucleus | 2.212 |
| potassium calcium-activated channel subfamily M alpha 1 | KCNMA1 | 3778 | Plasma Membrane | −1.726 |
| lysine demethylase 6A | KDM6A | 7403 | Nucleus | −1.511 |
| KLF transcription factor 2 | KLF2 | 10,365 | Nucleus | 2.261 |
| KLF transcription factor 9 | KLF9 | 687 | Nucleus | 1.803 |
| low density lipoprotein receptor | LDLR | 3949 | Plasma Membrane | −1.783 |
| leptin receptor | LEPR | 3953 | Plasma Membrane | −1.575 |
| lamin B1 | LMNB1 | 4001 | Nucleus | −1.862 |
| LDL receptor related protein associated protein 1 | LRPAP1 | 4043 | Plasma Membrane | −2.12 |
| LYN proto-oncogene, Src family tyrosine kinase | LYN | 4067 | Cytoplasm | −1.541 |
| MAPK activated protein kinase 2 | MAPKAPK2 | 9261 | Nucleus | 1.848 |
| matrix metallopeptidase 14 | MMP14 | 4323 | Extracellular Space | −1.874 |
| N-acylethanolamine acid amidase | NAAA | 27,163 | Cytoplasm | −2.184 |
| nuclear receptor coactivator 3 | NCOA3 | 8202 | Nucleus | −1.572 |
| NFE2 like bZIP transcription factor 2 | NFE2L2 | 4780 | Nucleus | −1.823 |
| nuclear receptor subfamily 3 group C member 1 | NR3C1 | 2908 | Nucleus | 2.214 |
| nuclear receptor subfamily 4 group A member 1 | NR4A1 | 3164 | Nucleus | 1.808 |
| nuclear receptor subfamily 4 group A member 2 | NR4A2 | 4929 | Nucleus | 4.047 |
| neuregulin 1 | NRG1 | 3084 | Plasma Membrane | −1.913 |
| O-linked N-acetylglucosamine (GlcNAc) transferase | OGT | 8473 | Cytoplasm | −2.058 |
| programmed cell death 4 | PDCD4 | 27,250 | Nucleus | 1.554 |
| platelet and endothelial cell adhesion molecule 1 | PECAM1 | 5175 | Plasma Membrane | 1.602 |
| phosphoinositide kinase, FYVE-type zinc finger containing | PIKFYVE | 200,576 | Cytoplasm | −1.529 |
| phospholipase A2 activating protein | PLAA | 9373 | Cytoplasm | −1.507 |
| peroxisome proliferator activated receptor alpha | PPARA | 5465 | Nucleus | 1.646 |
| peroxiredoxin 2 | PRDX2 | 7001 | Cytoplasm | −2.276 |
| perforin 1 | PRF1 | 5551 | Cytoplasm | 1.668 |
| protein kinase AMP-activated catalytic subunit alpha 1 | PRKAA1 | 5562 | Cytoplasm | −2.252 |
| prostaglandin E receptor 3 | PTGER3 | 5733 | Plasma Membrane | −1.539 |
| protein tyrosine phosphatase receptor type B | PTPRB | 5787 | Plasma Membrane | 2.444 |
| RAB7A, member RAS oncogene family | RAB7A | 7879 | Cytoplasm | 1.801 |
| Rac family small GTPase 1 | RAC1 | 5879 | Plasma Membrane | −2.019 |
| RUNX family transcription factor 1 | RUNX1 | 861 | Nucleus | −1.616 |
| sphingosine-1-phosphate receptor 2 | S1PR2 | 9294 | Plasma Membrane | −1.73 |
| Stratifin | SFN | 2810 | Cytoplasm | 1.7 |
| SNF2 histone linker PHD RING helicase | SHPRH | 257,218 | Nucleus | −1.812 |
| solute carrier family 37 member 4 | SLC37A4 | 2542 | Cytoplasm | 3.046 |
| solute carrier family 7 member 11 | SLC7A11 | 23,657 | Plasma Membrane | 1.721 |
| SMAD family member 4 | SMAD4 | 4089 | Nucleus | −2.153 |
| sphingomyelin phosphodiesterase 1 | SMPD1 | 6609 | Cytoplasm | −1.834 |
| suppressor of cytokine signaling 3 | SOCS3 | 9021 | Cytoplasm | 1.628 |
| superoxide dismutase 2 | SOD2 | 6648 | Cytoplasm | −2.242 |
| striated muscle enriched protein kinase | SPEG | 10,290 | Nucleus | 1.771 |
| Sarcospan | SSPN | 8082 | Plasma Membrane | 1.575 |
| signal transducer and activator of transcription 6 | STAT6 | 6778 | Nucleus | −1.571 |
| stanniocalcin 1 | STC1 | 6781 | Extracellular Space | −1.567 |
| SUZ12 polycomb repressive complex 2 subunit | SUZ12 | 23,512 | Nucleus | −2.001 |
| T-box transcription factor 3 | TBX3 | 6926 | Nucleus | −1.533 |
| transcription factor 3 | TCF3 | 6929 | Nucleus | 1.81 |
| transforming growth factor beta receptor 2 | TGFBR2 | 7048 | Plasma Membrane | −1.822 |
| TIA1 cytotoxic granule associated RNA binding protein | TIA1 | 7072 | Nucleus | −2.369 |
| transient receptor potential cation channel subfamily C member 1 | TRPC1 | 7220 | Plasma Membrane | −1.812 |
| TSC22 domain family member 3 | TSC22D3 | 1831 | Nucleus | 2.134 |
| twist family bHLH transcription factor 2 | TWIST2 | 117,581 | Nucleus | 1.652 |
| twinkle mtDNA helicase | TWNK | 56,652 | Cytoplasm | −2.459 |
| thioredoxin interacting protein | TXNIP | 10,628 | Cytoplasm | −1.567 |
| TYRO3 protein tyrosine kinase | TYRO3 | 7301 | Plasma Membrane | 1.905 |
| ubiquitin specific peptidase 16 | USP16 | 10,600 | Cytoplasm | −1.764 |
| zinc finger and BTB domain containing 14 | ZBTB14 | 7541 | Nucleus | −1.55 |
| ZFP36 ring finger protein | ZFP36 | 7538 | Nucleus | 1.827 |
| Entrez Gene Name | Symbol | Entrez Gene ID a | Location | Fold Change b |
|---|---|---|---|---|
| ATP binding cassette subfamily C member 1 | ABCC1 | 4363 | Plasma Membrane | −1.984 |
| ABL proto-oncogene 2, non-receptor tyrosine kinase | ABL2 | 27 | Cytoplasm | 1.744 |
| argonaute RISC catalytic component 2 | AGO2 | 27,161 | Cytoplasm | −1.588 |
| AKT serine/threonine kinase 3 | AKT3 | 10,000 | Cytoplasm | −2.393 |
| alkB homolog 1, histone H2A dioxygenase | ALKBH1 | 8846 | Cytoplasm | −1.83 |
| angiopoietin 1 | ANGPT1 | 284 | Extracellular Space | −1.869 |
| amyloid beta precursor protein | APP | 351 | Plasma Membrane | 1.506 |
| aryl hydrocarbon receptor nuclear translocator | ARNT | 405 | Nucleus | 1.749 |
| activating transcription factor 3 | ATF3 | 467 | Nucleus | 2.535 |
| activating transcription factor 6 | ATF6 | 22,926 | Cytoplasm | 1.56 |
| ATM serine/threonine kinase | ATM | 472 | Nucleus | −1.523 |
| BCL2 associated X, apoptosis regulator | BAX | 581 | Cytoplasm | −2.51 |
| BCL2 apoptosis regulator | BCL2 | 596 | Cytoplasm | −1.646 |
| BCL2 like 13 | BCL2L13 | 23,786 | Cytoplasm | −2.078 |
| BCL6 corepressor | BCOR | 54,880 | Nucleus | 1.656 |
| basic helix-loop-helix ARNT like 1 | BMAL1 | 406 | Nucleus | −1.562 |
| 3′(2′), 5′-bisphosphate nucleotidase 1 | BPNT1 | 10,380 | Nucleus | 1.565 |
| bromodomain containing 3 | BRD3 | 8019 | Nucleus | 2.089 |
| calmodulin regulated spectrin associated protein family member 2 | CAMSAP2 | 23,271 | Cytoplasm | −1.955 |
| cell division cycle and apoptosis regulator 1 | CCAR1 | 55,749 | Nucleus | −1.579 |
| C-C motif chemokine ligand 2 | CCL2 | 6347 | Extracellular Space | −1.655 |
| CD200 molecule | CD200 | 4345 | Plasma Membrane | 1.713 |
| cyclin dependent kinase 12 | CDK12 | 51,755 | Nucleus | 1.849 |
| cyclin dependent kinase 13 | CDK13 | 8621 | Nucleus | −1.555 |
| cAMP responsive element binding protein 1 | CREB1 | 1385 | Nucleus | −1.68 |
| cAMP responsive element modulator | CREM | 1390 | Nucleus | 1.601 |
| cathepsin V | CTSV | 1515 | Cytoplasm | 1.573 |
| cytoglobin | CYGB | 114,757 | Cytoplasm | 2.378 |
| disco interacting protein 2 homolog A | DIP2A | 23,181 | Nucleus | 1.549 |
| dystonin | DST | 667 | Plasma Membrane | 2.201 |
| dystrobrevin alpha | DTNA | 1837 | Plasma Membrane | 2.069 |
| early growth response 1 | EGR1 | 1958 | Nucleus | 1.786 |
| eukaryotic translation initiation factor 2 alpha kinase 4 | EIF2AK4 | 440,275 | Cytoplasm | 1.576 |
| elongator acetyltransferase complex subunit 1 | ELP1 | 8518 | Cytoplasm | −2.024 |
| endoglin | ENG | 2022 | Plasma Membrane | −1.629 |
| endothelial PAS domain protein 1 | EPAS1 | 2034 | Nucleus | −3.025 |
| EPH receptor B2 | EPHB2 | 2048 | Plasma Membrane | 1.591 |
| erythropoietin receptor | EPOR | 2057 | Plasma Membrane | 2.778 |
| endoplasmic reticulum to nucleus signaling 1 | ERN1 | 2081 | Cytoplasm | −1.588 |
| Fas cell surface death receptor | FAS | 355 | Plasma Membrane | 2.388 |
| fibroblast growth factor receptor 2 | FGFR2 | 2263 | Plasma Membrane | −1.515 |
| fibronectin 1 | FN1 | 2335 | Extracellular Space | −2.059 |
| glial cell derived neurotrophic factor | GDNF | 2668 | Extracellular Space | 1.512 |
| glycerol-3-phosphate dehydrogenase 2 | GPD2 | 2820 | Cytoplasm | −1.54 |
| histone deacetylase 4 | HDAC4 | 9759 | Nucleus | 1.563 |
| hes family bHLH transcription factor 1 | HES1 | 3280 | Nucleus | 5.645 |
| heat shock protein 90 beta family member 1 | HSP90B1 | 7184 | Cytoplasm | −1.51 |
| interferon gamma inducible protein 16 | IFI16 | 3428 | Nucleus | −1.892 |
| interleukin 1 receptor type 1 | IL1R1 | 3554 | Plasma Membrane | −2.05 |
| interleukin 6 | IL6 | 3569 | Extracellular Space | 1.777 |
| Jun proto-oncogene, AP-1 transcription factor subunit | JUN | 3725 | Nucleus | 1.827 |
| lysine demethylase 6A | KDM6A | 7403 | Nucleus | −2.947 |
| KIT ligand | KITLG | 4254 | Extracellular Space | −1.654 |
| KLF transcription factor 2 | KLF2 | 10,365 | Nucleus | 1.979 |
| leptin receptor | LEPR | 3953 | Plasma Membrane | −1.507 |
| lamin B1 | LMNB1 | 4001 | Nucleus | −2.167 |
| LDL receptor related protein associated protein 1 | LRPAP1 | 4043 | Plasma Membrane | −1.845 |
| LYN proto-oncogene, Src family tyrosine kinase | LYN | 4067 | Cytoplasm | −2.418 |
| mitogen-activated protein kinase 1 | MAPK1 | 5594 | Cytoplasm | −1.564 |
| MAPK activated protein kinase 2 | MAPKAPK2 | 9261 | Nucleus | 1.95 |
| MET proto-oncogene, receptor tyrosine kinase | MET | 4233 | Plasma Membrane | −2.447 |
| mitofusin 2 | MFN2 | 9927 | Cytoplasm | −1.554 |
| MIA SH3 domain ER export factor 3 | MIA3 | 375,056 | Cytoplasm | −1.621 |
| matrix metallopeptidase 14 | MMP14 | 4323 | Extracellular Space | −1.7 |
| nuclear receptor coactivator 3 | NCOA3 | 8202 | Nucleus | −2.462 |
| neuronal growth regulator 1 | NEGR1 | 257,194 | Plasma Membrane | −1.799 |
| NFE2 like bZIP transcription factor 2 | NFE2L2 | 4780 | Nucleus | −1.85 |
| nuclear transcription factor Y subunit alpha | NFYA | 4800 | Nucleus | −1.914 |
| nicotinamide nucleotide transhydrogenase | NNT | 23,530 | Cytoplasm | −1.592 |
| notch receptor 2 | NOTCH2 | 4853 | Plasma Membrane | 1.741 |
| nuclear receptor subfamily 3 group C member 1 | NR3C1 | 2908 | Nucleus | 2.526 |
| nuclear receptor subfamily 4 group A member 1 | NR4A1 | 3164 | Nucleus | 2.282 |
| nuclear receptor subfamily 4 group A member 2 | NR4A2 | 4929 | Nucleus | 5.217 |
| O-linked N-acetylglucosamine (GlcNAc) transferase | OGT | 8473 | Cytoplasm | 1.511 |
| programmed cell death 4 | PDCD4 | 27,250 | Nucleus | 2.085 |
| phosphatidylinositol 3-kinase catalytic subunit type 3 | PIK3C3 | 5289 | Cytoplasm | 1.887 |
| phosphoinositide-3-kinase regulatory subunit 1 | PIK3R1 | 5295 | Cytoplasm | 1.557 |
| DNA polymerase kappa | POLK | 51,426 | Nucleus | 1.562 |
| cytochrome p450 oxidoreductase | POR | 5447 | Cytoplasm | −2.999 |
| peroxiredoxin 2 | PRDX2 | 7001 | Cytoplasm | 1.62 |
| perforin 1 | PRF1 | 5551 | Cytoplasm | 1.888 |
| protein kinase AMP-activated catalytic subunit alpha 1 | PRKAA1 | 5562 | Cytoplasm | −1.683 |
| presenilin 2 | PSEN2 | 5664 | Cytoplasm | −1.611 |
| prostaglandin E receptor 2 | PTGER2 | 5732 | Plasma Membrane | 1.877 |
| protein tyrosine phosphatase non-receptor type 11 | PTPN11 | 5781 | Cytoplasm | −1.714 |
| protein tyrosine phosphatase receptor type B | PTPRB | 5787 | Plasma Membrane | 1.544 |
| peroxidasin | PXDN | 7837 | Extracellular Space | 1.827 |
| RB transcriptional corepressor like 1 | RBL1 | 5933 | Nucleus | −1.614 |
| RAR related orphan receptor A | RORA | 6095 | Nucleus | −1.609 |
| RUNX family transcription factor 1 | RUNX1 | 861 | Nucleus | −1.563 |
| sphingosine-1-phosphate receptor 2 | S1PR2 | 9294 | Plasma Membrane | −1.619 |
| sarcoglycan beta | SGCB | 6443 | Plasma Membrane | −2.825 |
| SNF2 histone linker PHD RING helicase | SHPRH | 257,218 | Nucleus | −1.917 |
| solute carrier family 1 member 1 | SLC1A1 | 6505 | Plasma Membrane | −1.692 |
| solute carrier family 7 member 11 | SLC7A11 | 23,657 | Plasma Membrane | 1.719 |
| striated muscle enriched protein kinase | SPEG | 10,290 | Nucleus | 1.96 |
| secreted phosphoprotein 1 | SPP1 | 6696 | Extracellular Space | −1.573 |
| signal transducer and activator of transcription 6 | STAT6 | 6778 | Nucleus | −1.638 |
| stanniocalcin 1 | STC1 | 6781 | Extracellular Space | −1.546 |
| small VCP interacting protein | SVIP | 258,010 | Cytoplasm | −1.696 |
| transforming growth factor beta 2 | TGFB2 | 7042 | Extracellular Space | −1.584 |
| transforming growth factor beta receptor 2 | TGFBR2 | 7048 | Plasma Membrane | −1.801 |
| thrombospondin 1 | THBS1 | 7057 | Extracellular Space | −1.66 |
| TIA1 cytotoxic granule associated RNA binding protein | TIA1 | 7072 | Nucleus | −2.305 |
| toll like receptor 4 | TLR4 | 7099 | Plasma Membrane | −2.167 |
| tumor protein p53 | TP53 | 7157 | Nucleus | −1.518 |
| thioredoxin | TXN | 7295 | Cytoplasm | 1.692 |
| thioredoxin interacting protein | TXNIP | 10,628 | Cytoplasm | −2.163 |
| ubiquitination factor E4B | UBE4B | 10,277 | Cytoplasm | −1.591 |
| WASH complex subunit 1 | WASHC1 | 100,287,171 | Cytoplasm | −3.587 |
| ZFP36 ring finger protein | ZFP36 | 7538 | Nucleus | 1.942 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.G.; George, N.P.; Hwang, J.S.; Park, S.; Kim, M.O.; Lee, S.H.; Lee, G. Human Bone Marrow-Derived Mesenchymal Stem Cell Applications in Neurodegenerative Disease Treatment and Integrated Omics Analysis for Successful Stem Cell Therapy. Bioengineering 2023, 10, 621. https://doi.org/10.3390/bioengineering10050621
Kim SG, George NP, Hwang JS, Park S, Kim MO, Lee SH, Lee G. Human Bone Marrow-Derived Mesenchymal Stem Cell Applications in Neurodegenerative Disease Treatment and Integrated Omics Analysis for Successful Stem Cell Therapy. Bioengineering. 2023; 10(5):621. https://doi.org/10.3390/bioengineering10050621
Chicago/Turabian StyleKim, Seok Gi, Nimisha Pradeep George, Ji Su Hwang, Seokho Park, Myeong Ok Kim, Soo Hwan Lee, and Gwang Lee. 2023. "Human Bone Marrow-Derived Mesenchymal Stem Cell Applications in Neurodegenerative Disease Treatment and Integrated Omics Analysis for Successful Stem Cell Therapy" Bioengineering 10, no. 5: 621. https://doi.org/10.3390/bioengineering10050621
APA StyleKim, S. G., George, N. P., Hwang, J. S., Park, S., Kim, M. O., Lee, S. H., & Lee, G. (2023). Human Bone Marrow-Derived Mesenchymal Stem Cell Applications in Neurodegenerative Disease Treatment and Integrated Omics Analysis for Successful Stem Cell Therapy. Bioengineering, 10(5), 621. https://doi.org/10.3390/bioengineering10050621