Feature Extraction Based on Local Histogram with Unequal Bins and a Recurrent Neural Network for the Diagnosis of Kidney Diseases from CT Images
Abstract
1. Introduction
- The use of gradients and histograms with asymmetric intervals to extract features to classify kidney cancer subtypes accurately;
- The use of feature extraction before applying the DL model to reduce the dimensionality of the input data compared to the conventional methods;
- The reduction in the dimensions of the input data to increase the training speed and reduce the complexity of the used DL model.
2. Literature Review
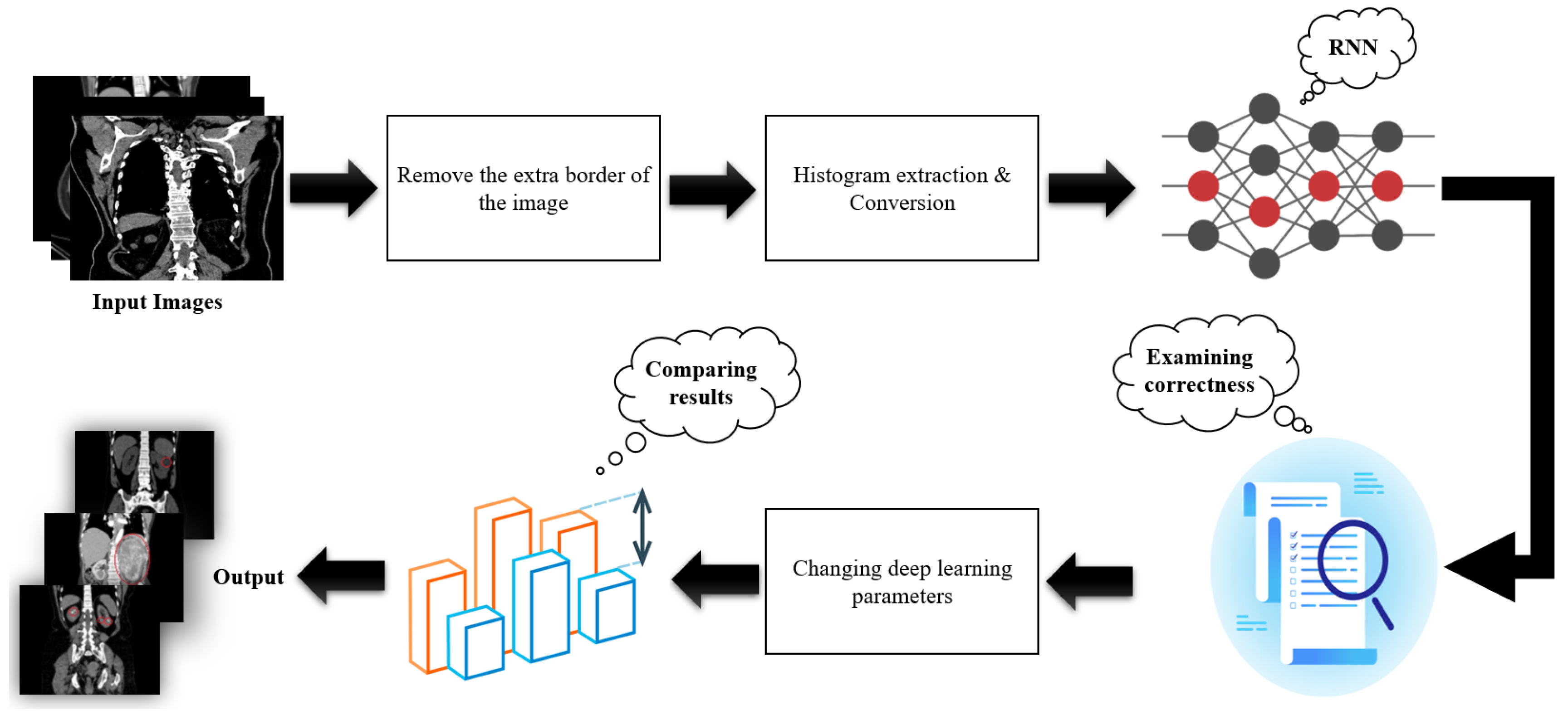
3. Materials and Methods
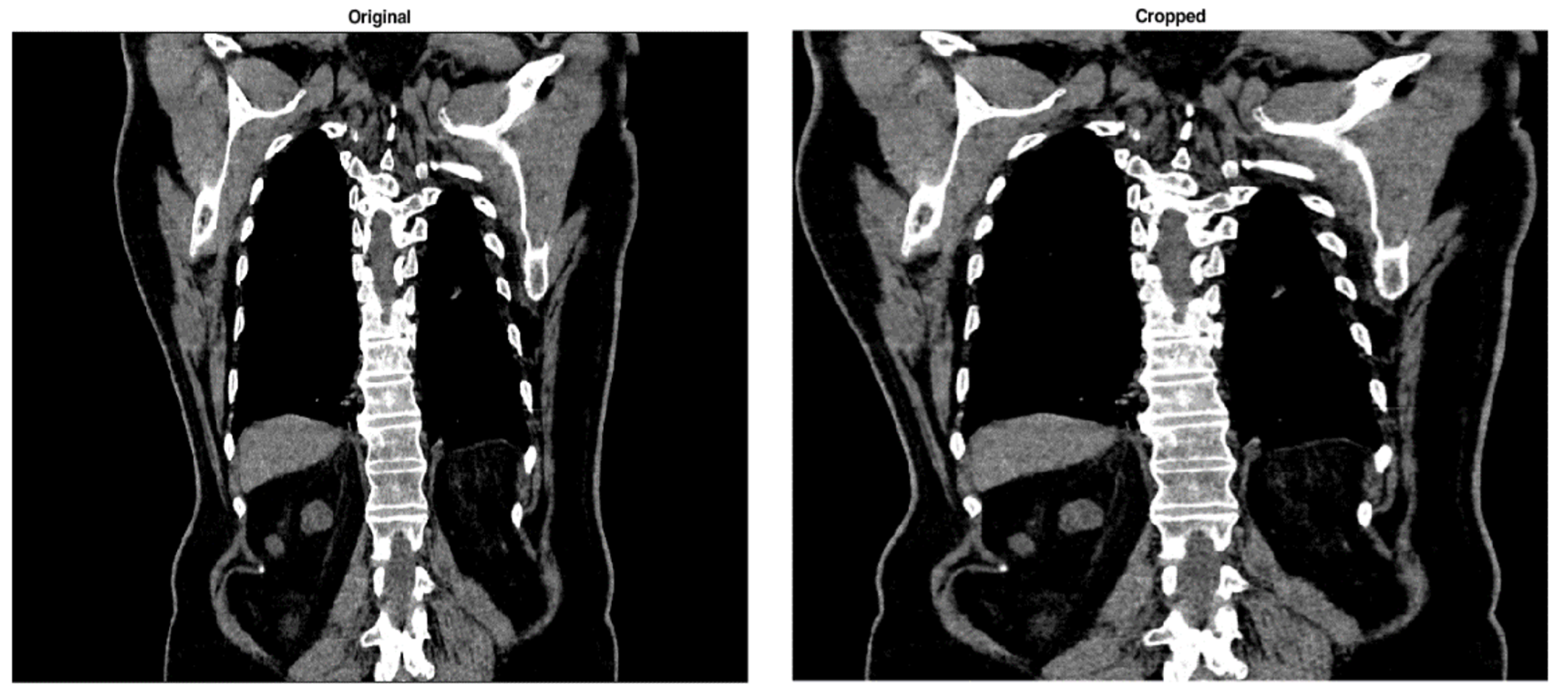
3.1. Image Black Border Removal
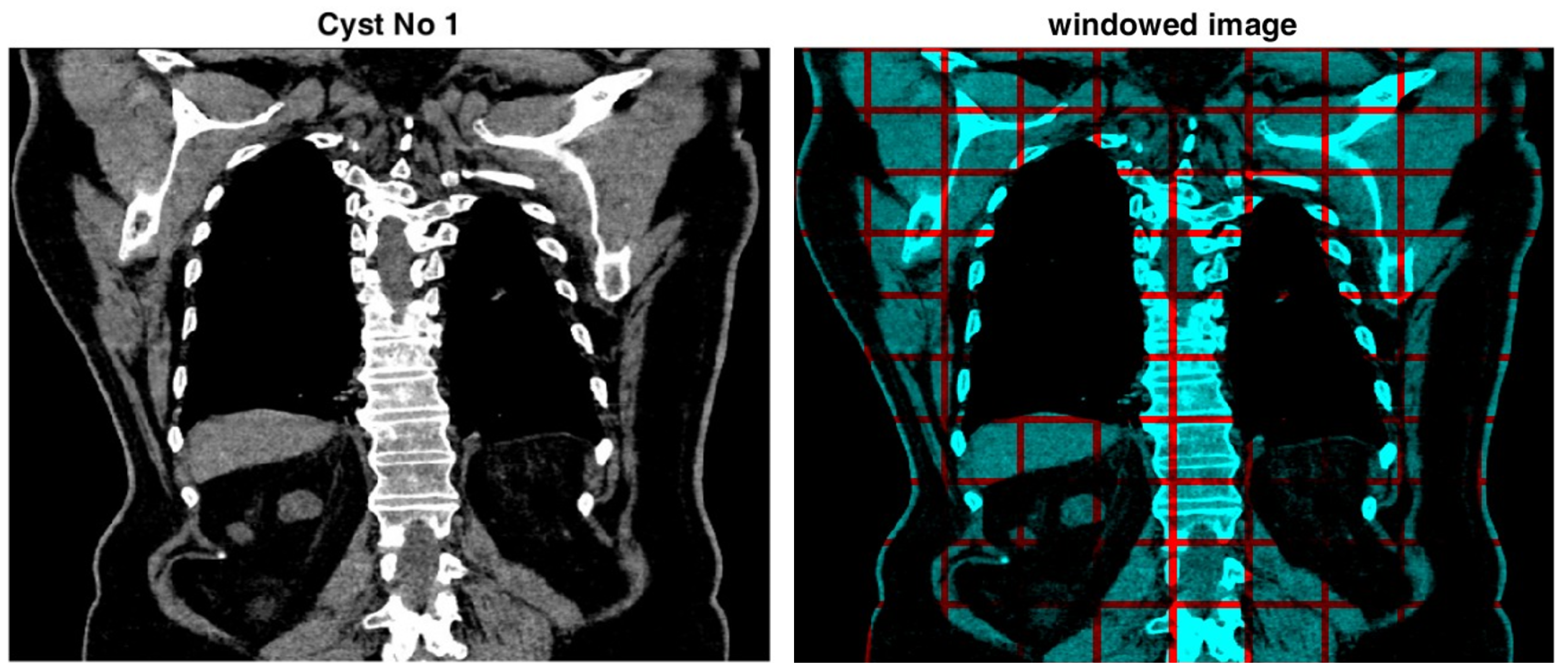
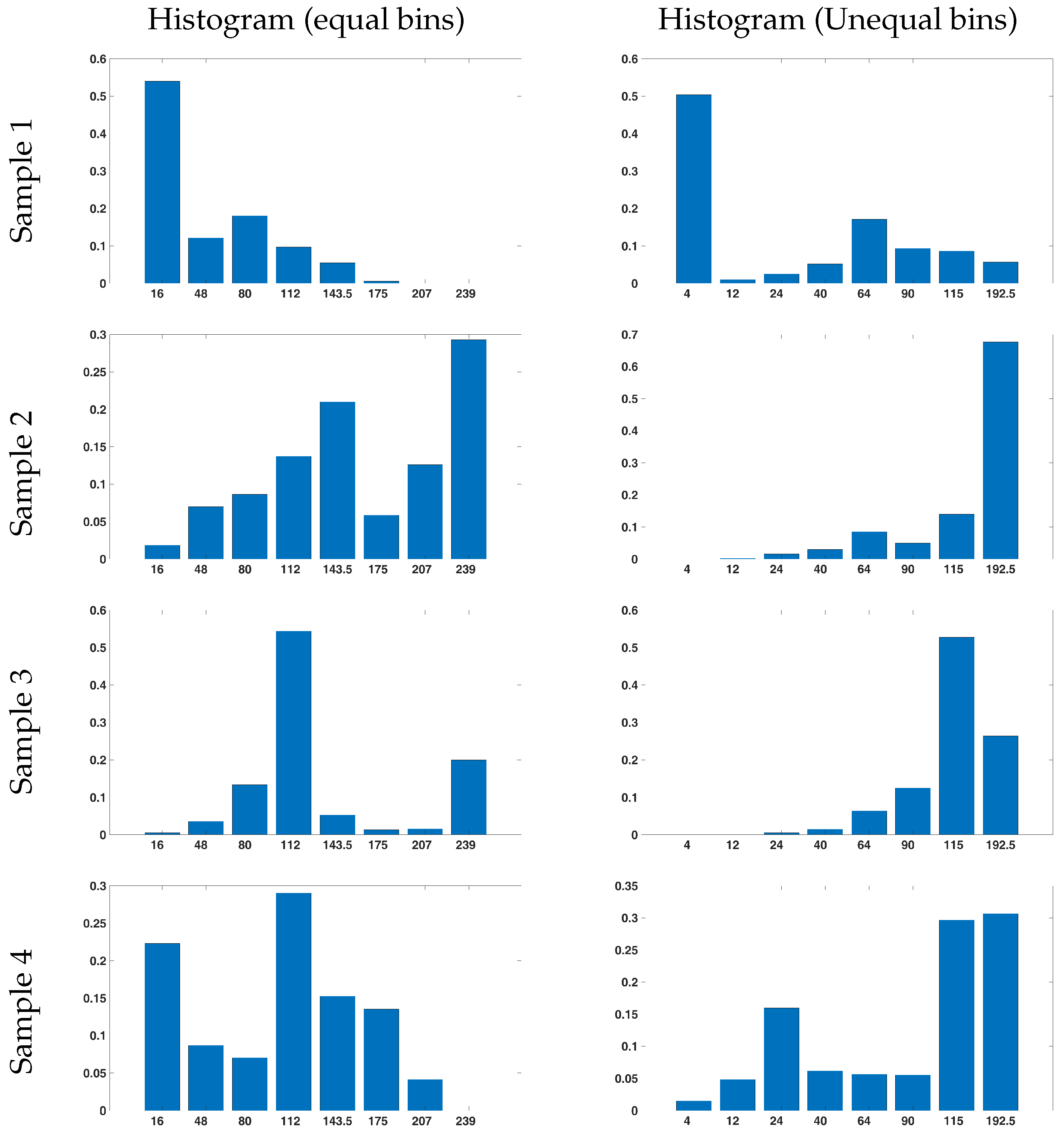
3.2. Local Histogram with Asymmetric Intervals
3.3. Image Gradient Histogram with Asymmetric Intervals
3.4. Recurrent Neural Network
4. Results and Discussion
4.1. Evaluation Metrics
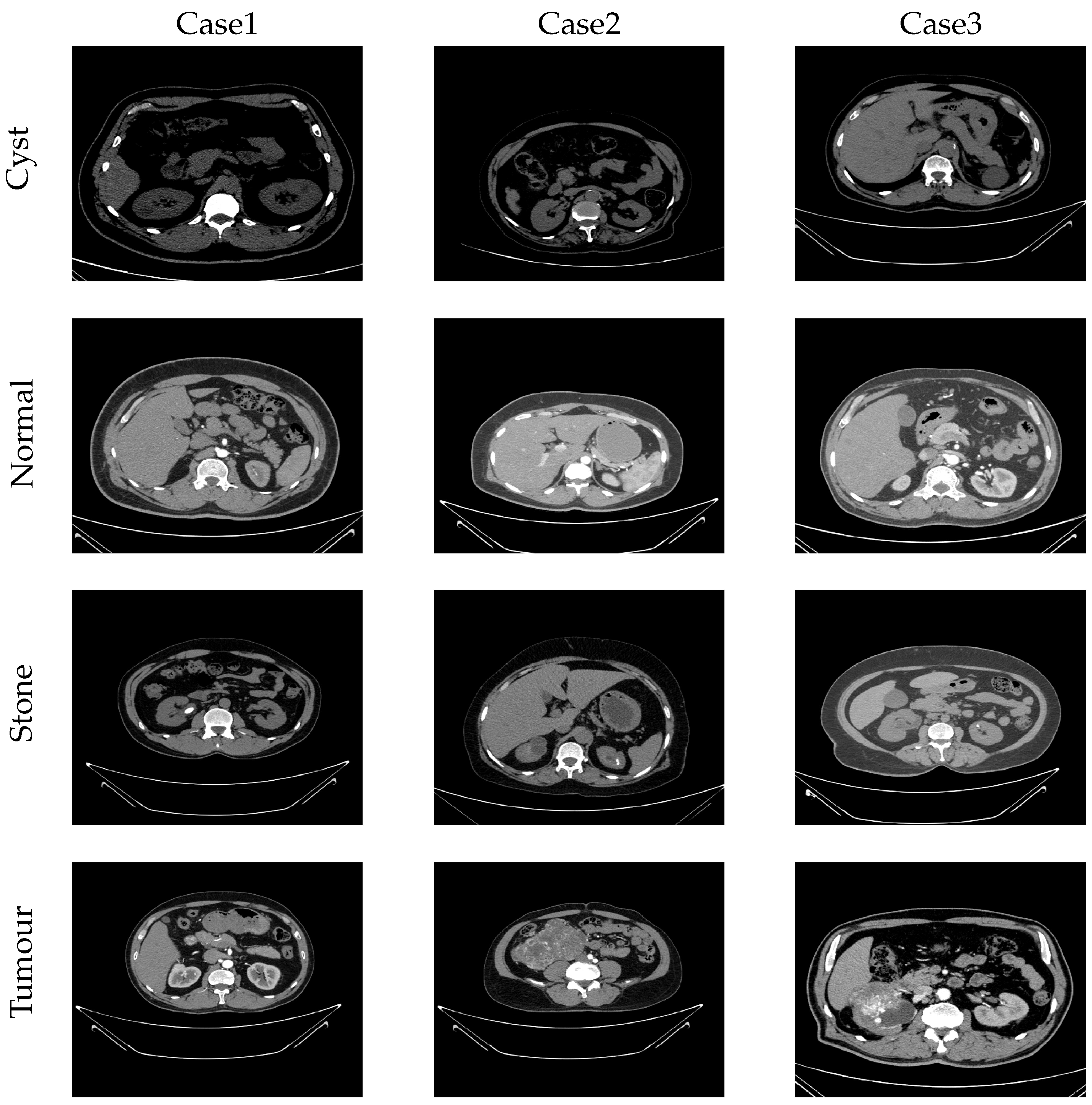
4.2. Used Dataset
4.3. Experimental Results
4.3.1. Selection of Asymmetric Intervals of the Intensity Histogram
4.3.2. Selection of Asymmetric Intervals for the Gradient Histogram
4.4. Experimental Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fund, W.C.R. Kidney Cancer Statistics. 2019. Available online: https://www.wcrf.org/dietandcancer/cancer-trends/kidney-cancer-statistics (accessed on 27 November 2019).
- Society, A.C. What Is Kidney Cancer? 2017. Available online: https://www.cancer.org/cancer/kidney-cancer/about/what-is-kidney-cancer (accessed on 27 November 2019).
- Rossi, S.H.; Klatte, T.; Usher-Smith, J.; Stewart, G.D. Epidemiology and screening for renal cancer. World J. Urol. 2018, 36, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Znaor, A.; Lortet-Tieulent, J.; Laversanne, M.; Jemal, A.; Bray, F. International variations and trends in renal cell carcinoma incidence and mortality. Eur. Urol. 2015, 67, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Shuch, B.; Amin, A.; Armstrong, A.J.; Eble, J.N.; Ficarra, V.; Lopez-Beltran, A.; Martignoni, G.; Rini, B.I.; Kutikov, A. Understanding pathologic variants of renal cell carcinoma: Distilling therapeutic opportunities from biologic complexity. Eur. Urol. 2015, 67, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed]
- Islam, M. Ct KIDNEY DATASET: Normal-Cyst-Tumor and Stone. Kaggle.com. 2021. Available online: https://www.kaggle.com/datasets/nazmul0087/ct-kidney-dataset-normal-cyst-tumor-and-stone/code (accessed on 18 February 2024).
- Bergeron, A. The pulmonologist’s point of view on lung infiltrates in haematological malignancies. Diagn. Interv. Imaging 2013, 94, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Candemir, S.; Jaeger, S.; Palaniappan, K.; Musco, J.P.; Singh, R.K.; Xue, Z.; Karargyris, A.; Antani, S.; Thoma, G.; McDonald, C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 2013, 33, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Nath, M.; Tripathi, P.; Saini, K. Analysis and identification of kidney stone using K th nearest neighbour (KNN) and support vector machine (SVM) classification techniques. Pattern Recognit. Image Anal. 2017, 27, 574–580. [Google Scholar] [CrossRef]
- Khalifa, F.; Gimel’farb, G.; El-Ghar, M.A.; Sokhadze, G.; Manning, S.; McClure, P.; Ouseph, R.; El-Baz, A. A new deformable model-based segmentation approach for accurate extraction of the kidney from abdominal CT images. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; pp. 3393–3396. [Google Scholar] [CrossRef]
- Wolz, R.; Chu, C.; Misawa, K.; Fujiwara, M.; Mori, K.; Rueckert, D. Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans. Med. Imaging 2013, 32, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Gu, J.; Chen, Y.; Liu, W.; Tang, L.; Shu, H.; Toumoulin, C. Automatic kidney segmentation in CT images based on multi-atlas image registration. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 5538–5541. [Google Scholar] [CrossRef]
- Zhao, E.; Liang, Y.; Fan, H. Contextual information-aided kidney segmentation in CT sequences. Opt. Commun. 2013, 290, 55–62. [Google Scholar] [CrossRef]
- Shehata, M.; Khalifa, F.; Soliman, A.; Alrefai, R.; Abou El-Ghar, M.; Dwyer, A.C.; Ouseph, R.; El-Baz, A. A level set-based framework for 3D kidney segmentation from diffusion MR images. In Proceedings of the 2015 IEEE International Conference on Image Processing (ICIP), Quebec City, QC, Canada, 27–30 September 2015; pp. 4441–4445. [Google Scholar] [CrossRef]
- Khalifa, F.; Soliman, A.; Takieldeen, A.; Shehata, M.; Mostapha, M.; Shaffie, A.; Ouseph, R.; Elmaghraby, A.; El-Baz, A. Kidney segmentation from CT images using a 3D NMF-guided active contour model. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 432–435. [Google Scholar] [CrossRef]
- Skalski, A.; Heryan, K.; Jakubowski, J.; Drewniak, T. Kidney segmentation in ct data using hybrid level-set method with ellipsoidal shape constraints. Metrol. Meas. Syst. 2017, 24, 101–112. [Google Scholar] [CrossRef]
- Aksakalli, I.; Kaçdioğlu, S.; Hanay, Y.S. Kidney X-ray images classification using machine learning and deep learning methods. Balk. J. Electr. Comput. Eng. 2021, 9, 144–151. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning (PMLR), Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Kolesnikov, A.; Beyer, L.; Zhai, X.; Puigcerver, J.; Yung, J.; Gelly, S.; Houlsby, N. Big transfer (bit): General visual representation learning. In Proceedings of the Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020, Proceedings, Part V 16; Springer: Berlin/Heidelberg, Germany, 2020; pp. 491–507. [Google Scholar] [CrossRef]
- Guo, M.H.; Liu, Z.N.; Mu, T.J.; Hu, S.M. Beyond self-attention: External attention using two linear layers for visual tasks. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 5436–5447. [Google Scholar] [CrossRef]
- Hassani, A.; Walton, S.; Shah, N.; Abuduweili, A.; Li, J.; Shi, H. Escaping the big data paradigm with compact transformers. arXiv 2021, arXiv:2104.05704. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Yang, G.; Li, G.; Pan, T.; Kong, Y.; Wu, J.; Shu, H.; Luo, L.; Dillenseger, J.L.; Coatrieux, J.L.; Tang, L.; et al. Automatic segmentation of kidney and renal tumor in ct images based on 3d fully convolutional neural network with pyramid pooling module. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3790–3795. [Google Scholar] [CrossRef]
- Haghighi, M.; Warfield, S.K.; Kurugol, S. Automatic renal segmentation in DCE-MRI using convolutional neural networks. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1534–1537. [Google Scholar] [CrossRef]
- Mehta, P.; Sandfort, V.; Gheysens, D.; Braeckevelt, G.J.; Berte, J.; Summers, R.M. Segmenting the kidney on ct scans via crowdsourcing. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 829–832. [Google Scholar] [CrossRef]
- Fu, X.; Liu, H.; Bi, X.; Gong, X. Deep-learning-based CT imaging in the quantitative evaluation of chronic kidney diseases. J. Healthc. Eng. 2021, 2021, 3774423. [Google Scholar] [CrossRef]
- da Cruz, L.B.; Araújo, J.D.L.; Ferreira, J.L.; Diniz, J.O.B.; Silva, A.C.; de Almeida, J.D.S.; de Paiva, A.C.; Gattass, M. Kidney segmentation from computed tomography images using deep neural network. Comput. Biol. Med. 2020, 123, 103906. [Google Scholar] [CrossRef] [PubMed]
- Sudharson, S.; Kokil, P. An ensemble of deep neural networks for kidney ultrasound image classification. Comput. Methods Programs Biomed. 2020, 197, 105709. [Google Scholar] [CrossRef]
- Islam, M.N.; Hasan, M.; Hossain, M.K.; Alam, M.G.R.; Uddin, M.Z.; Soylu, A. Vision transformer and explainable transfer learning models for auto detection of kidney cyst, stone and tumor from CT-radiography. Sci. Rep. 2022, 12, 11440. [Google Scholar] [CrossRef] [PubMed]
- Bayram, A.F.; Gurkan, C.; Budak, A.; Karataş, H. A detection and prediction model based on deep learning assisted by explainable artificial intelligence for kidney diseases. Avrupa Bilim Teknol. Derg. 2022, 40, 67–74. [Google Scholar] [CrossRef]
- Asif, S.; Yi, W.; Si, J.; Ain, Q.U.; Yi, Y.; Hou, J. Modeling a Fine-Tuned Deep Convolutional Neural Network for Diagnosis of Kidney Diseases from CT Images. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; pp. 2571–2576. [Google Scholar] [CrossRef]
- Bhandari, M.; Yogarajah, P.; Kavitha, M.S.; Condell, J. Exploring the Capabilities of a Lightweight CNN Model in Accurately Identifying Renal Abnormalities: Cysts, Stones, and Tumors, Using LIME and SHAP. Appl. Sci. 2023, 13, 3125. [Google Scholar] [CrossRef]



| Total | Train | Test | |
|---|---|---|---|
| Cyst | 3709 | 1000 | 2709 |
| Normal | 5077 | 1000 | 4077 |
| Stone | 1377 | 1000 | 377 |
| Tumour | 2283 | 1000 | 1283 |
| Total | 12,446 | 4000 | 8446 |
| Symmetric Interval | 0 | 32 | 64 | 96 | 128 | 159 | 191 | 223 | 255 |
| Suggested Range | 0 | 8 | 16 | 32 | 48 | 80 | 100 | 130 | 255 |
| Symmetric Interval | 0 | 32 | 64 | 96 | 128 | 159 | 191 | 223 | 255 |
| Suggested Range | 0 | 4 | 8 | 12 | 16 | 24 | 48 | 96 | 255 |
| Train | Test | |||||||
|---|---|---|---|---|---|---|---|---|
| Cyst | Normal | Stone | Tumour | Cyst | Normal | Stone | Tumour | |
| Cyst | 1000 | 0 | 0 | 0 | 2709 | 0 | 0 | 0 |
| Normal | 2 | 998 | 0 | 0 | 3 | 4074 | 0 | 0 |
| Stone | 0 | 2 | 998 | 0 | 0 | 9 | 368 | 0 |
| Tumour | 0 | 0 | 7 | 993 | 0 | 0 | 13 | 1271 |
| Train | Test | |||||||
|---|---|---|---|---|---|---|---|---|
| Cyst | Normal | Stone | Tumour | Cyst | Normal | Stone | Tumour | |
| Cyst | 1000 | 0 | 0 | 0 | 2707 | 0 | 0 | 2 |
| Normal | 12 | 945 | 0 | 43 | 12 | 4013 | 19 | 33 |
| Stone | 0 | 9 | 991 | 0 | 0 | 17 | 360 | 0 |
| Tumour | 0 | 0 | 8 | 992 | 0 | 0 | 12 | 1271 |
| Train | Test | |||||||
|---|---|---|---|---|---|---|---|---|
| Cyst | Normal | Stone | Tumour | Cyst | Normal | Stone | Tumour | |
| Cyst | 1000 | 0 | 0 | 0 | 2709 | 0 | 0 | 0 |
| Normal | 2 | 998 | 0 | 0 | 2 | 4074 | 1 | 0 |
| Stone | 0 | 3 | 997 | 0 | 0 | 1 | 360 | 0 |
| Tumour | 0 | 0 | 3 | 997 | 0 | 0 | 5 | 1278 |
| Train | Test | |||||||
|---|---|---|---|---|---|---|---|---|
| Cyst | Normal | Stone | Tumour | Cyst | Normal | Stone | Tumour | |
| Cyst | 808 | 0 | 192 | 0 | 2131 | 0 | 578 | 0 |
| Normal | 1 | 989 | 10 | 0 | 0 | 4066 | 11 | 0 |
| Stone | 2 | 13 | 985 | 0 | 0 | 6 | 371 | 0 |
| Tumour | 9 | 1 | 22 | 968 | 22 | 15 | 59 | 1187 |
| Model | Accuracy | Class | Precision | Recall | F1 Score | MCC |
|---|---|---|---|---|---|---|
| YOLOv7 [35] | — | Cyst | 0.892 | 0.633 | 0.74 | 0.673 |
| Normal | — | — | — | — | ||
| Stone | 0.819 | 0.855 | 0.836 | 0.816 | ||
| Tumour | 0.936 | 1 | 0.966 | 0.960 | ||
| Average | 0.882 | 0.829 | 0.854 | 0.648 | ||
| EANet [34] | 77.02% | Cyst | 0.593 | 1 | 0.745 | 0.788 |
| Normal | 0.896 | 0.848 | 0.871 | 0.616 | ||
| Stone | 0.845 | 0.495 | 0.624 | 0.821 | ||
| Tumour | 0.93 | 0.777 | 0.847 | 0.994 | ||
| Swin Transformer [34] | 99.30% | Cyst | 0.996 | 0.996 | 0.996 | 0.981 |
| Normal | 0.996 | 0.981 | 0.988 | 0.983 | ||
| Stone | 0.981 | 0.989 | 0.985 | 0.996 | ||
| Tumour | 0.993 | 1 | 0.996 | 0.923 | ||
| CCT [34] | 96.54% | Cyst | 0.968 | 0.923 | 0.945 | 0.970 |
| Normal | 0.989 | 0.975 | 0.982 | 0.966 | ||
| Stone | 0.94 | 1 | 0.969 | 0.956 | ||
| Tumour | 0.964 | 0.964 | 0.964 | 0.974 | ||
| VGG16 [34] | 98.20% | Cyst | 0.996 | 0.968 | 0.982 | 0.965 |
| Normal | 0.985 | 0.973 | 0.979 | 0.974 | ||
| Stone | 0.966 | 0.988 | 0.977 | 0.986 | ||
| Tumour | 0.982 | 0.996 | 0.989 | 0.596 | ||
| Inception v3 [34] | 61.60% | Cyst | 0.645 | 0.826 | 0.724 | 0.465 |
| Normal | 0.584 | 0.898 | 0.708 | 0.459 | ||
| Stone | 0.568 | 0.462 | 0.509 | 0.412 | ||
| Tumour | 0.76 | 0.295 | 0.425 | 0.566 | ||
| Resnet50 [34] | 73.80% | Cyst | 0.735 | 0.641 | 0.685 | 0.625 |
| Normal | 0.77 | 0.79 | 0.78 | 0.684 | ||
| Stone | 0.745 | 0.692 | 0.717 | 0.706 | ||
| Tumour | 0.706 | 0.827 | 0.762 | 0.673 | ||
| Deep CNN [36] | 99.25% | Cyst | 0.97 | 1 | 0.98 | 1 |
| Normal | 1 | 1 | 1 | 0.994 | ||
| Stone | 1 | 0.99 | 1 | 0.988 | ||
| Tumour | 1 | 0.98 | 0.99 | 1 | ||
| Lightweight CNN [37] | 99.52% | Cyst | 0.994 | 0.999 | 0.998 | 0.995 |
| Normal | 0.995 | 0.997 | 0.997 | 0.993 | ||
| Stone | 0.997 | 0.979 | 0.988 | 0.986 | ||
| Tumour | 0.993 | 0.995 | 0.995 | 0.993 | ||
| Proposed method | 99.89% | Cyst | 0.999 | 1 | 1 | 0.999 |
| Normal | 1 | 0.999 | 1 | 0.999 | ||
| Stone | 0.984 | 0.997 | 0.991 | 0.990 | ||
| Tumour | 1 | 0.996 | 0.998 | 0.998 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gharahbagh, A.A.; Hajihashemi, V.; Machado, J.J.M.; Tavares, J.M.R.S. Feature Extraction Based on Local Histogram with Unequal Bins and a Recurrent Neural Network for the Diagnosis of Kidney Diseases from CT Images. Bioengineering 2024, 11, 220. https://doi.org/10.3390/bioengineering11030220
Gharahbagh AA, Hajihashemi V, Machado JJM, Tavares JMRS. Feature Extraction Based on Local Histogram with Unequal Bins and a Recurrent Neural Network for the Diagnosis of Kidney Diseases from CT Images. Bioengineering. 2024; 11(3):220. https://doi.org/10.3390/bioengineering11030220
Chicago/Turabian StyleGharahbagh, Abdorreza Alavi, Vahid Hajihashemi, José J. M. Machado, and João Manuel R. S. Tavares. 2024. "Feature Extraction Based on Local Histogram with Unequal Bins and a Recurrent Neural Network for the Diagnosis of Kidney Diseases from CT Images" Bioengineering 11, no. 3: 220. https://doi.org/10.3390/bioengineering11030220
APA StyleGharahbagh, A. A., Hajihashemi, V., Machado, J. J. M., & Tavares, J. M. R. S. (2024). Feature Extraction Based on Local Histogram with Unequal Bins and a Recurrent Neural Network for the Diagnosis of Kidney Diseases from CT Images. Bioengineering, 11(3), 220. https://doi.org/10.3390/bioengineering11030220










