Age Encoded Adversarial Learning for Pediatric CT Segmentation
Abstract
1. Introduction
2. Methodology
2.1. Generative Adversarial Networks (GANs)
2.2. CFG-SegNet
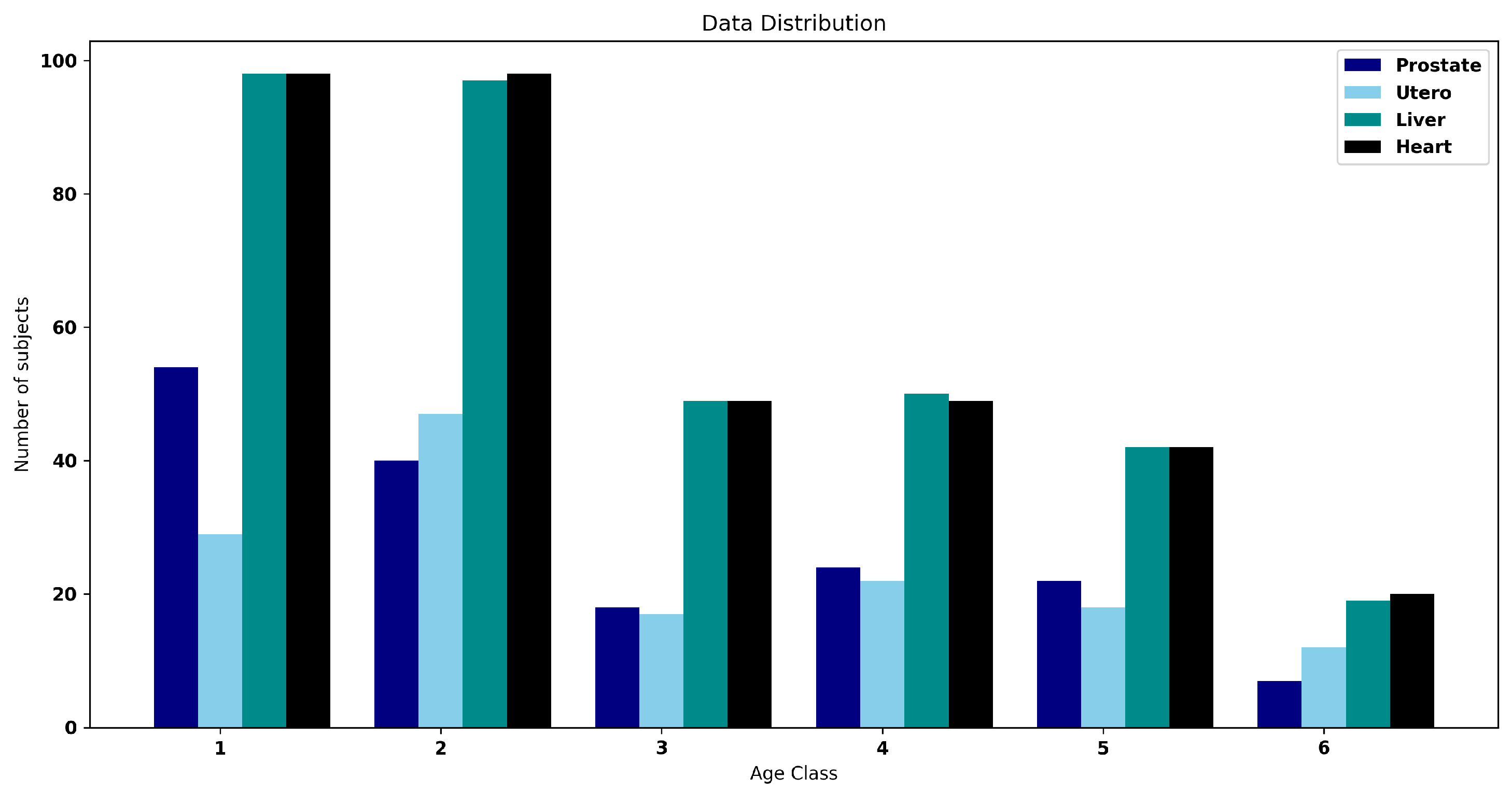
3. Dataset
4. Experiment
4.1. Implementation Details
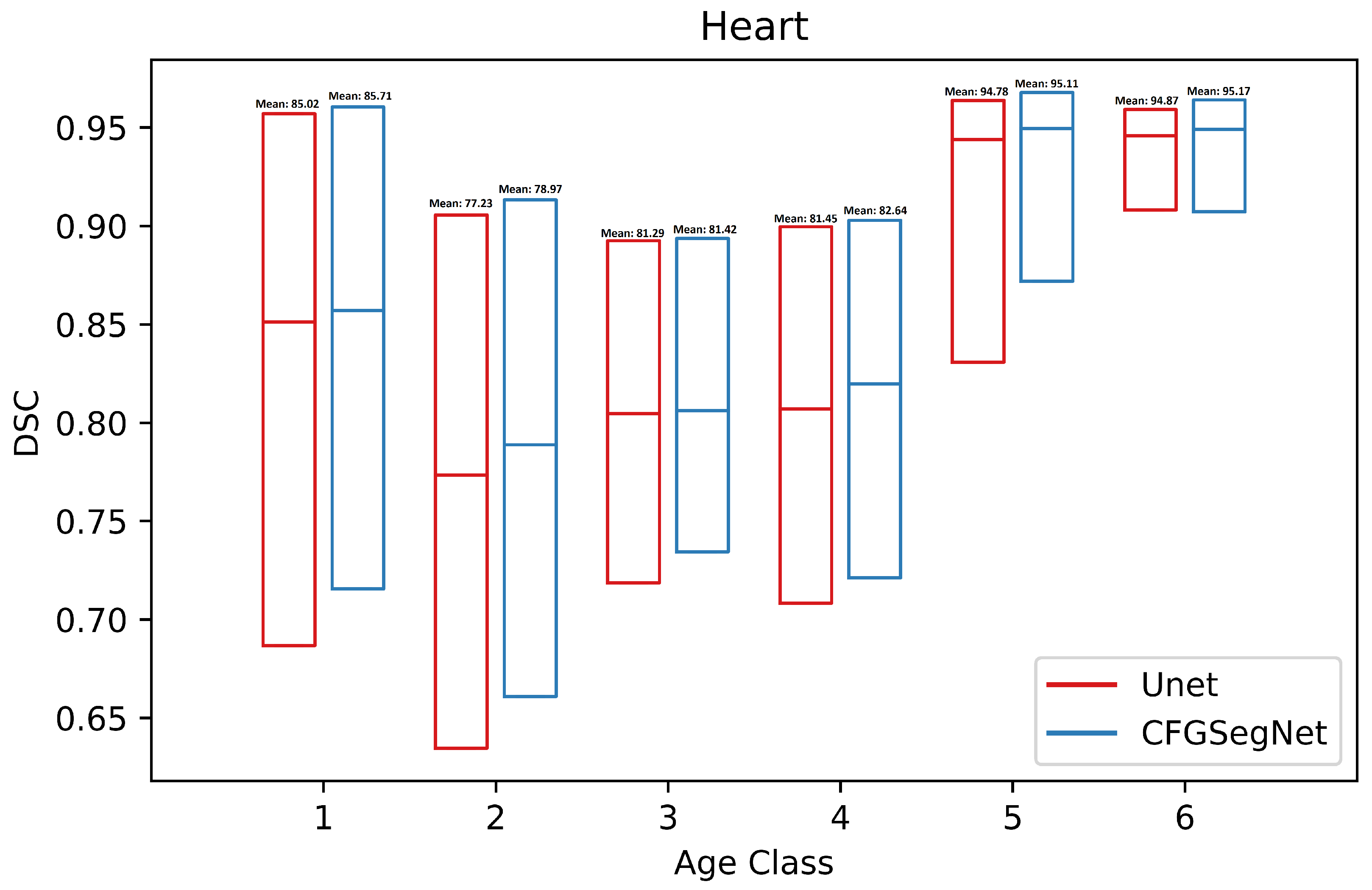
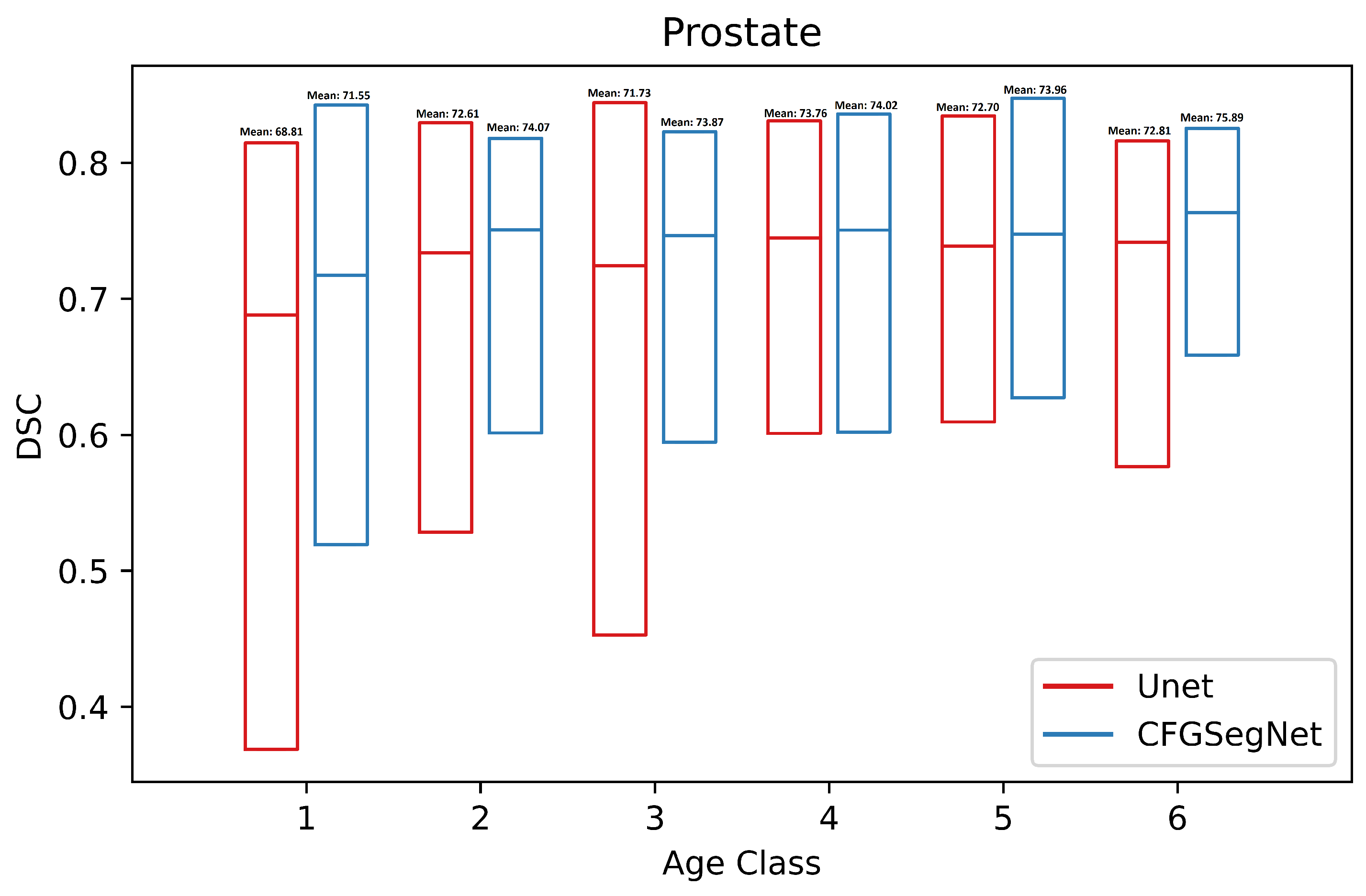
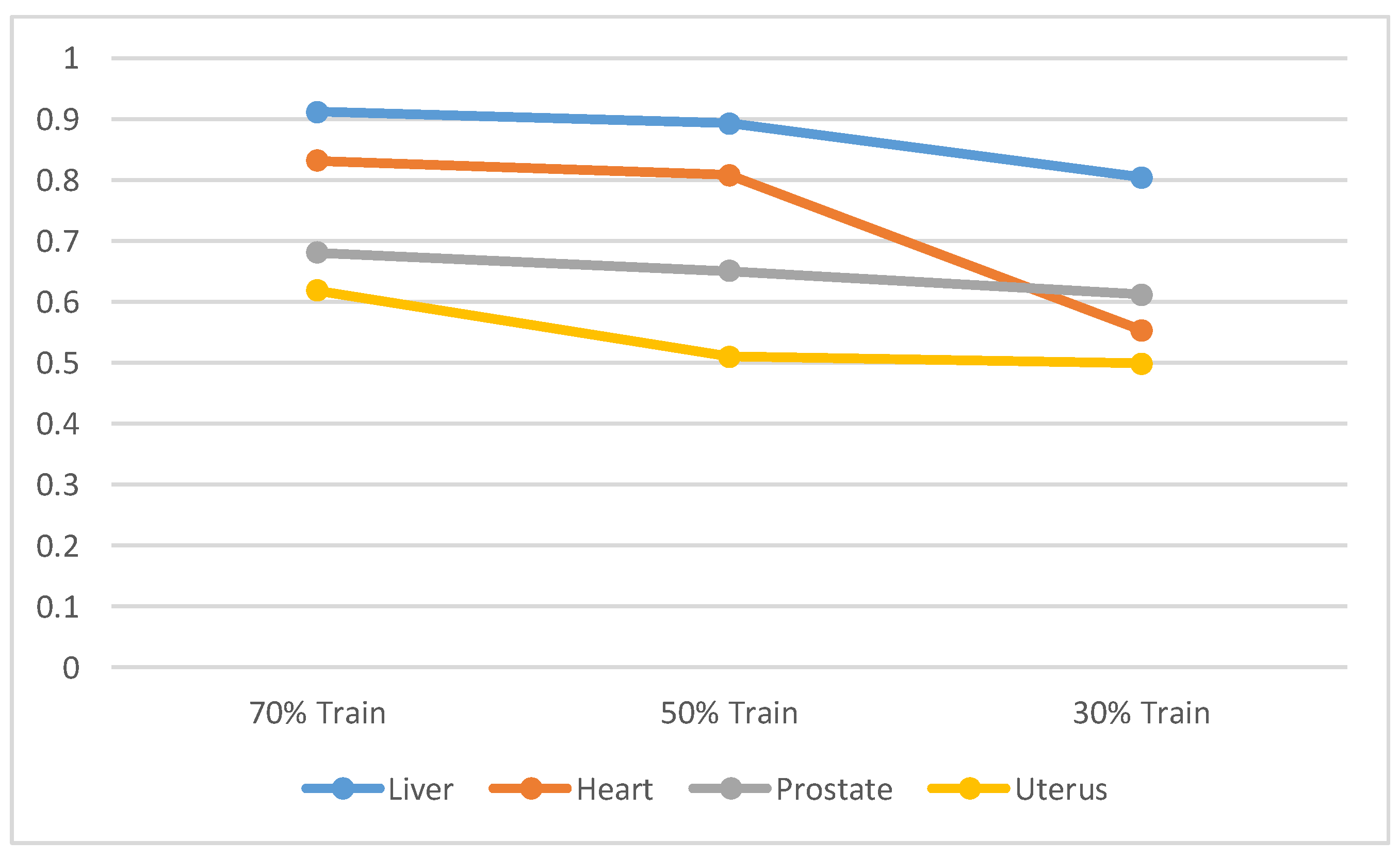
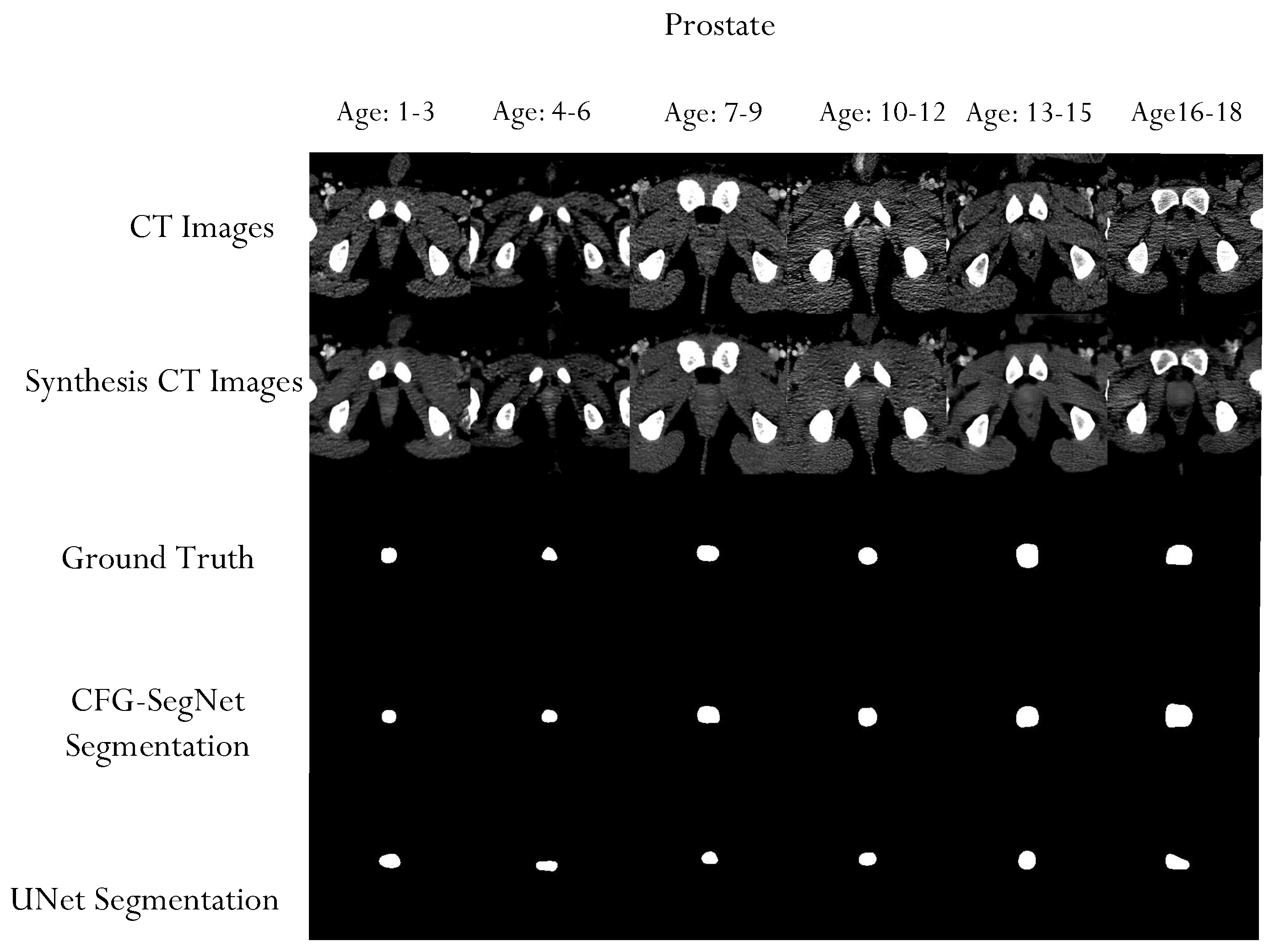
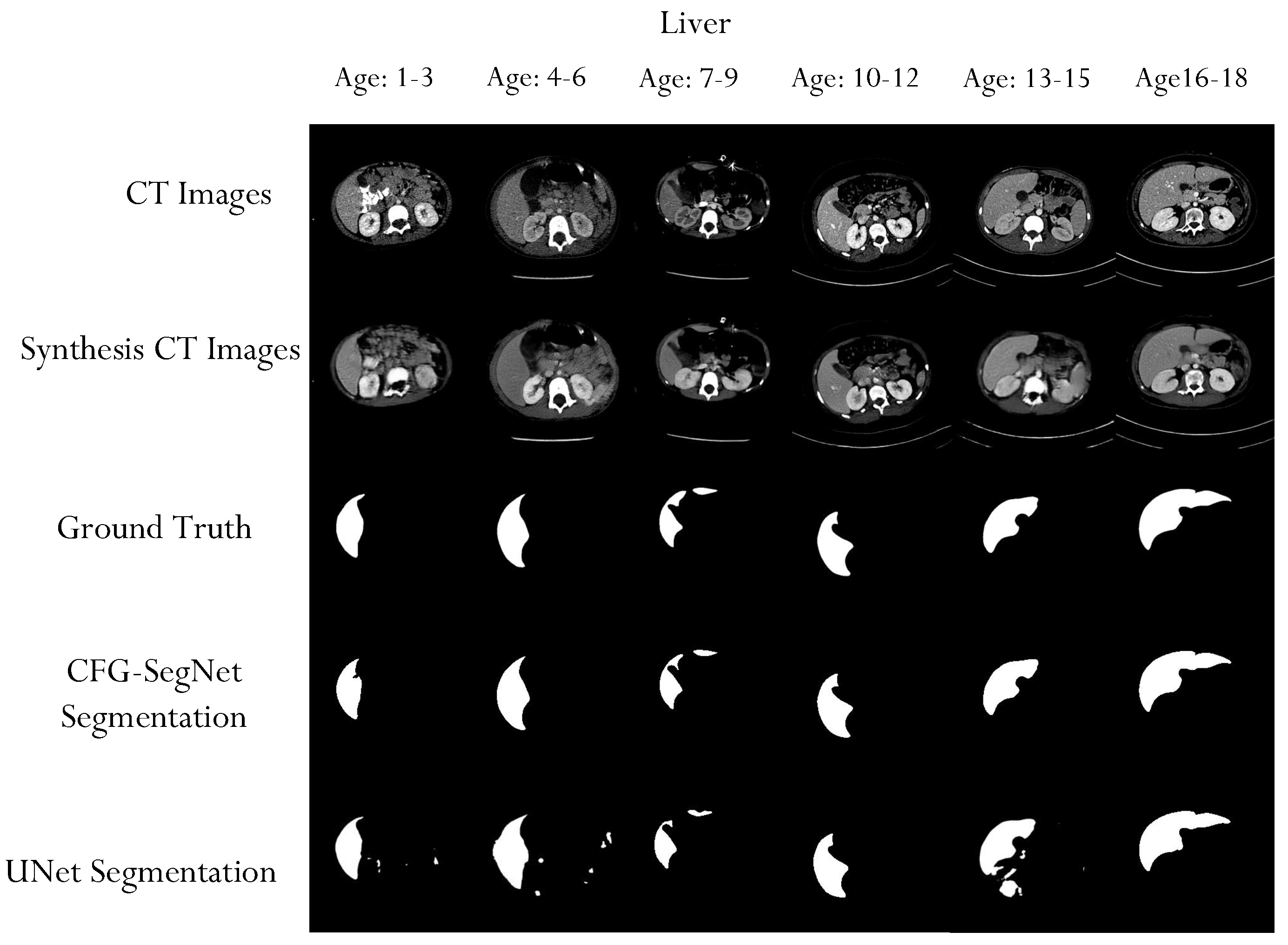
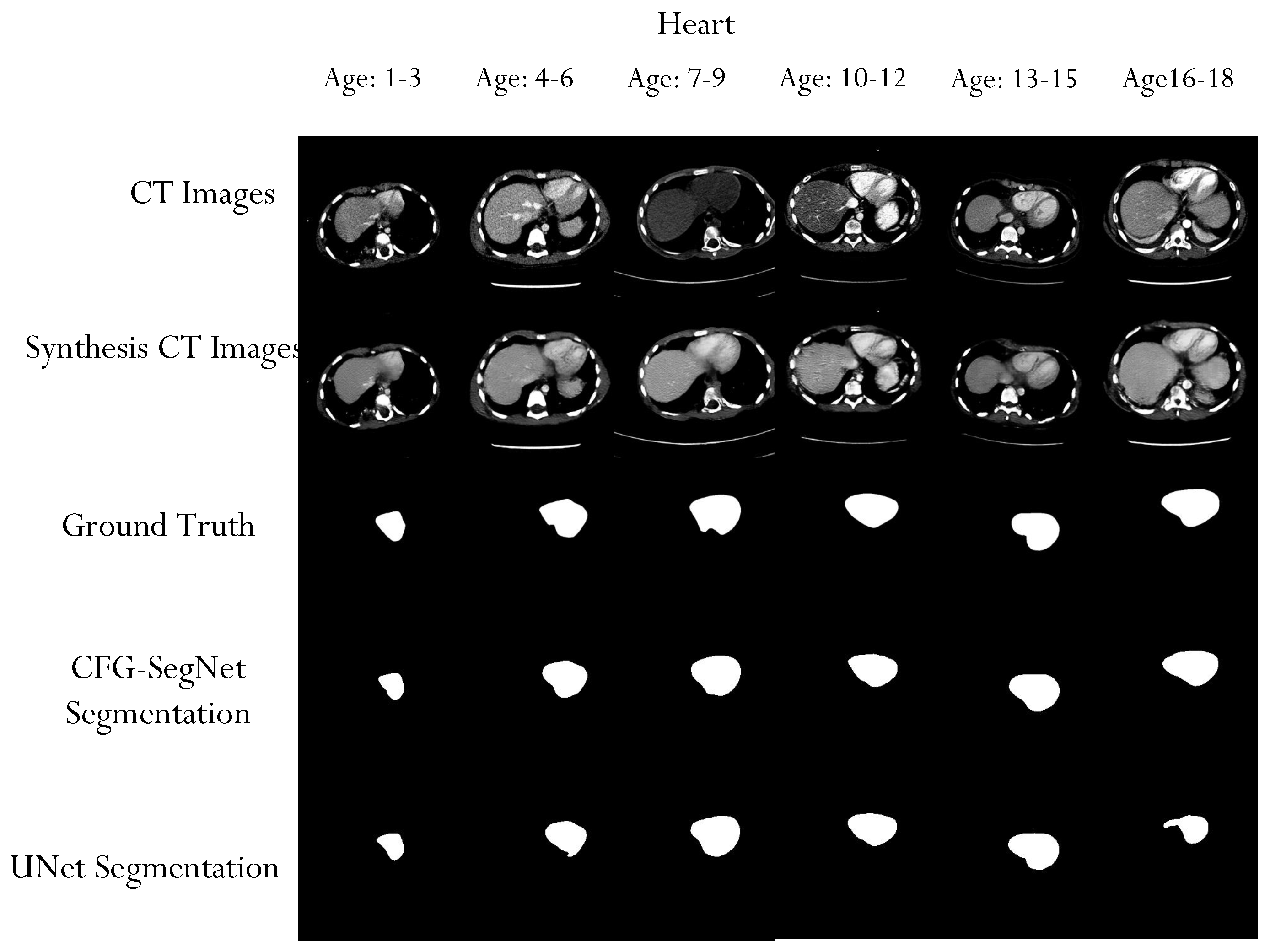
4.2. Segmentation Performance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ali, M.; Magee, D.; Dasgupta, U. Signal Processing Overview of Ultrasound Systems for Medical Imaging; SPRAB12; Texas Instruments: Dallas, TX, USA, 2008; Volume 55. [Google Scholar]
- Foomani, F.H.; Anisuzzaman, D.; Niezgoda, J.; Niezgoda, J.; Guns, W.; Gopalakrishnan, S.; Yu, Z. Synthesizing time-series wound prognosis factors from electronic medical records using generative adversarial networks. J. Biomed. Inform. 2022, 125, 103972. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Islam, M.T.; Siddique, B.N.K.; Rahman, S.; Jabid, T. Image recognition with deep learning. In Proceedings of the 2018 International Conference on Intelligent Informatics and Biomedical Sciences (ICIIBMS), Bangkok, Thailand, 21–24 October 2018; Volume 3, pp. 106–110. [Google Scholar]
- Malekzadeh, M.; Hajibabaee, P.; Heidari, M.; Zad, S.; Uzuner, O.; Jones, J.H. Review of Graph Neural Network in Text Classification. In Proceedings of the 2021 IEEE 12th Annual Ubiquitous Computing, Electronics Mobile Communication Conference (UEMCON), New York, NY, USA, 1–4 December 2021; pp. 0084–0091. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, N.; Das, I.; Maulik, U. Understanding deep learning techniques for image segmentation. ACM Comput. Surv. (CSUR) 2019, 52, 1–35. [Google Scholar] [CrossRef]
- Gheshlaghi, S.H.; Dehzangi, O.; Dabouei, A.; Amireskandari, A.; Rezai, A.; Nasrabadi, N.M. Efficient OCT Image Segmentation Using Neural Architecture Search. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Virtual Conference, 25–28 October 2020; pp. 428–432. [Google Scholar]
- Liu, X.; Li, K.W.; Yang, R.; Geng, L.S. Review of Deep Learning Based Automatic Segmentation for Lung Cancer Radiotherapy. Front. Oncol. 2021, 11, 2599. [Google Scholar] [CrossRef] [PubMed]
- Ayache, N.; Duncan, J. 20th anniversary of the medical image analysis journal (MedIA). Med. Image Anal. 2016, 33, 1–3. [Google Scholar]
- Liu, J.; Malekzadeh, M.; Mirian, N.; Song, T.A.; Liu, C.; Dutta, J. Artificial intelligence-based image enhancement in pet imaging: Noise reduction and resolution enhancement. PET Clin. 2021, 16, 553–576. [Google Scholar] [CrossRef] [PubMed]
- Gheshlaghi, S.H.; Kan, C.N.E.; Ye, D.H. Breast Cancer Histopathological Image Classification with Adversarial Image Synthesis. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine &Biology Society (EMBC), Virtual Meeting, 1–5 November 2021; IEEE: New York, NY, USA, 2021; pp. 3387–3390. [Google Scholar]
- Gadermayr, M.; Heckmann, L.; Li, K.; Bähr, F.; Müller, M.; Truhn, D.; Merhof, D.; Gess, B. Image-to-image translation for simplified MRI muscle segmentation. Front. Radiol 2021, 1, 664444. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.S. Patterns in paediatric CT use: An international and epidemiological perspective. J. Med. Imaging Radiat. Oncol. 2011, 55, 107–109. [Google Scholar]
- Rehani, M.M.; Berry, M. Radiation doses in computed tomography: The increasing doses of radiation need to be controlled. BMJ 2000, 320, 593–594. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, H.; Loew, M.; Ko, H. COVID-19 CT image synthesis with a conditional generative adversarial network. IEEE J. Biomed. Health Inform. 2020, 25, 441–452. [Google Scholar] [CrossRef]
- Li, L.; Wei, M.; Liu, B.; Atchaneeyasakul, K.; Zhou, F.; Pan, Z.; Kumar, S.A.; Zhang, J.Y.; Pu, Y.; Liebeskind, D.S.; et al. Deep learning for hemorrhagic lesion detection and segmentation on brain CT images. IEEE J. Biomed. Health Inform. 2020, 25, 1646–1659. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical image computing and computer-assisted intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; Proceedings, Part II 19. Springer: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar]
- Gu, Z.; Cheng, J.; Fu, H.; Zhou, K.; Hao, H.; Zhao, Y.; Zhang, T.; Gao, S.; Liu, J. Ce-net: Context encoder network for 2d medical image segmentation. IEEE Trans. Med. Imaging 2019, 38, 2281–2292. [Google Scholar] [CrossRef] [PubMed]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 fourth international conference on 3D vision (3DV), Stanford, CA, USA, 25–28 October 2016; IEEE: New York, NY, USA, 2016; pp. 565–571. [Google Scholar]
- Schmidt, T.G.; Wang, A.S.; Coradi, T.; Haas, B.; Star-Lack, J. Accuracy of patient-specific organ dose estimates obtained using an automated image segmentation algorithm. J. Med. Imaging 2016, 3, 043502. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jackson, P.; Hardcastle, N.; Dawe, N.; Kron, T.; Hofman, M.S.; Hicks, R.J. Deep learning renal segmentation for fully automated radiation dose estimation in unsealed source therapy. Front. Oncol. 2018, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Fang, Y.; Yang, X. Multi-organ Segmentation Network with Adversarial Performance Validator. arXiv 2022, arXiv:2204.07850. [Google Scholar]
- Okada, T.; Linguraru, M.G.; Hori, M.; Suzuki, Y.; Summers, R.M.; Tomiyama, N.; Sato, Y. Multi-organ segmentation in abdominal CT images. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; IEEE: New York, NY, USA, 2012; pp. 3986–3989. [Google Scholar]
- Tong, N.; Gou, S.; Niu, T.; Yang, S.; Sheng, K. Self-paced DenseNet with boundary constraint for automated multi-organ segmentation on abdominal CT images. Phys. Med. Biol. 2020, 65, 135011. [Google Scholar] [CrossRef] [PubMed]
- Balagopal, A.; Kazemifar, S.; Nguyen, D.; Lin, M.H.; Hannan, R.; Owrangi, A.; Jiang, S. Fully automated organ segmentation in male pelvic CT images. Phys. Med. Biol. 2018, 63, 245015. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic Multi-Organ Segmentation on Abdominal CT With Dense V-Networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef]
- Alsamadony, K.L.; Yildirim, E.U.; Glatz, G.; Bin Waheed, U.; Hanafy, S.M. Deep Learning Driven Noise Reduction for Reduced Flux Computed Tomography. Sensors 2021, 21, 1921. [Google Scholar] [CrossRef]
- Wang, J.; Perez, L. The effectiveness of data augmentation in image classification using deep learning. Convolutional Neural Netw. Vis. Recognit 2017, 11, 1–8. [Google Scholar]
- Nahian, S.; Paheding, S.; Colin, E.; Vijay, D. U-Net and its variants for medical image segmentation: Theory and applications. arXiv 2011, arXiv:2011.01118. [Google Scholar]
- Kan, C.N.E.; Gilat-Schmidt, T.; Ye, D.H. Enhancing reproductive organ segmentation in pediatric CT via adversarial learning. In Proceedings of the Medical Imaging 2021: Image Processing. International Society for Optics and Photonics, Online, 1 February 2021; Volume 11596, p. 1159612. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Advances in Neural Information Processing Systems. In Proceedings of the 28th Annual Conference on Neural Information Processing Systems 2014, Montreal, QC, Canada, 8–13 December 2014; Volume 27. [Google Scholar]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative adversarial networks: An overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Jordan, P.; Adamson, P.M.; Bhattbhatt, V.; Beriwal, S.; Shen, S.; Radermecker, O.; Bose, S.; Strain, L.S.; Offe, M.; Fraley, D.; et al. Pediatric chest-abdomen-pelvis and abdomen-pelvis CT images with expert organ contours. Med. Phys. 2022, 49, 3523–3528. [Google Scholar] [CrossRef] [PubMed]
- Adamson, P.M.; Bhattbhatt, V.; Principi, S.; Beriwal, S.; Strain, L.S.; Offe, M.; Wang, A.S.; Vo, N.J.; Gilat Schmidt, T.; Jordan, P. Evaluation of a V-Net autosegmentation algorithm for pediatric CT scans: Performance, generalizability, and application to patient-specific CT dosimetry. Med. Phys. 2022, 49, 2342–2354. [Google Scholar] [CrossRef]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 6023–6032. [Google Scholar]
- Qadri, S.F.; Ahmad, M.; Ai, D.; Yang, J.; Wang, Y. Deep belief network based vertebra segmentation for CT images. In Proceedings of the Image and Graphics Technologies and Applications: 13th Conference on Image and Graphics Technologies and Applications, IGTA 2018, Beijing, China, 8–10 April 2018; Revised Selected Papers 13. Springer: Berlin/Heidelberg, Germany, 2018; pp. 536–545. [Google Scholar]
- Ahmad, M.; Ai, D.; Xie, G.; Qadri, S.F.; Song, H.; Huang, Y.; Wang, Y.; Yang, J. Deep belief network modeling for automatic liver segmentation. IEEE Access 2019, 7, 20585–20595. [Google Scholar] [CrossRef]
- Chen, X.; Meng, Y.; Zhao, Y.; Williams, R.; Vallabhaneni, S.R.; Zheng, Y. Learning unsupervised parameter-specific affine transformation for medical images registration. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part IV 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 24–34. [Google Scholar]
- Strittmatter, A.; Schad, L.R.; Zöllner, F.G. Deep learning-based affine medical image registration for multimodal minimal-invasive image-guided interventions—A comparative study on generalizability. Z. FÜR Med. Phys. 2023; in press. [Google Scholar] [CrossRef]




| Unet | CFG-SegNet | ||
|---|---|---|---|
| Augmentation/Preprocessing | - | CutMix | Affine Transformations |
| Liver | |||
| Heart | |||
| Prostate | |||
| Uterus | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gheshlaghi, S.H.; Kan, C.N.E.; Schmidt, T.G.; Ye, D.H. Age Encoded Adversarial Learning for Pediatric CT Segmentation. Bioengineering 2024, 11, 319. https://doi.org/10.3390/bioengineering11040319
Gheshlaghi SH, Kan CNE, Schmidt TG, Ye DH. Age Encoded Adversarial Learning for Pediatric CT Segmentation. Bioengineering. 2024; 11(4):319. https://doi.org/10.3390/bioengineering11040319
Chicago/Turabian StyleGheshlaghi, Saba Heidari, Chi Nok Enoch Kan, Taly Gilat Schmidt, and Dong Hye Ye. 2024. "Age Encoded Adversarial Learning for Pediatric CT Segmentation" Bioengineering 11, no. 4: 319. https://doi.org/10.3390/bioengineering11040319
APA StyleGheshlaghi, S. H., Kan, C. N. E., Schmidt, T. G., & Ye, D. H. (2024). Age Encoded Adversarial Learning for Pediatric CT Segmentation. Bioengineering, 11(4), 319. https://doi.org/10.3390/bioengineering11040319






