1. Introduction
Chest X-ray (CXR) examination is a widely used examination in clinical diagnosis. However, training a qualified doctor who can understand X-ray images is expensive. In many developing countries, ensuring every local hospital has a doctor who can read and understand a medical X-ray imaging is difficult. Therefore, using artificial intelligence (AI) models [
1,
2,
3,
4] to simulate the knowledge of diagnosis experts and using them in local hospitals is a solution that has been considered. Ensuring the generality of AI models is the most crucial factor in completing this system.
An AI-based diagnosis system is considered a multi-label classification problem [
5,
6]. Initial research uses transfer learning to train a deep learning model for the classification task. This method involves reusing or adapting a model trained on one task to improve performance on a related task. Instead of starting from scratch, transfer learning leverages knowledge acquired from a source task to enhance learning on the target task. The knowledge from the source task is represented by parameters in the model, which is called a backbone. This approach is particularly beneficial when there are limited labeled data available for the target task or when training a model from scratch is computationally expensive. In chest X-ray (CXR) image analysis, popular backbone architectures used for transfer learning include AlexNet [
7], GoogLeNet [
8], DenseNet121 [
9], and ResNet [
10]. AlexNet, introduced in 2012, is a relatively simple network with five convolutional neural network (CNN) layers but a large number of parameters (60 million). GoogLeNet, introduced in 2014, employs inception blocks to capture multi-scale features and has a deeper architecture compared to AlexNet, with fewer parameters (6.7 million). ResNet utilizes residual learning, where each layer learns a residual mapping with respect to the input by incorporating skip connections, resulting in various depths, such as ResNet-18, ResNet-50, ResNet-101, and ResNet-152. DenseNet introduces dense connectivity patterns, where each layer is connected to every other layer within a dense block, facilitating feature reuse and information flow throughout the network. While the backbones work well in most vision-based applications, CXR image-based diagnosis has its own challenges that must be adequately addressed.
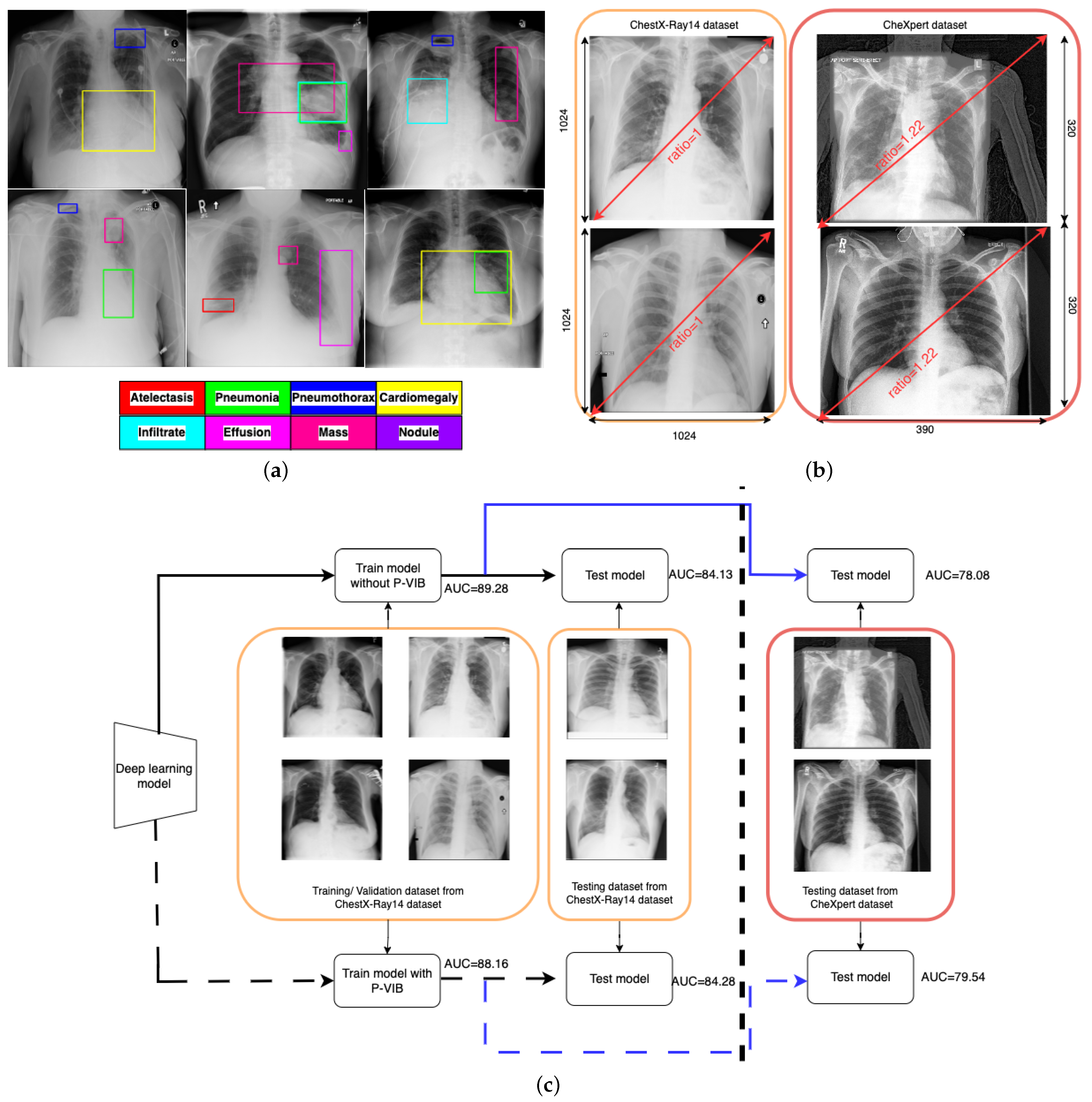
One major challenge of CXR image-based diagnosis is that multiple lesion areas exist in a CXR image, as shown in
Figure 1a. In the figure, each box represents a lesion area, and the color corresponds to its respective pathology. This phenomenon occurs because one latent reason may cause several pathologies. The pathology co-occurrence is considered an intrinsic correlation among multiple diseases, and the correlation is modeled by interaction among non-local regions [
11]. According to the review of Guo et al. [
12], the attention module is a successful solution to learning non-local features. The attention module is a mechanism that allows neural networks to focus on specific parts of the input data (such as words in a sentence or regions in an image) while performing a task. Inspired by the human visual system, the key idea behind the attention module is to assign different weights to different parts of the input data dynamically, allowing the model to attend to the most relevant information selectively. In thoracic disease classification [
13,
14,
15], attention modules help lesion areas in a chest X-ray image interact together and extract a better feature.
In addition to the attention mechanisms, advantage techniques [
15,
16] were used to enhance accuracy. Typically, an advanced classifier comprises three main components: the backbone, neck, and head. The backbone is responsible for extracting features from an input image, while the neck enhances these features, and the head makes the final classification prediction. In ConsultNet [
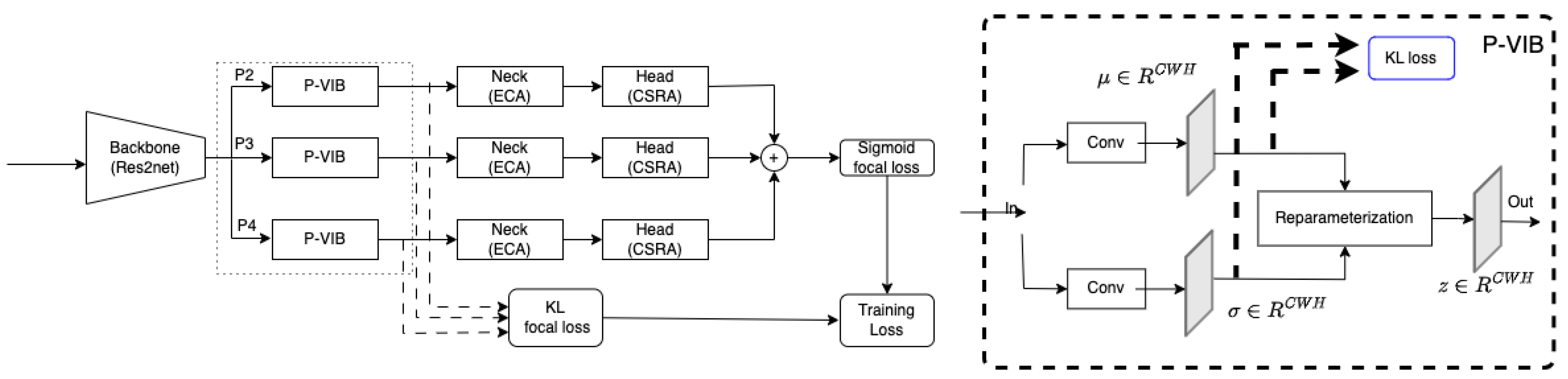
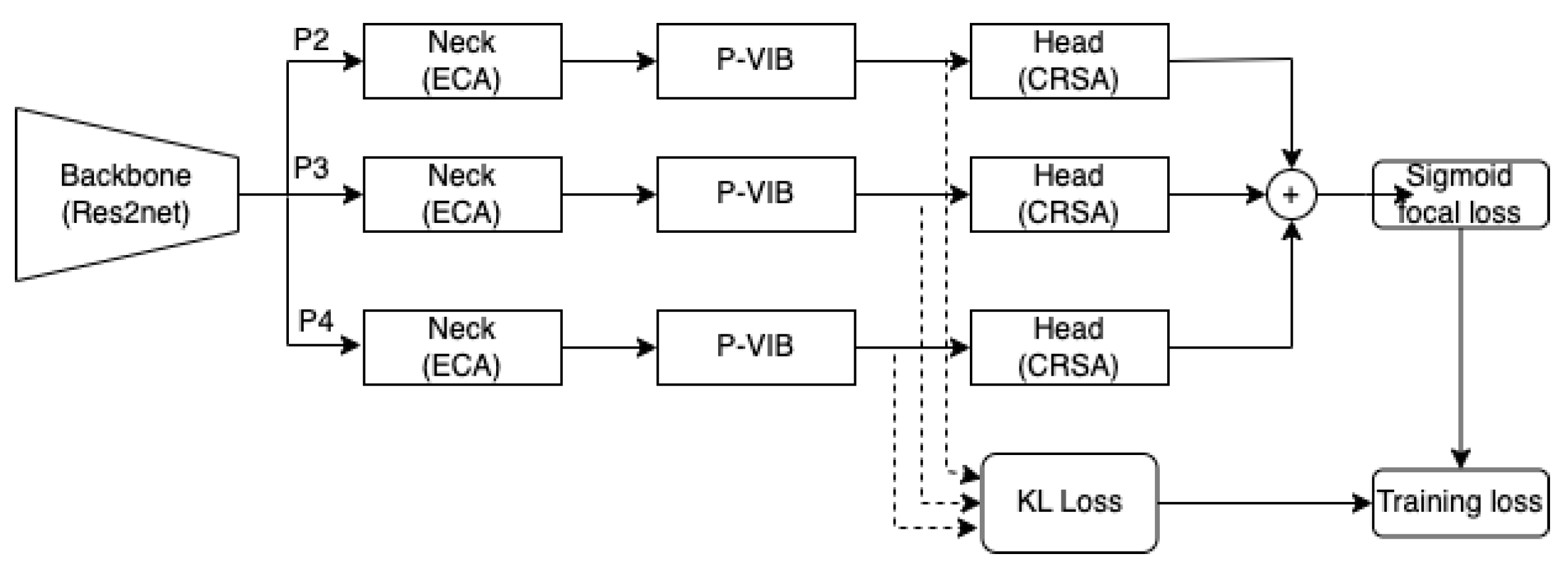
15], the backbone is DenseNet121, while the neck consists of a two-branch network comprising a feature selector and a feature integrator, and the head comprises a fully connected layer. Specifically, the feature integrator is an attention-based layer designed to capture non-local information, while the feature selector is a variational selective information bottleneck (VSIB) aimed at selecting crucial disease-specific features based on their importance. A vanilla variational information bottleneck (VIB) introduces a probabilistic model aimed at approximating the posterior distribution of latent variables given input data, thereby enhancing the model’s generalization capabilities on unseen data. The VSIB module in ConsultNet [
15] builds upon the conventional VIB but introduces a novel selection mechanism. This mechanism generates an importance matrix to specify the significance of each element of the input feature, eliminating the need to approximate the posterior distribution for these critical features. Recently, MLRFNet [
16] has greatly improved in CXR image-based diagnosis. This method utilizes Res2Net [
17] as the backbone, the ECA module [
18] as the neck, and the CSRA module [
19] as the head. The ECA module, functioning as an attention mechanism, focuses on capturing non-local features. Meanwhile, the CSRA module serves as the multi-label classification head. Additionally, MLRFNet employs a multi-scale approach to resolve classification challenges, thereby enhancing accuracy. While many promising results have been reported, deploying these models in the operation phase is still an open question. X-ray machines at different medical facilities will vary in calibration and operation. The domain gap can degrade the accuracy of the AI model.
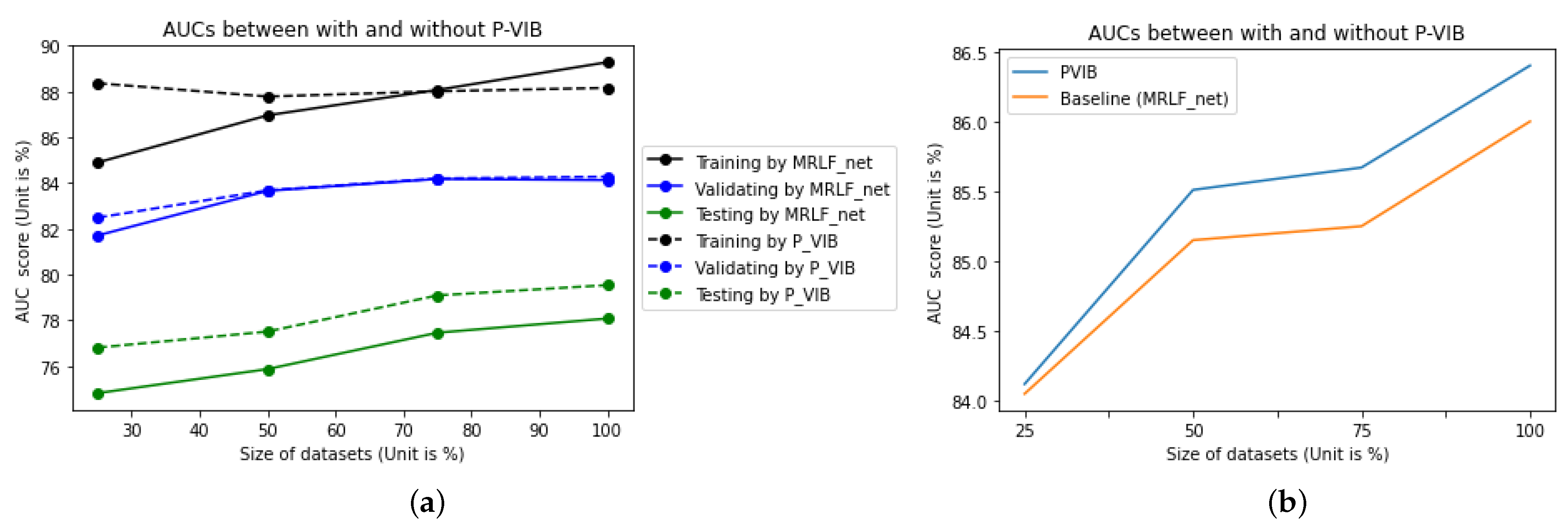
Figure 1b illustrates the domain gap between the ChestX-Ray14 [
5] and ChestExpert [
6] datasets. In the ChestX-Ray14 dataset, all images have a standardized resolution, with dimensions of 1024 rows and 1024 columns, resulting in a width-to-height ratio of 1.0. Conversely, most images in the ChestExpert dataset have dimensions of 320 rows and 390 columns, leading to a width-to-height ratio of 1.22. During regular training processes, input images are typically resized to specific ratios, such as
or
. This resizing approach may not significantly impact the ChestX-Ray14 data due to the consistent width-to-height ratio across both training and original images. However, it could introduce distortions into the ChestExpert dataset, where the original width-to-height ratio has been altered. Additionally, the position of lung regions differs between the two datasets. In the ChestX-Ray14 dataset, the lung region is consistently aligned at the center of the image. Conversely, in the ChestExpert dataset, the lung region is not consistently centered, resulting in some redundancies on the right side of the images. This analysis suggests that the ChestX-Ray14 dataset is better prepared and cleaner compared to the ChestExpert dataset.
In real-world applications, deep learning models are trained and validated using meticulously curated datasets. However, during operational deployment, testing images may differ substantially from those used in training. To simulate such scenarios, we utilize the ChestX-Ray14 dataset for training and validation while employing the ChestExpert dataset for testing purposes. As depicted by the solid line in
Figure 1c, the area under the curve (AUC) is 89.28% and 84.13% for the training and validation datasets, respectively. However, due to the domain gap illustrated in
Figure 1b, there is a significant reduction in AUC for the testing dataset. Following the blue solid line, the testing AUC is only 78.08%.
To improve accuracy in the operation phase, this paper proposes a patch-level feature selection technique. This method’s core idea is to use a variational information bottleneck (VIB) [
20] to select important features at a patch level. Given an input image with dimensions
, the backbone extracts features at dimensions
, where
and
are smaller than
W and
H, respectively. Hence, each position on the feature map captures information within a small
window. In conventional VIB, features are flattened before applying the re-parameterization trick. In this scenario, the feature selection is applied globally on the image. In our method, feature selection is involved at every position within a feature map in a
local region (or a patch-level). In addition, unlike the conventional VIB approach [
20] or ConsultNet [
15], where VIB-based features serve as the input for a classification head, our method can extract VIB-based features at any position in a CNN network, not just at the classification head. Instead of flattening all features and using the VIB method [
20] to sample a new latent feature, we apply a convolution block to learn sampling parameters for every position in a feature map. This design allows only important pixels to respond on a feature map, and the model will select important patches in an image. In addition, the innovative design enables seamless integration of the P-VIB module into various CNN-based networks at any position. Consequently, we incorporate it into MLRFNet [
16] to enhance accuracy. With the help of the proposed module, our method can work better in various scenarios. In detail, implementing the P-VIB module leads to an improvement in the testing AUC to 79.54%, while the evaluation AUC remains stable at 84.28%. This outcome is depicted by the dashed line in
Figure 1c.
In summary, the main contributions of this article are as follows:
We made a connection between VIB and MLRFNet and proposed a patch-level feature selection to enhance disease diagnosis models based on MLRFNet [
16] architecture.
In the standard scenarios (training and testing datasets are from the same dataset) as defined by Wang et al. [
5], the proposed method showed an improvement. If the training dataset and testing dataset have some domain gap, the proposed method avoids overfitting on the training dataset and improves the performance of the testing dataset.
Unlike conventional VIB [
15,
20], which is usually used for the input of a classification head, the proposed P-VIB could be used at any position in a network and can be successfully integrated with many different lung disease classification methods.
4. Dataset and Experiment Setting
ChestX-Ray14 [
5] is a well-known dataset for thoracic disease classification. It comprises 112,120 frontal-view X-ray images of 30,805 (collected from the years 1992 to 2015) unique patients with 14 common disease labels, text-mined from the radiological reports via NLP techniques. It expands on ChestX-Ray8 by adding six additional thorax diseases: edema, emphysema, fibrosis, pleural thinning, and hernia. If one image has no pathologies, it is considered a No Finding. In addition, each image is assigned one or more of the 14 pathologies, and 880 images have been annotated with 984 labeled bounding boxes for 8 pathologies. The training dataset, testing dataset, and evaluation dataset have been defined in Wang et al. [
5], which make it a standard setting used by many research works. Detail of the pathologies and their abbreviations are shown in
Table 1.
Motivated by the observation, this work also uses the ChestX-Ray14 [
5] dataset to evaluate the proposed P-VIB network. We evaluate performance on all 14 labels. For fairness, the dataset split in the comparative experiments strictly follows the official splitting standards of the dataset published by Wang et al. [
5].
To evaluate the generalization of the model, which is trained by ChestX-Ray14 [
5], we also use the testing dataset from another dataset. For this paper, CheXpert [
6] was selected for the evaluation. The dataset contains 224,316 X-ray scans of 65,240 patients, with 14 observations extracted from the medical reports. Each observation is assigned a positive label in our experiments. Because the 14 pathologies in ChestX-Ray14 [
5] are not the same as the 14 pathologies in CheXpert [
6] dataset, only 5 observations are selected for examination. These observations are Atelectasis, Cardiomegaly, Consolidation, Edema, and Pleural Effusion, as recommended by Irvin et al. [
6].
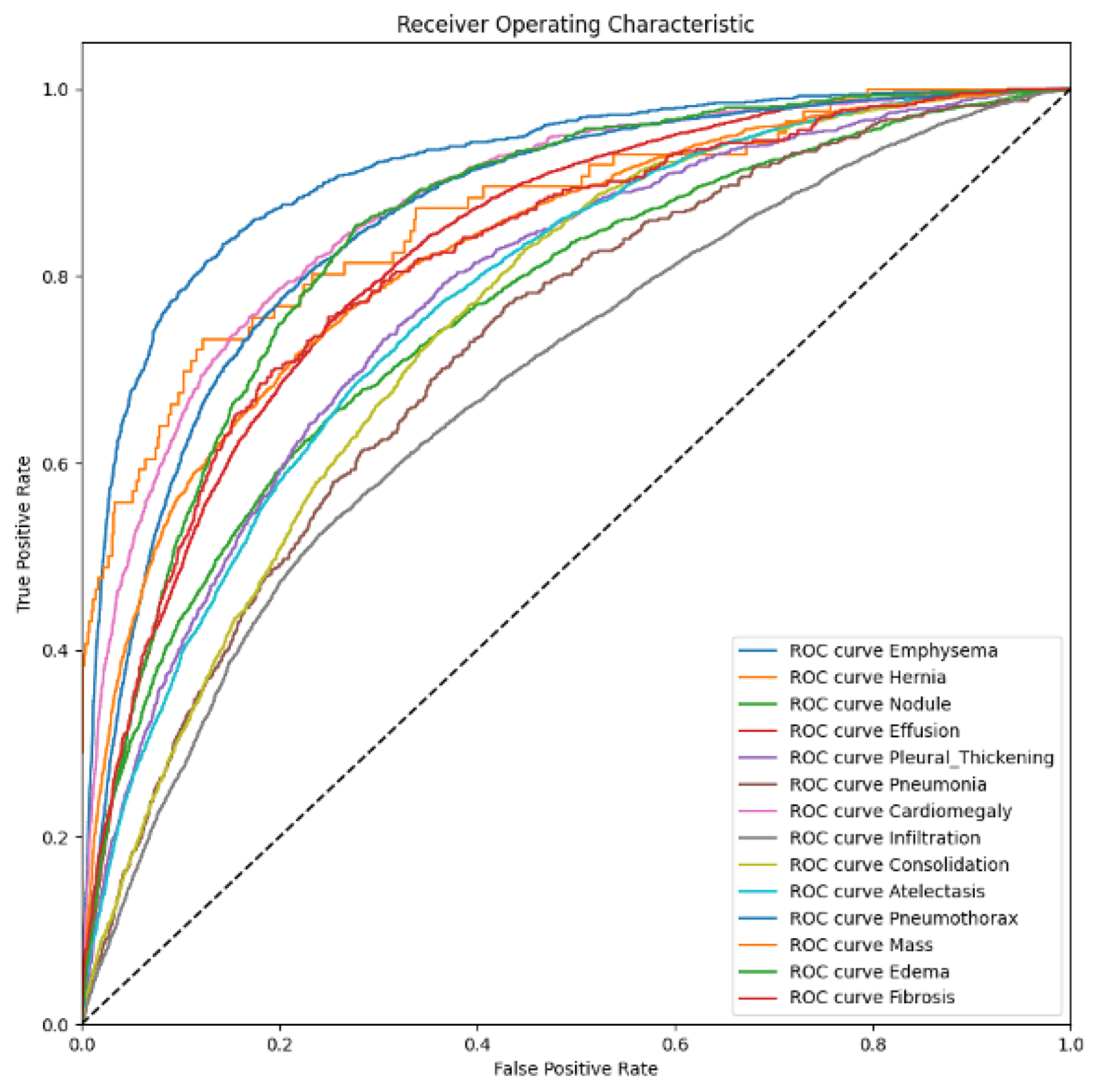
A thoracic disease diagnosis model is evaluated by the area under the curve (AUC) of the receiver operating characteristic (ROC). This curve is a set of (sensitive and specificity) pairs, and the area under the curve is better than accuracy in terms of evaluating the confidence of a classification task. A smaller AUC represents the performance of a random classifier, and a greater AUC would correspond to a perfect classifier (e.g., with a classification error rate equivalent to zero). Originally, the AUC-ROC curve was only for binary classification problems. However, it can be extended to multi-class classification problems using the One vs. All technique. The average of AUC represents a unique evaluation metric in a multi-label classification setting.
We follow the hyperparameter setting in MRLFNet [
16] to train the model. Details of the hyperparameter settings can be found in
Table 2. However, our patch size is set as 64 due to the availability of hardware.
In all experiments, early stopping is used to select the best model. This technique applies an evaluation process that estimates the AUC metric on the evaluation dataset for every training epoch. The highest AUC is stored during the training phase. If the AUC in a current epoch is higher than the recent highest AUC, the model at the current epoch is considered the best model, and the current AUC updates the highest AUC. Because the model is trained to fit with the training dataset, the training loss and training AUC will be better at later epochs. Hence, an overfitting may occur at later epochs when the model is trained too long. Applying early stopping allows us to select the best model for the evaluation dataset instead of the last model that may be overfitted to training data.