Synergizing Deep Learning-Enabled Preprocessing and Human–AI Integration for Efficient Automatic Ground Truth Generation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Microscopy Imaging
2.1.2. Dataset Arrangement
- Cohort 1: 11 sections from 11 mice, with expert ground truth.
- Cohort 2: 45 sections from 9 additional mice.
- Cohort 3: 44 sections from the same 9 mice as Cohort 2.
2.1.3. Deep Learning Model
2.2. Methods
2.2.1. Semantic Preprocessing (SP)
2.2.2. Bootstrapped Semantic Preprocessing (BSP)
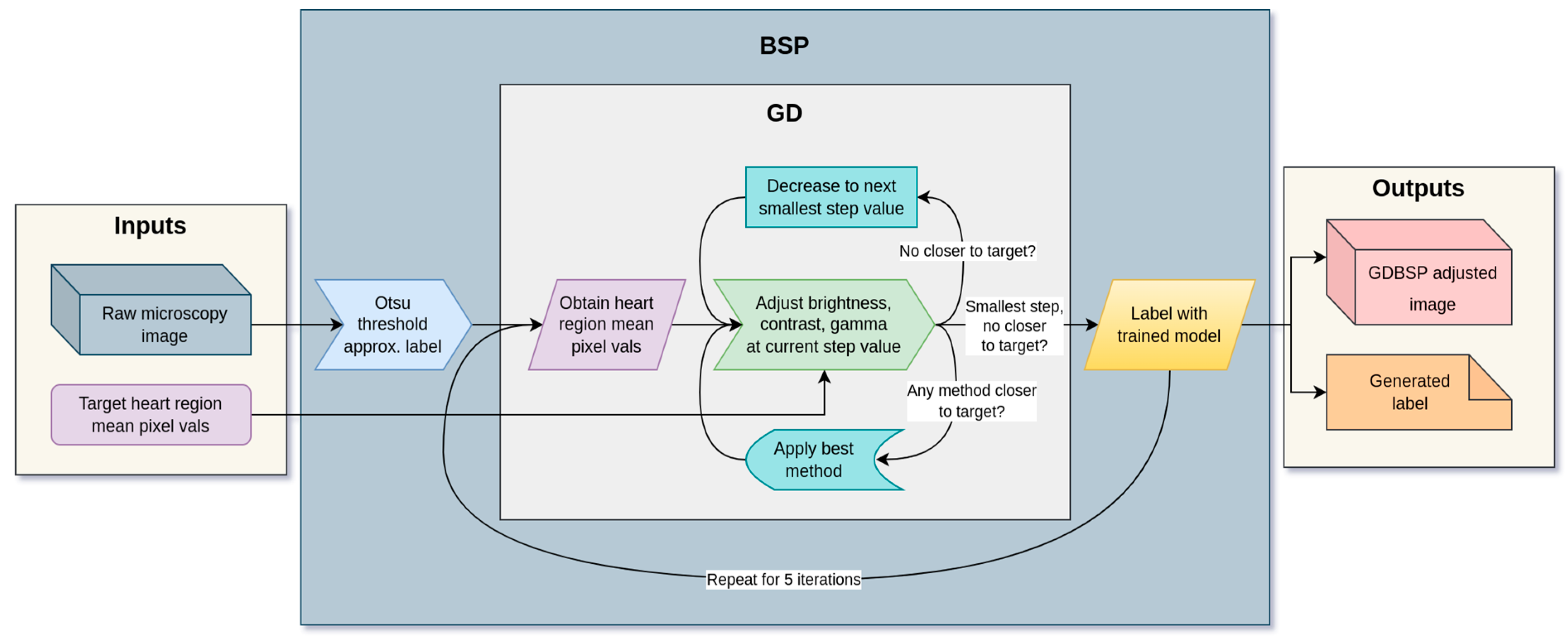
2.2.3. Gradient Descent BSP (GDBSP)
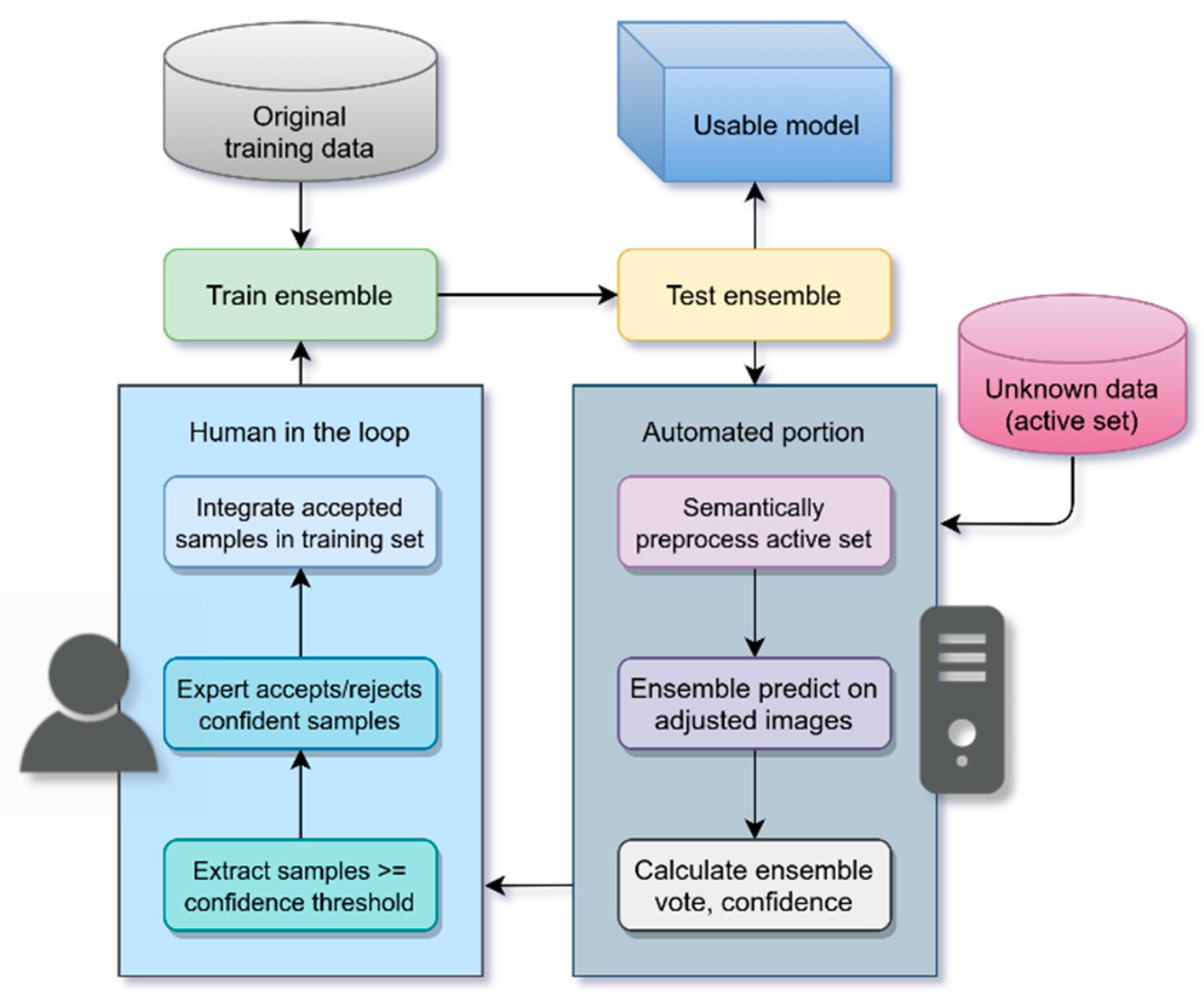
2.2.4. Active Deep Learning (ADL)
2.2.5. Experiments
- (1)
- The six-fold ensembles are trained; any accepted sections are included in the training set for all folds.
- (2)
- Each ensemble is tested for cross-validation.
- (3)
- The least overfit fold (lowest test Dice) ensemble preprocesses active set sections using the given method, predicts, and then votes on, the final composite prediction maps.
- (4)
- With ground truths as the reference, record Dice accuracy, confidence per-section confidence, and mean confidence.
- (5)
- A human expert evaluates active set sections that reach at least 97% confidence, deciding to accept or reject the training set.
- (6)
- Accepted samples are integrated into the training set for the next iteration.
- (7)
- The four-fold ensembles are trained; each training set is expanded with any accepted sections.
- (8)
- Each ensemble is tested for cross-validation.
- (9)
- The least overfit (lowest test Dice) fold ensemble is used to preprocess active set sections using the given method, predicts, and then votes on, the final composite prediction maps.
- (10)
- Record per-section confidence and mean confidence on the active set.
- (11)
- A human expert chooses a confidence threshold that produces roughly one hour of work, or approximately 12 sections. Sections above the threshold are evaluated for acceptance or rejection.
- (12)
- Accepted samples are integrated into the training set for the next iteration.
3. Results
3.1. Cohort 1 Experiment
3.2. Cohorts 2 and 3 Experiment
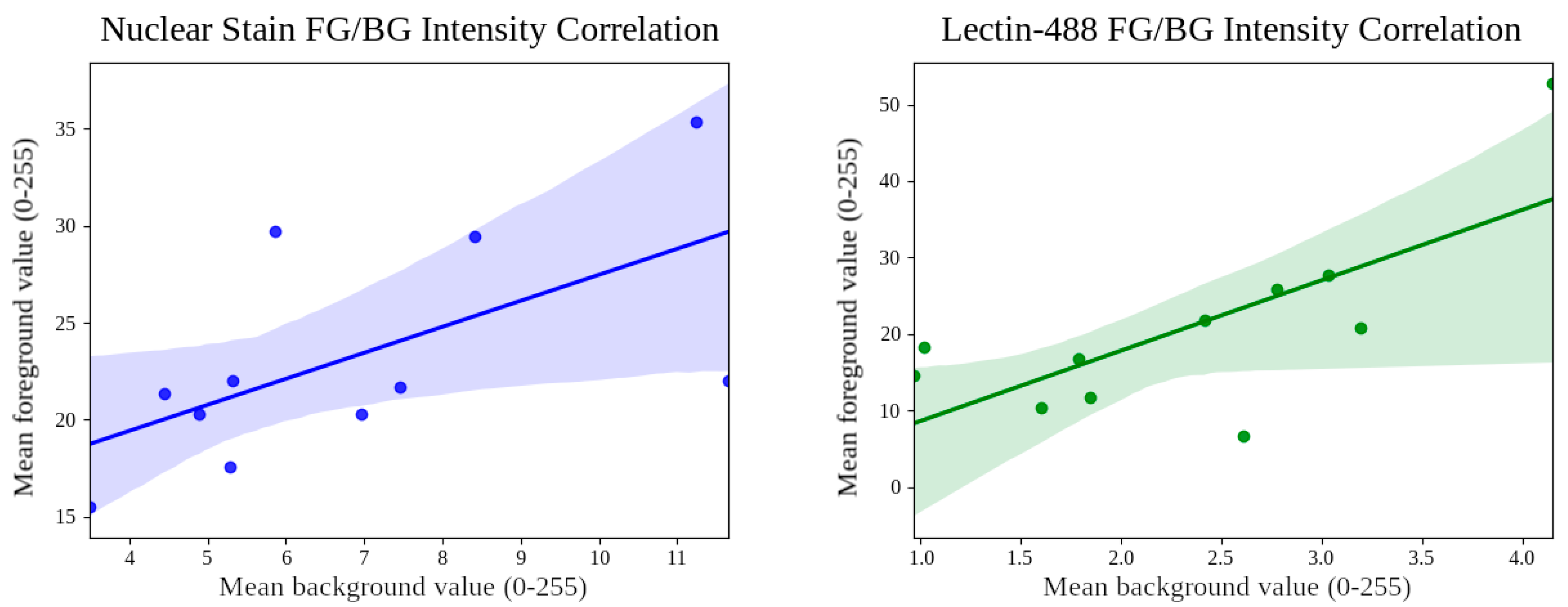
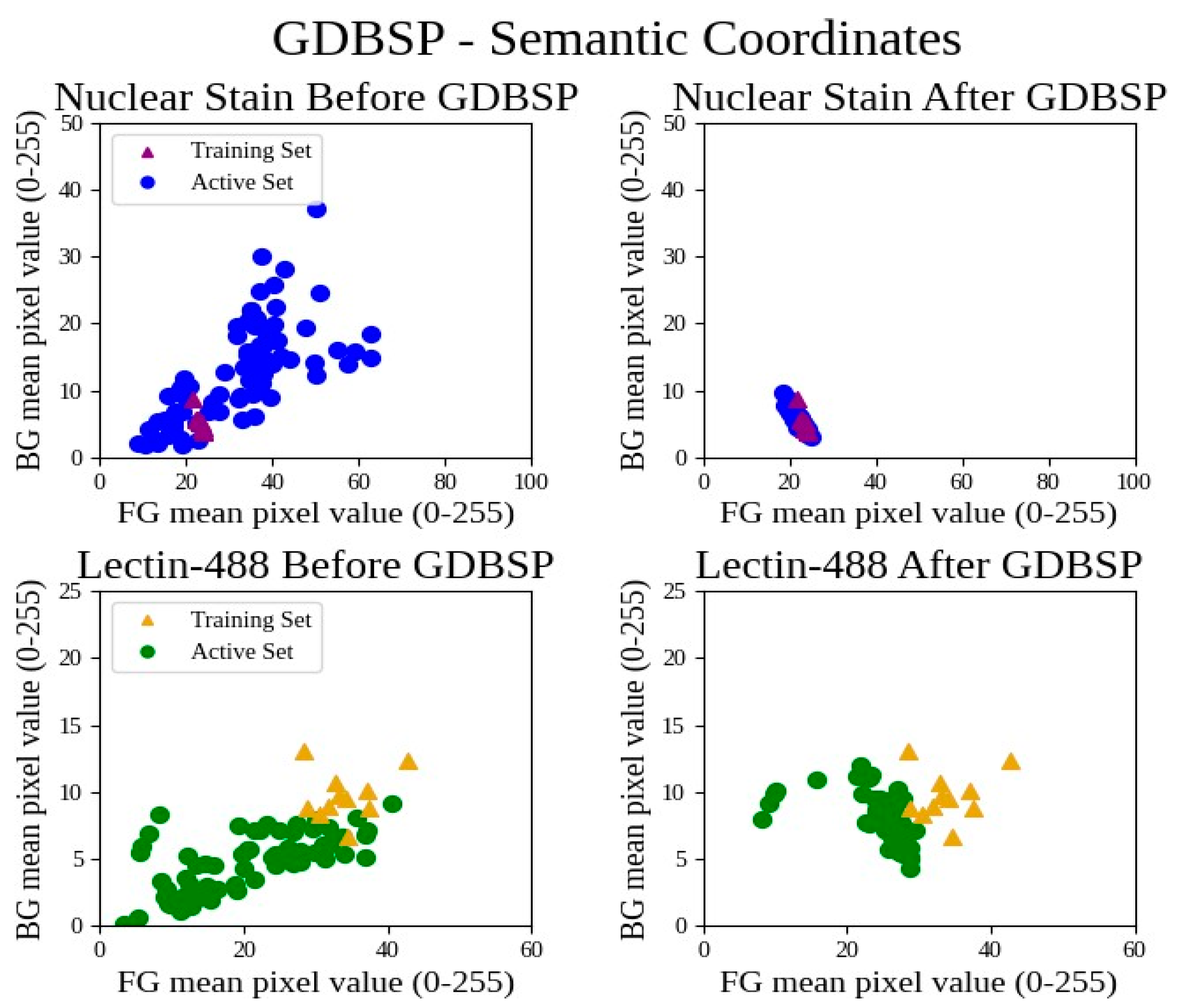
3.3. GDBSP Preprocessing
4. Discussion
5. Conclusions, Limitations, and Future Work
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Maki, S.; Furuya, T.; Inoue, M.; Shiga, Y.; Inage, K.; Eguchi, Y.; Orita, S.; Ohtori, S. Machine Learning and Deep Learning in Spinal Injury: A Narrative Review of Algorithms in Diagnosis and Prognosis. J. Clin. Med. 2024, 13, 705. [Google Scholar] [CrossRef]
- Alnasser, T.N.; Abdulaal, L.; Maiter, A.; Sharkey, M.; Dwivedi, K.; Salehi, M.; Garg, P.; Swift, A.J.; Alabed, S. Advancements in cardiac structures segmentation: A comprehensive systematic review of deep learning in CT imaging. Front. Cardiovasc. Med. 2024, 11, 1323461. [Google Scholar] [CrossRef]
- Alongi, P.; Arnone, A.; Vultaggio, V.; Fraternali, A.; Versari, A.; Casali, C.; Arnone, G.; DiMeco, F.; Vetrano, I.G. Artificial Intelligence Analysis Using MRI and PET Imaging in Gliomas: A Narrative Review. Cancers 2024, 16, 407. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jager, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.; Saha, A.; Campbell, C.J.V.; Nejat, P.; Lokker, C.; Norgan, A.P. Seeing the random forest through the decision trees. Supporting learning health systems from histopathology with machine learning models: Challenges and opportunities. J. Pathol. Inform. 2024, 15, 100347. [Google Scholar] [CrossRef]
- Labrada, A.; Barkana, B.D. A Comprehensive Review of Computer-Aided Models for Breast Cancer Diagnosis Using Histopathology Images. Bioengineering 2023, 10, 1289. [Google Scholar] [CrossRef]
- Amber, M.A.; Simpson, L.; Bakas, S.; Bilello, M.; Farahani, K.; van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Srinidhi, C.L.; Ciga, O.; Martel, A.L. Deep neural network models for computational histopathology: A survey. Med. Image Anal. 2020, 67, 101813. [Google Scholar] [CrossRef]
- Albarqouni, S.; Baur, C.; Achilles, F.; Belagiannis, V.; Demirci, S.; Navab, N. AggNet: Deep Learning From Crowds for Mitosis Detection in Breast Cancer Histology Images. IEEE Trans. Med. Imaging 2016, 35, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Irshad, H.; Oh, E.Y.; Schmolze, D.; Quintana, L.M.; Collins, L.; Tamimi, R.M.; Beck, A.H. Crowdsourcing scoring of immunohistochemistry images: Evaluating Performance of the Crowd and an Automated Computational Method. Sci. Rep. 2017, 7, 43286. [Google Scholar] [CrossRef] [PubMed]
- Amgad, M.; Elfandy, H.; Hussein, H.; Atteya, L.A.; Elsebaie, M.A.T.; Elnasr, L.S.A.; Sakr, R.A.; Salem, H.S.E.; Ismail, A.F.; Saad, A.M.; et al. Structured crowdsourcing enables convolutional segmentation of histology images. Bioinformatics 2019, 35, 3461–3467. [Google Scholar] [CrossRef] [PubMed]
- Kaissis, G.A.; Makowski, M.R.; Rückert, D.; Braren, R.F. Secure, privacy-preserving and federated machine learning in medical imaging. Nat. Mach. Intell. 2020, 2, 305–311. [Google Scholar] [CrossRef]
- Nettrour, J.F.; Burch, M.B.; Bal, B.S. Patients, pictures, and privacy: Managing clinical photographs in the smartphone era. Arthroplast. Today 2018, 5, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Ahn, D.K.; Unde, B.; Gage, H.D.; Carr, J.J. Patient-controlled sharing of medical imaging data across unaffiliated healthcare organizations. J. Am. Med. Inform. Assoc. 2013, 20, 157–163. [Google Scholar] [CrossRef][Green Version]
- Zamzmi, G.; Rajaraman, S.; Hsu, L.Y.; Sachdev, V.; Antani, S. Real-time echocardiography image analysis and quantification of cardiac indices. Med. Image Anal. 2022, 80, 102438. [Google Scholar] [CrossRef] [PubMed]
- van Ginneken, B.; Katsuragawa, S.; Romeny, B.M.T.H.; Doi, K.; Viergever, M.A. Automatic detection of abnormalities in chest radiographs using local texture analysis. IEEE Trans. Med. Imaging 2002, 21, 139–149. [Google Scholar] [CrossRef]
- Masulli, F.; Schenone, A. A fuzzy clustering based segmentation system as support to diagnosis in medical imaging. Artif. Intell. Med. 1999, 16, 129–147. [Google Scholar] [CrossRef]
- Pham, D.L.; Xu, C.; Prince, J.L. Current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2000, 2, 315–337. [Google Scholar] [CrossRef]
- Xiong, G.; Zhou, X.; Degterev, A.; Ji, L.; Wong, S.T. Automated neurite labeling and analysis in fluorescence microscopy images. Cytom. Part A 2006, 69A, 494–505. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, C.; Nie, D.; Lin, D.; Zhu, Y.; Chen, C.; Zhao, L.; Wu, X.; Dongye, M.; Xu, F.; et al. Deep learning from “passive feeding” to “selective eating” of real-world data. NPJ Digit. Med. 2020, 3, 143. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, Y.; Obata, T.; Kershaw, J.; Sakaki, H.; Urushihata, T.; Omatsu, T.; Kishimoto, R.; Higashi, T. The Utility of Applying Various Image Preprocessing Strategies to Reduce the Ambiguity in Deep Learning-based Clinical Image Diagnosis. Magn. Reson. Med. Sci. 2020, 19, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Sathiyaseelan, R.; Ravi, K.; Ramamoorthy, R.; Chennaiah, M.P. Haemorrhage diagnosis in colour fundus images using a fast-convolutional neural network based on a modified U-Net. Network 2024. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.B.; Shinde, R.K.; Oh, S.; Kwon, K.C.; Kim, N. A Systematic Review and Identification of the Challenges of Deep Learning Techniques for Undersampled Magnetic Resonance Image Reconstruction. Sensors 2024, 24, 753. [Google Scholar] [CrossRef]
- Cohn, D.; Atlas, L.; Ladner, R. Improving generalization with active learning. Mach. Learn. 1994, 15, 201–221. [Google Scholar] [CrossRef]
- Li, Y.-F.; Zhou, Z.-H. Towards Making Unlabeled Data Never Hurt. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 175–188. [Google Scholar] [PubMed]
- Raczkowska, A.; Mozejko, M.; Zambonelli, J.; Szczurek, E. ARA: Accurate, reliable and active histopathological image classification framework with Bayesian deep learning. Sci. Rep. 2019, 9, 14347. [Google Scholar] [CrossRef]
- Sourati, J.; Gholipour, A.; Dy, J.G.; Kurugol, S.; Warfield, S.K. Active Deep Learning with Fisher Information for Patch-wise Semantic Segmentation. Deep. Learn. Med. Image Anal. Multimodal Learn. Clin. Decis. Support 2018, 11045, 83–91. [Google Scholar]
- DeVries, T.; Taylor, G.W. Leveraging uncertainty estimates for predicting segmentation quality. arXiv 2018, arXiv:1807.00502. [Google Scholar]
- Huang, G.; Li, Y.; Pleiss, G.; Liu, Z.; Hopcroft, J.E.; Weinberger, K.Q. Snapshot Ensembles: Train 1, get M for free. arXiv 2017, arXiv:1704.00109. [Google Scholar]
- Solaguren-Beascoa, A. Active Learning in Machine Learning. 2020. [Google Scholar]
- Alahmari, S.; Goldgof, D.; Hall, L.; Dave, P.; Phoulady, H.A.; Mouton, P. Iterative deep learning based unbiased stereology with human-in-the-loop. In Proceedings of the 17th IEEE International Conference on Machine Learning and Applications (ICMLA), Orlando, FL, USA, 17–20 December 2018. [Google Scholar]
- Alahmari, S.S.; Goldgof, D.; Hall, L.O.; Mouton, P.R. Automatic Cell Counting using Active Deep Learning and Unbiased Stereology. In Proceedings of the 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC), Bari, Italy, 6–9 October 2019. [Google Scholar]
- Efron, B. Bootstrap Methods: Another Look at the Jackknife. Ann. Stat. 1979, 7, 1–26. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the IEEE Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019, 32, 8024–8035. [Google Scholar]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. In The Handbook of Brain Theory and Neural Networks; Arbib, M.A., Ed.; MIT Press: Cambridge, MA, USA, 1995. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Zeiler, M.D. ADADELTA: An adaptive learning rate method. arXiv 2012, arXiv:1212.5701. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Sgdr: Stochastic gradient descent with warm restarts. arXiv 2016, arXiv:1608.03983. [Google Scholar]
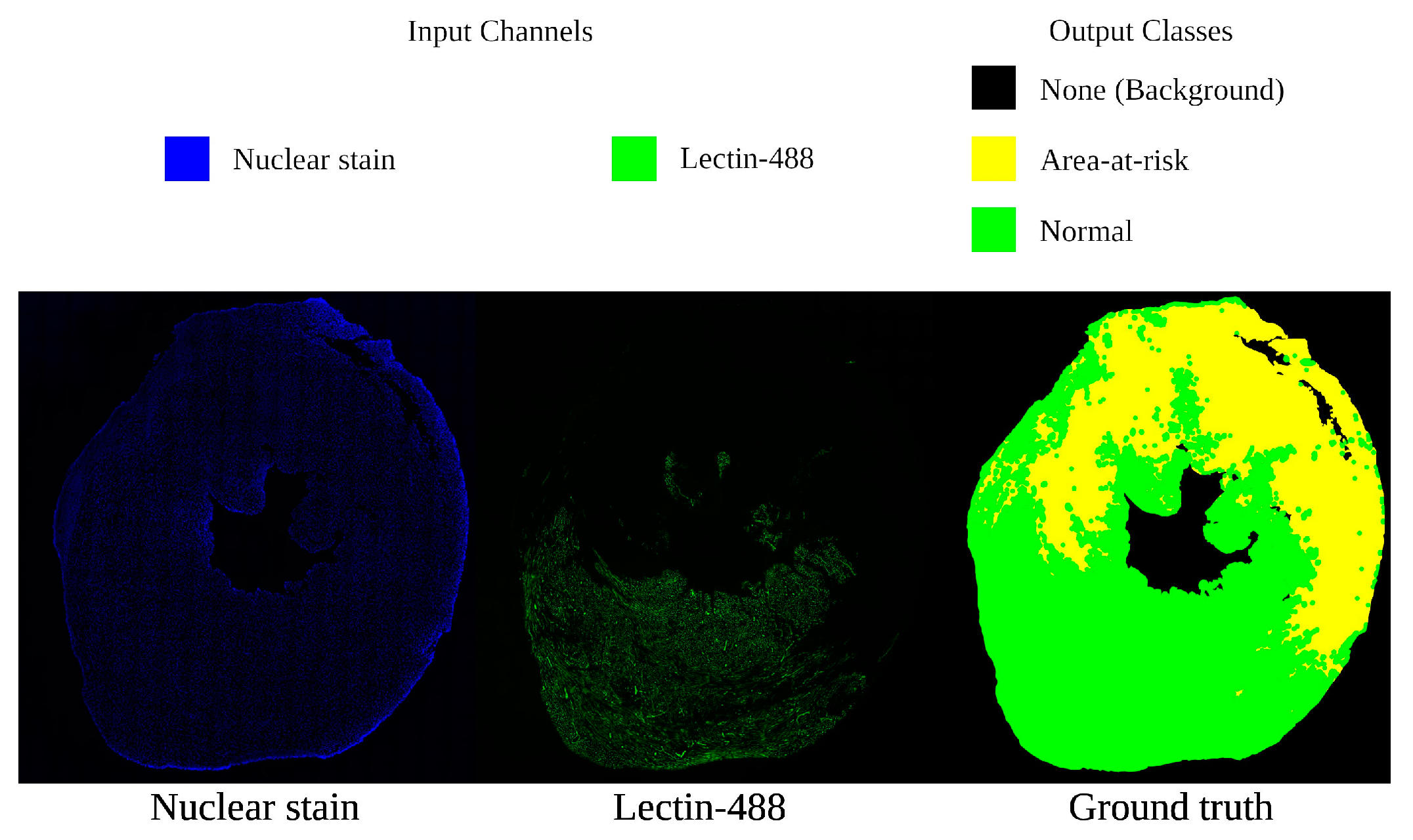
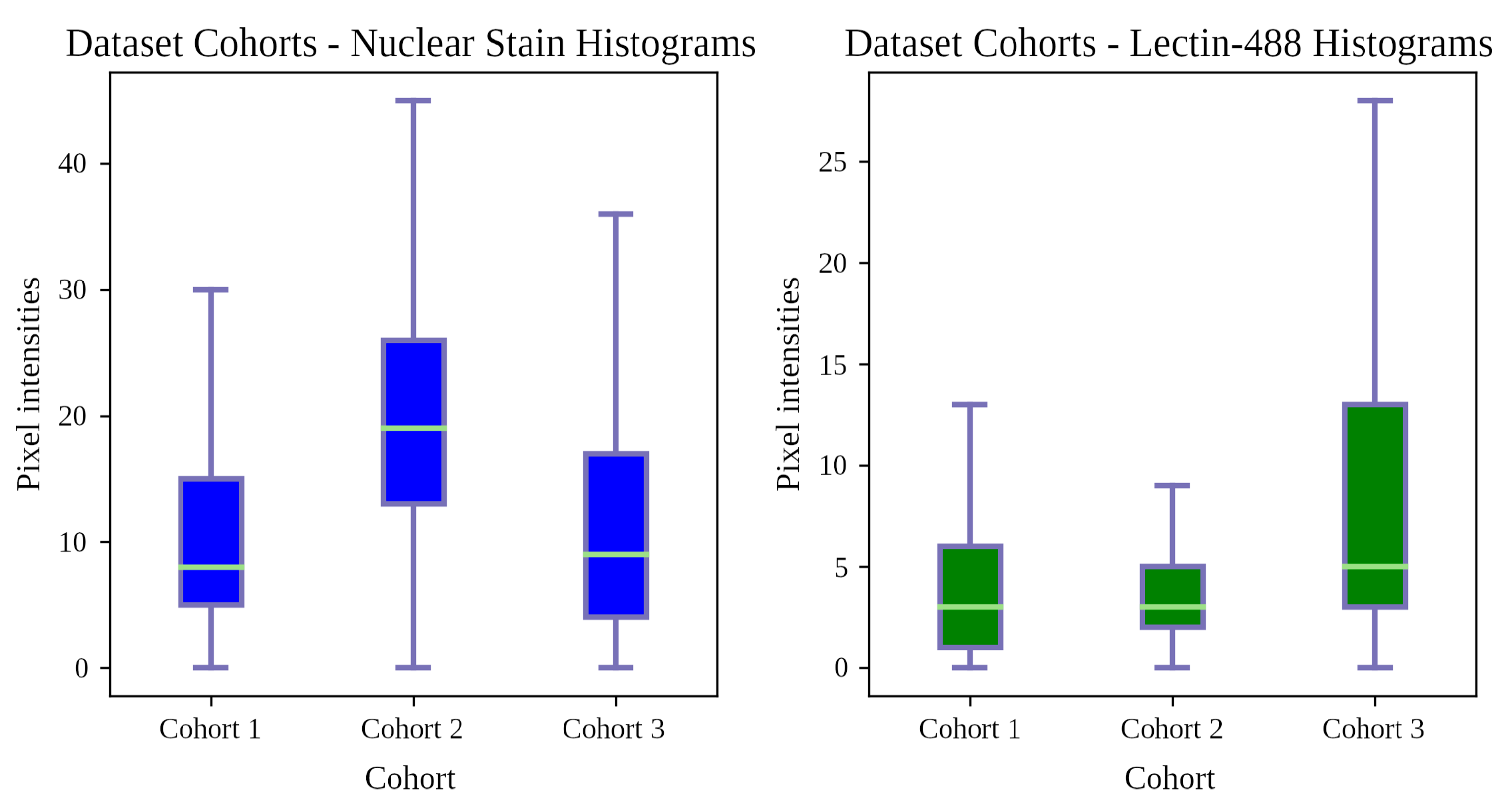
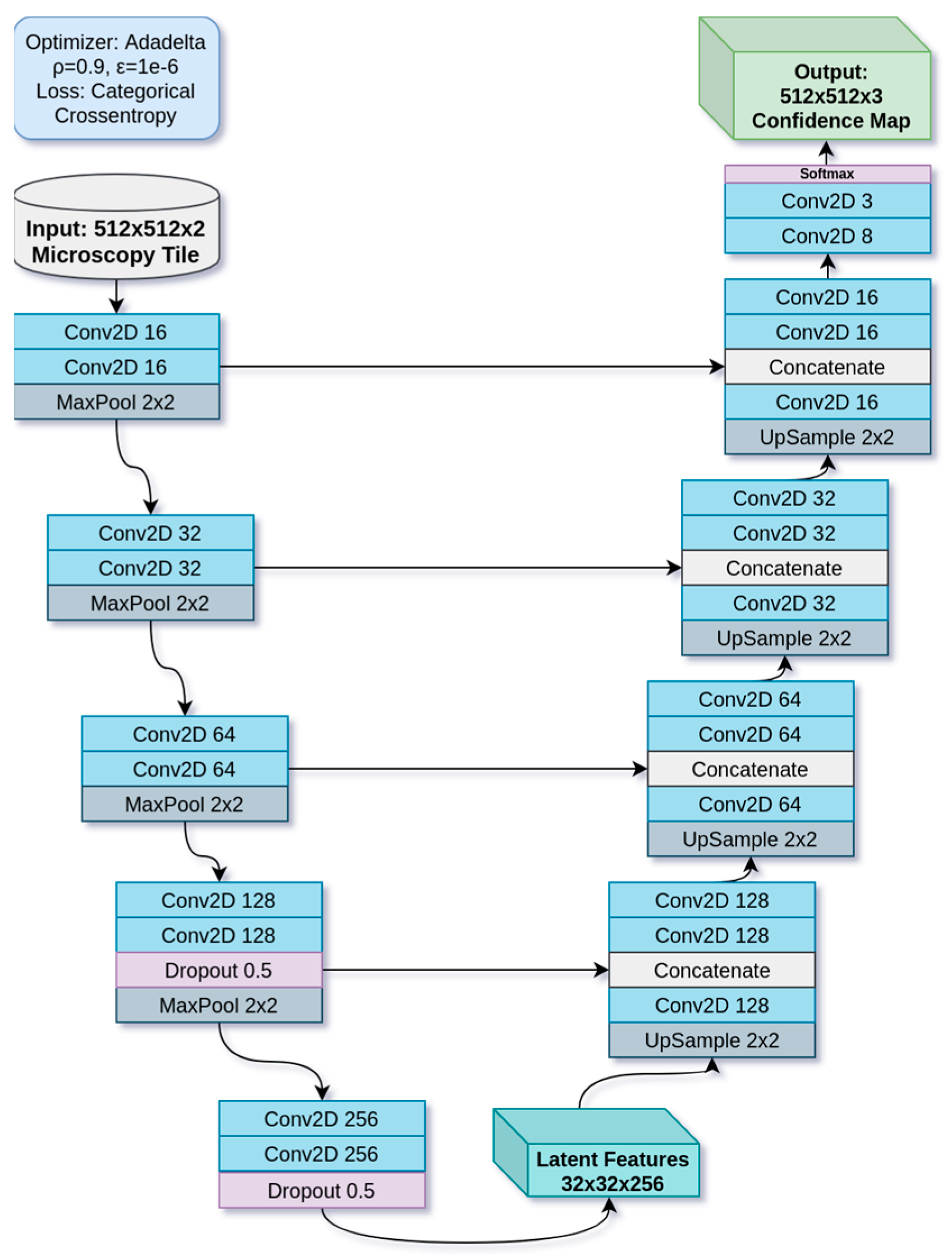
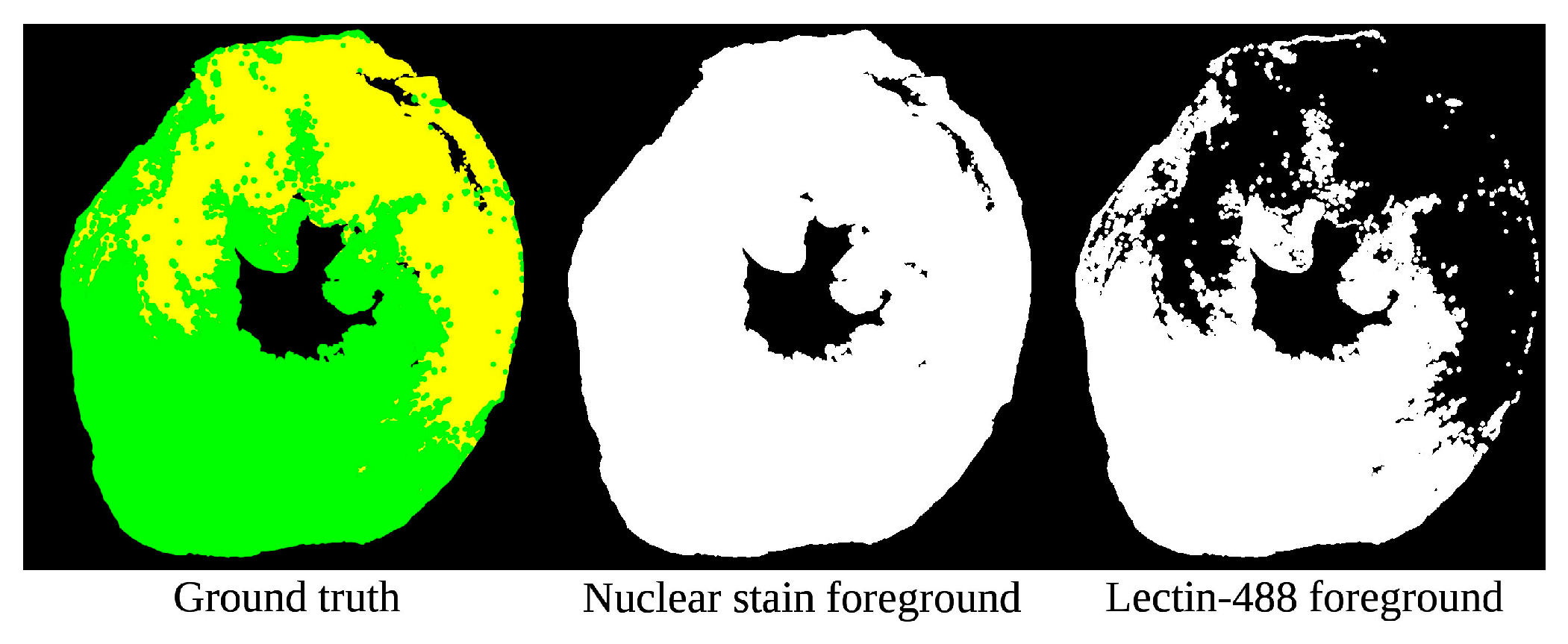
| Data Subset | Number of Samples |
|---|---|
| Cohort 1 | 4513 (with ground truth) |
| Cohort 2 | 10,604 |
| Cohort 3 | 10,684 |
| Active set total (cohorts 2 + 3) | 21,288 |
| Total | 25,801 |
| Preprocessing | Iteration | Mean 6-Fold Test Dice | Active Set Mean Dice | Active Set Mean Confidence | 97% Conf Threshold Samples | Accepted Samples | Dataset Size after Acceptance |
|---|---|---|---|---|---|---|---|
| Raw | 1 | 87.8% | 89.4% | 76.7% | 0 | 0 | 100% |
| DoG | 1 | 40.9% | 40.7% | 68.3% | 0 | 0 | 100% |
| HE | 1 | 83.9% | 78.9% | 86.7% | 0 | 0 | 100% |
| AHE | 1 | 90.1% | 94.6% | 92.2% | 0 | 0 | 100% |
| HM | 1 | 79.4% | 79.0% | 65.9% | 0 | 0 | 100% |
| 1 | 91.8% | 91.5% | 96.7% | 777 | 399 | 116% | |
| BSP | 2 | 93.5% | 92.2% | 97.7% | 1618 | 778 | 147% |
| 3 | 93.7% | 94.5% | 97.4% | 840 | 462 | 166% | |
| GDBSP | 1 | 82.2% | 53.0% | 92.4% | 0 | 0 | 100% |
| Preprocessing | Iteration | Mean Active Conf. | Chosen Threshold | % Samples Accepted | Cum. Accepted Samples | Cum. Expert Time (h) | Cum. Manual Expert Time (h) | Dataset Size |
|---|---|---|---|---|---|---|---|---|
| Raw | 1 | 80.4% | 90.0% | 0% | 0 | 0.0 | 100% | |
| DoG | 1 | 63.5% | 90.0% | 0 | 0.0 | 100% | ||
| HE | 1 | 83.4% | 90.0% | 0 | 0.0 | 100% | ||
| 1 | 89.1% | 93.0% | 69% | 1411 | 0.6 | 14 | 131% | |
| AHE | 2 | 94.1% | 97.5% | 100% | 4881 | 1.7 | 40 | 208% |
| 3 | 93.9% | 97.7% | 100% | 7767 | 2.6 | 62 | 272% | |
| 1 | 95.2% | 97.5% | 48% | 831 | 0.5 | 10 | 118% | |
| HM | 2 | 92.7% | 96.0% | 55% | 2271 | 1.1 | 22 | 150% |
| 3 | 93.3% | 96.5% | 47% | 3171 | 1.5 | 30 | 170% | |
| 1 | 92.3% | 96.0% | 43% | 584 | 0.5 | 10 | 113% | |
| BSP | 2 | 96.0% | 98.0% | 49% | 1861 | 1.1 | 22 | 141% |
| 3 | 95.5% | 98.0% | 44% | 3060 | 1.6 | 32 | 168% | |
| 1 | 96.1% | 98.0% | 92% | 2748 | 0.9 | 22 | 161% | |
| GDBSP | 2 | 96.6% | 98.5% | 93% | 5804 | 1.9 | 44 | 229% |
| 3 | 96.5% | 98.3% | 90% | 8374 | 2.7 | 64 | 286% |
| Iterations | Cumulative Accepted Sections | % Active Set Accepted | Dataset Size | Cumulative Expert Time (h) | Equivalent Manual Expert Time (h) | Expert Time Saved |
|---|---|---|---|---|---|---|
| 10 | 82 | 92% | 845% | 7.2 | 164 | 96% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Collazo, C.; Vargas, I.; Cara, B.; Weinheimer, C.J.; Grabau, R.P.; Goldgof, D.; Hall, L.; Wickline, S.A.; Pan, H. Synergizing Deep Learning-Enabled Preprocessing and Human–AI Integration for Efficient Automatic Ground Truth Generation. Bioengineering 2024, 11, 434. https://doi.org/10.3390/bioengineering11050434
Collazo C, Vargas I, Cara B, Weinheimer CJ, Grabau RP, Goldgof D, Hall L, Wickline SA, Pan H. Synergizing Deep Learning-Enabled Preprocessing and Human–AI Integration for Efficient Automatic Ground Truth Generation. Bioengineering. 2024; 11(5):434. https://doi.org/10.3390/bioengineering11050434
Chicago/Turabian StyleCollazo, Christopher, Ian Vargas, Brendon Cara, Carla J. Weinheimer, Ryan P. Grabau, Dmitry Goldgof, Lawrence Hall, Samuel A. Wickline, and Hua Pan. 2024. "Synergizing Deep Learning-Enabled Preprocessing and Human–AI Integration for Efficient Automatic Ground Truth Generation" Bioengineering 11, no. 5: 434. https://doi.org/10.3390/bioengineering11050434
APA StyleCollazo, C., Vargas, I., Cara, B., Weinheimer, C. J., Grabau, R. P., Goldgof, D., Hall, L., Wickline, S. A., & Pan, H. (2024). Synergizing Deep Learning-Enabled Preprocessing and Human–AI Integration for Efficient Automatic Ground Truth Generation. Bioengineering, 11(5), 434. https://doi.org/10.3390/bioengineering11050434








