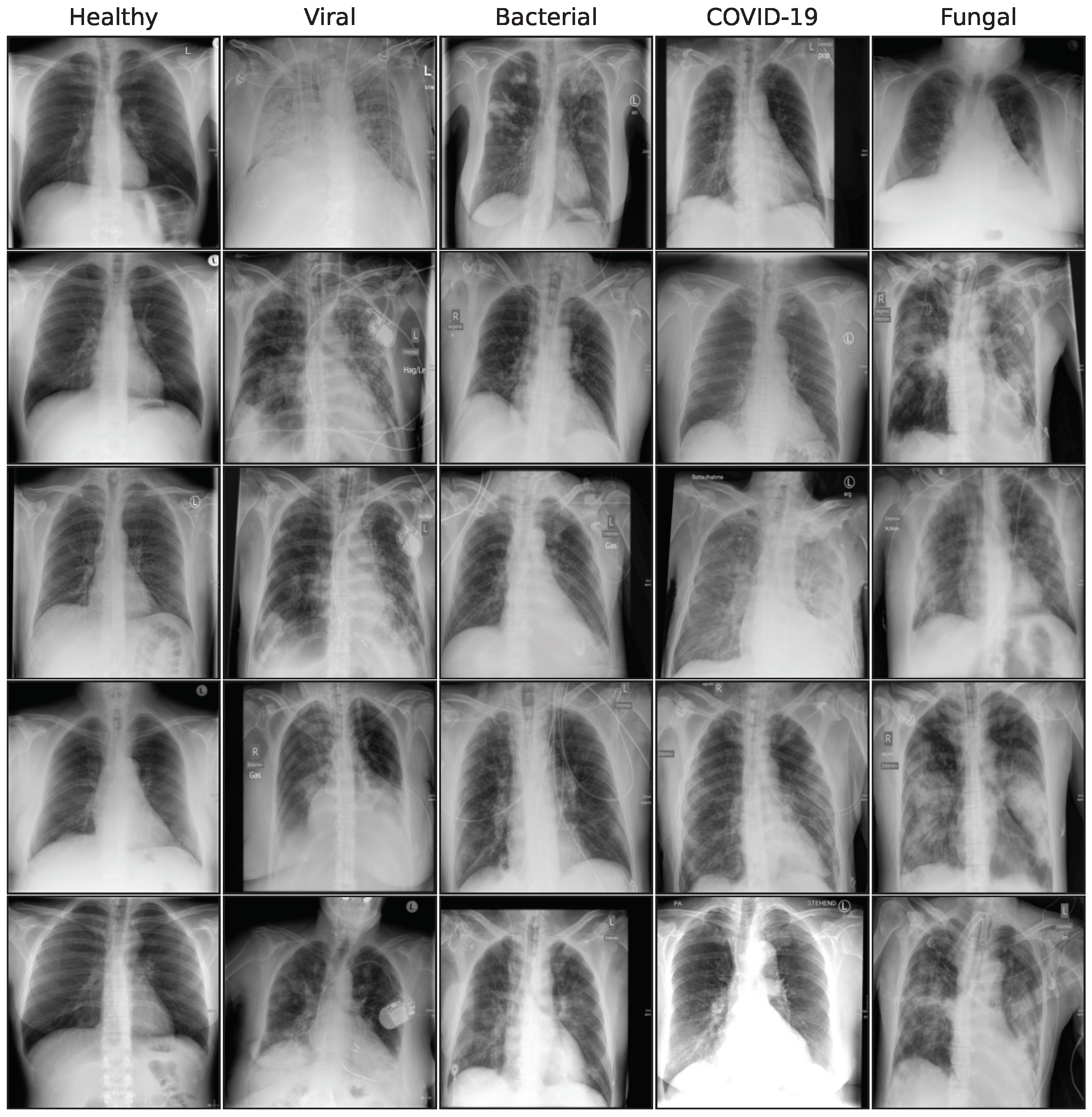
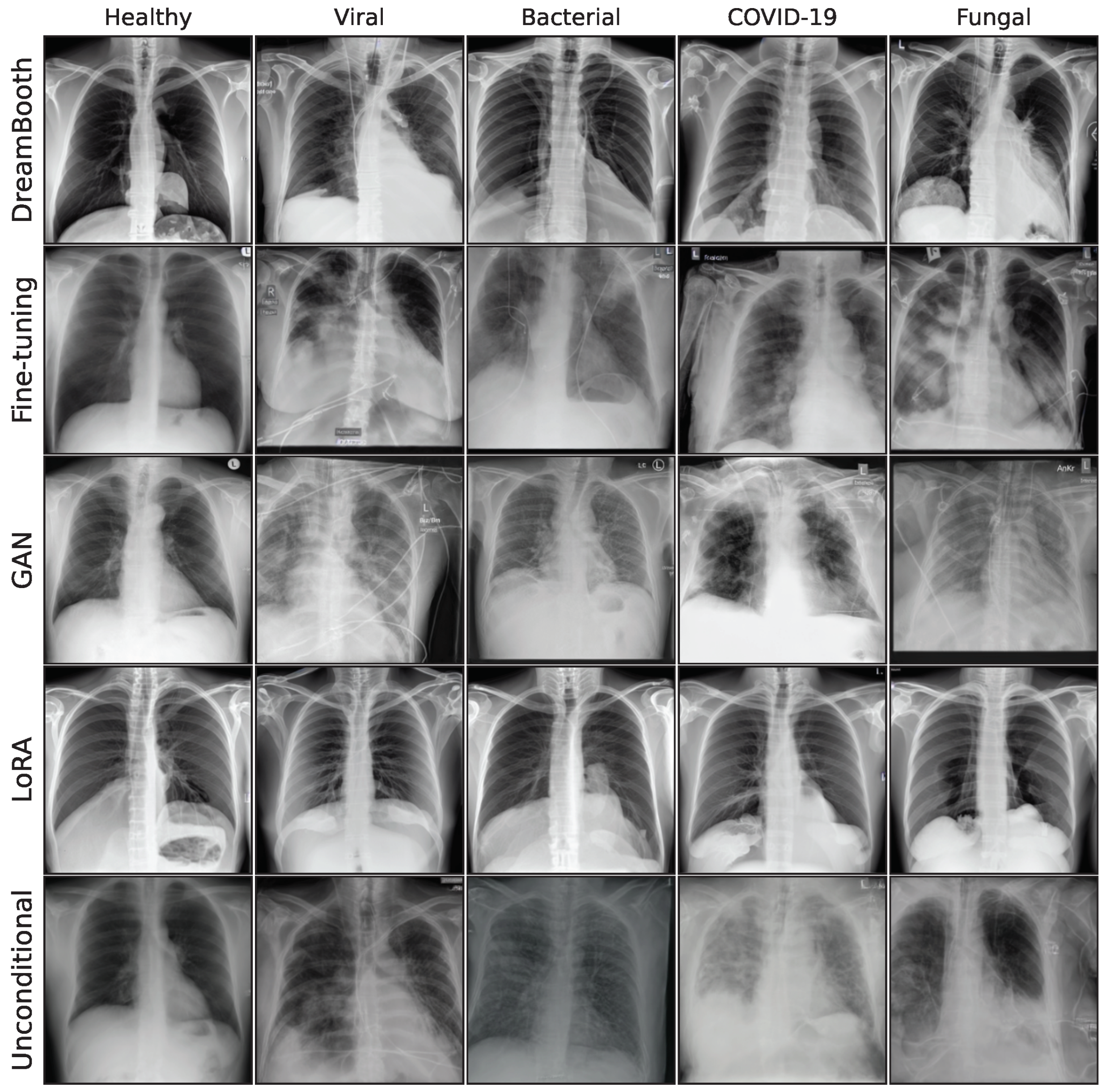
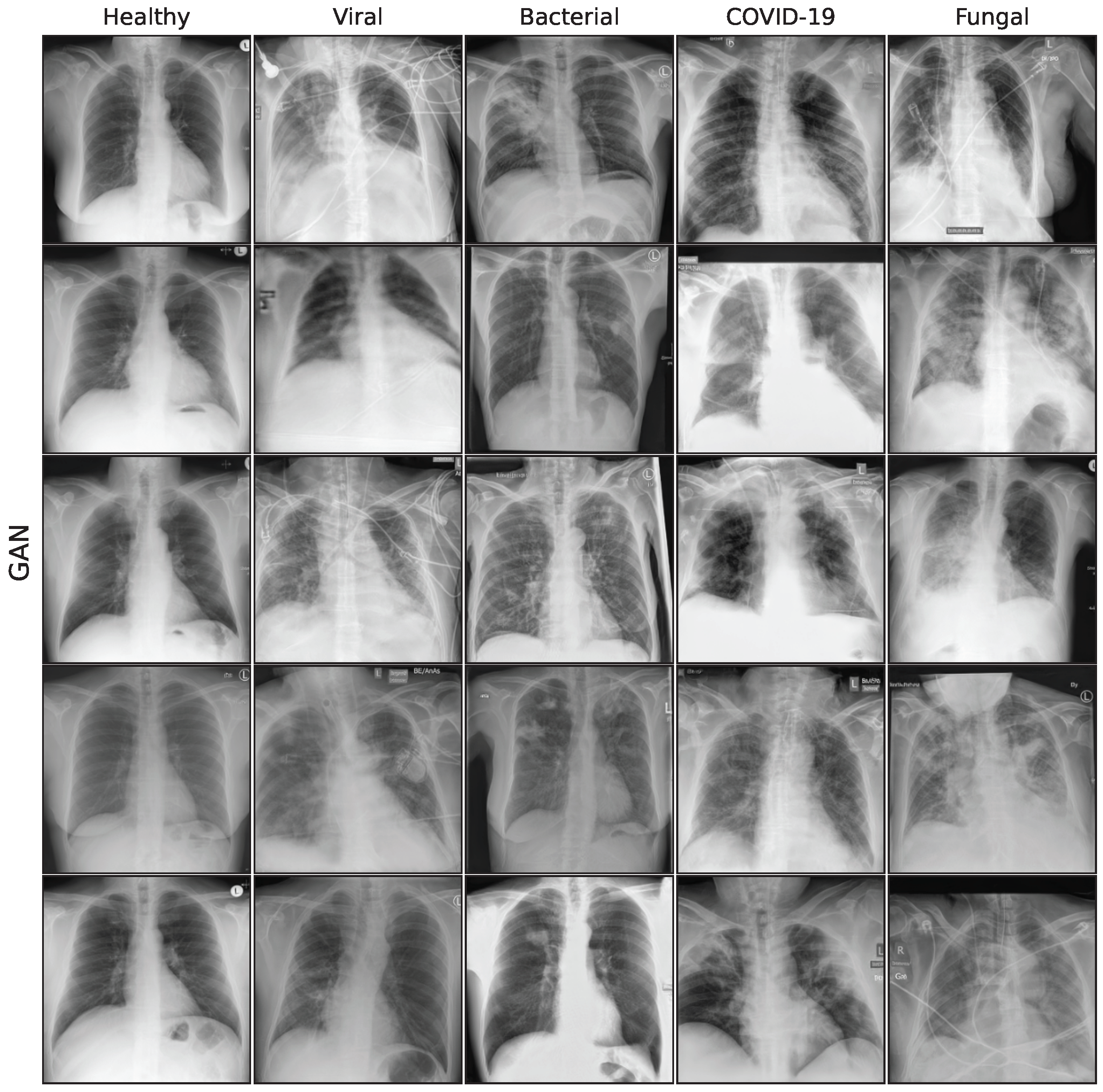
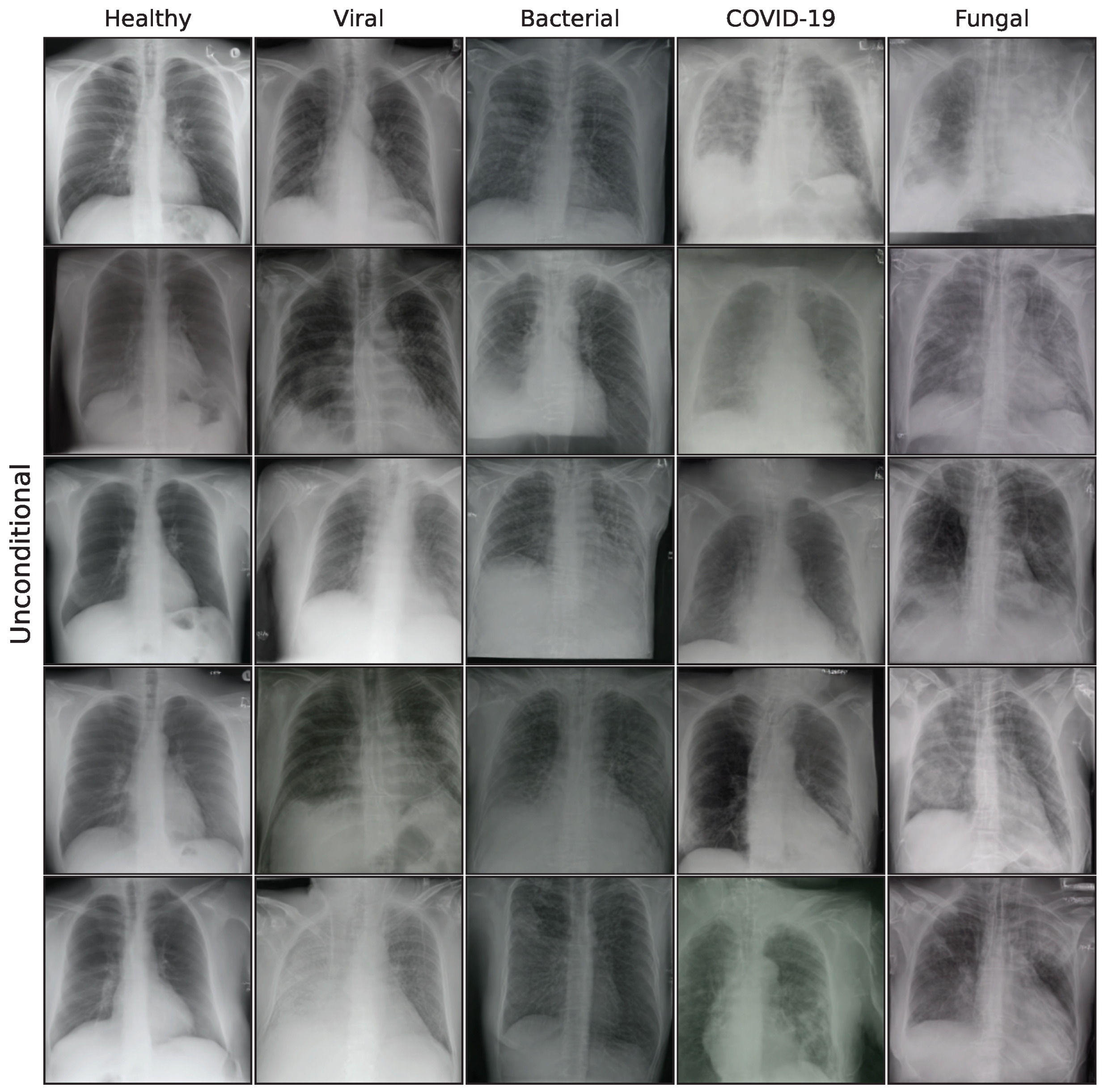
1. Introduction
The necessity for swift and dependable patient screening emerged as a key lesson from the COVID-19 pandemic. The development of machine learning models for aiding early pandemic clinical decisions is crucial, reducing diagnosis time and assisting emergency medical personnel [
1]. However, a significant challenge in rapidly creating models for new infectious diseases is the limited access to high-quality data. This constraint is a common issue in the medical field, often stemming from privacy concerns [
2] and high data acquisition costs. In radiology, all imaging modalities are affected equally (including X-rays, computed tomography, and magnetic resonance imaging), as well as various organ systems and diseases. Recently, as part of the COVID-19 pandemic, inflammatory changes in the lungs have come into focus, as these are of great importance in everyday life and have an impact on patients’ lives as well as hospital capacities [
3]. Hence, it is imperative to effectively utilize machine learning models under scarce data conditions.
While methods like transfer learning and self-/semi-supervised learning exist, the performance of deep learning models is notably influenced by the data quantity, as shown theoretically [
4,
5] and empirically [
6,
7,
8]. This study exemplifies such a scenario within the medical domain, focusing on a limited dataset. An analysis on chest X-ray (CXR) images pertaining to four different pneumonia causes is conducted, along with healthy patient images, with as few as 74 images for viral/non-COVID-19 cases. The objective is to leverage generative models to achieve reliable predictions despite the constraints of limited data. To date, using generative models for synthetic data augmentation on limited data is an under-explored research area. Although generative models are commonly used for larger datasets in medical imaging with a reported increase in performance [
9,
10,
11,
12,
13], we do not see the same rigorous research towards scarce data scenarios, where such approaches would be most helpful. We aim to close this gap and initiate the discussion in this area.
This study provides a comprehensive evaluation of diffusion and Generative Adversarial Network (GAN)-based learning approaches, specifically aiming at improving performance of the downstream classification task of predicting COVID-19, other viral pneumonia, fungal pneumonia, bacterial pneumonia, and healthy cases on 1082 CXR images. We examine five different generative approaches and provide quantitative and medical assessments of image quality, diversity, and plausibility. The artificially generated images are used for synthetic data augmentation, where we measure the impact on performance for five different classification models. Additionally, varying amounts of synthetic images are added to the training data to further increase robustness of the evaluation. Although some generative approaches outperform our baseline models by a substantial amount, this study does not show an improvement in classification performance on average over all architectures and configurations. Despite that, class-specific metrics like precision, recall, and F1-score show a substantial improvement by using synthetic images, emphasizing the data rebalancing effect for the less frequent classes. This holds true when compared to a simple oversampling approach. Although we report better average classification improvement on this dataset in a previous study [
14], this study deliberately does not utilize additional domain knowledge in the process, using only simple prompts for text-conditioned models and a non-domain specific text encoder instead.
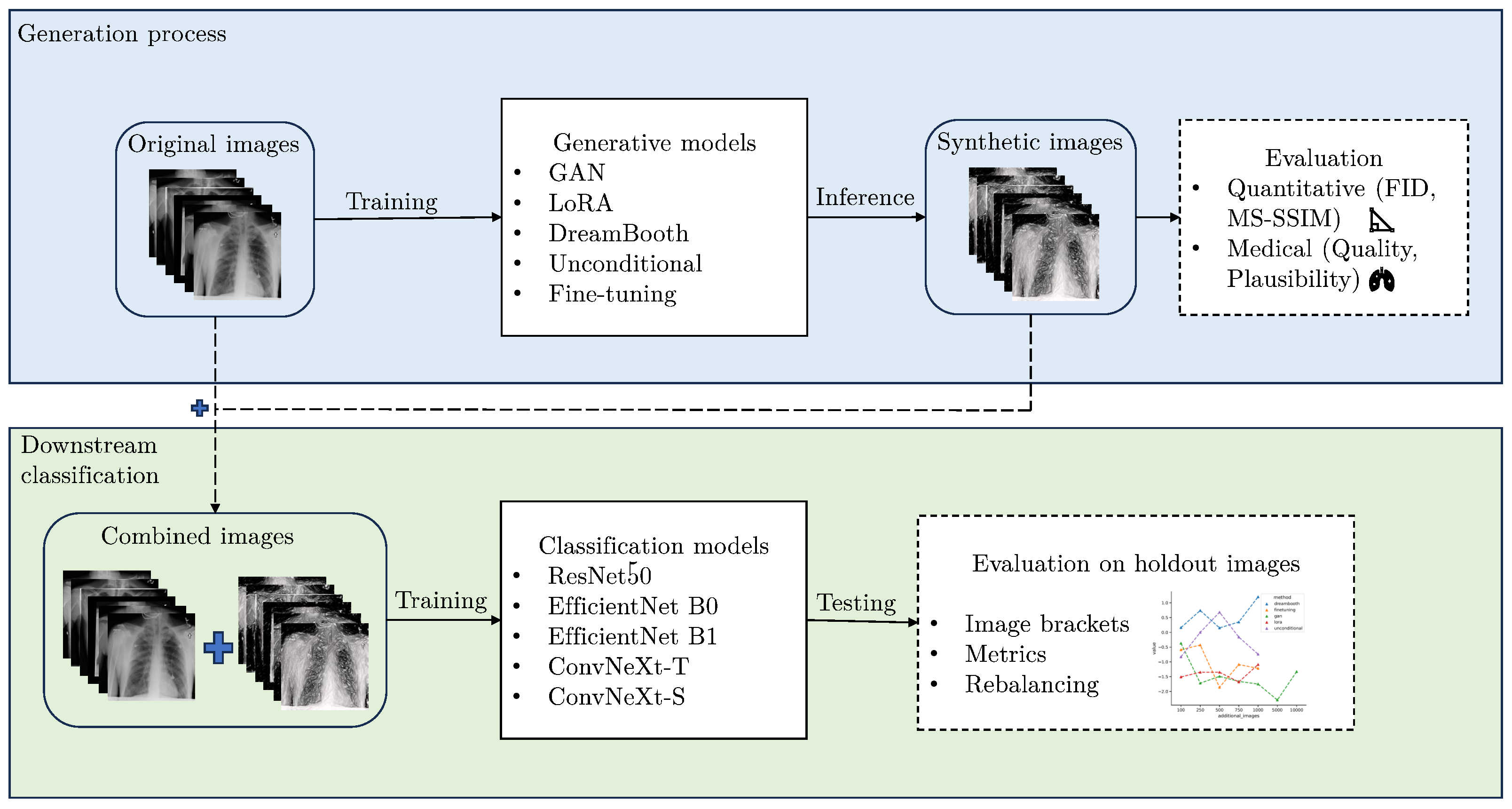
Figure 1 shows a schematic representation of our research approach. The code for this work can be found at:
https://github.com/dschaudt42/synthetic_pneumonia (accessed on 30 November 2023). The 70,000 synthetic images produced in this work are available at:
https://huggingface.co/datasets/dschaudt42/synthetic_pneumonia (accessed on 30 November 2023).
In summary, the main contributions of our work are:
A comprehensive evaluation of diffusion and GAN-based learning approaches on a limited pneumonia X-ray dataset, testing the applicability of generative models in a scarce data scenario.
Quantitative and medical assessments of image quality, diversity, and plausibility for synthetically generated images and show large gaps between the demonstrated generative approaches.
Evaluation of synthetic data augmentation on a downstream classification task in a comprehensive manner, examining multiple classification architectures and additional image brackets for robust results. This study shows that higher quality images, as perceived by metrics and experts, do not necessarily lead to better classification performance. Furthermore, synthetic images can improve class-specific metrics substantially due to a data rebalancing effect, while aggregated performance metrics do not benefit in most cases.
2. Related Work
Generating high quality synthetic images is a field that gained a lot of traction with the inception of the GAN model [
15]. In medical imaging, synthetic images have been used to translate between image modalities [
16,
17,
18], enable sharing of privacy-protected data [
19,
20], and improve deep learning models on diverse downstream tasks [
16,
19,
21,
22,
23].
Al Khalil et al. [
24] propose a conditional GAN model, which increases segmentation performance on cardiac magnetic resonance images. The performance increase is especially noticeable when real and synthetic images are combined during training. Prasanna Das et al. [
25] propose a conditional flow model to generate chest CT images and validate their approach by synthetic data augmentation for a downstream classification task of detecting COVID-19. Several GAN-based models have been proposed to improve performance on downstream COVID-19 tasks [
9,
10,
11].
Recently, diffusion-based models have shown improved performances over GAN-based architectures in many domains [
26,
27,
28]. He et al. [
12] show that using synthetic data from large text-to-image models is a valid approach to downstream image recognition tasks, but do not consider smaller, more domain-specific datasets, like the ones presented in this work. Pinaya et al. [
29] use latent diffusion models to generate synthetic brain MRI images, conditioned on the covariates age, sex, and brain structure volumes and compare them to inferior GAN-based baselines.
Chambon et al. [
30] provide experiments on fine-tuning the different components of a Stable Diffusion model to secure domain-adaption for chest X-ray images. They found that the pre-trained variational autoencoder and the CLIP text encoder have a sufficient domain-adaption capabilities for chest X-ray images, and that fine-tuning the U-Net component is critical to improve image quality. Since we want to exclude further domain knowledge from our generation process, we also chose this approach of only fine-tuning the U-Net, while using simple, class-specific prompts. In a follow up work, the authors fine-tune the CLIP text encoder and report improved performance on a downstream classification task [
23].
Müller-Franzes et al. [
31] compare their latent denoising diffusion model, Medfusion, to GAN models on multiple medical imaging datasets of fundoscopy images, radiographs, and histopathology images. Packhäuser et al. [
20] compare a PGGAN [
32] model with a latent diffusion model on the ChestX-ray14 dataset [
33] with a focus on privacy-enhancing sampling. Both found that classification performance does not increase when using the synthetic data, but found that the diffusion model generates higher quality images than the GAN model.
Most of the existing literature uses large amounts of samples to train generative models, even in the medical imaging domain with large image collections, sometimes with over 100,000 samples. Although there is undeniable evidence that generative models and deep learning models in general perform better with more samples [
4,
5,
6,
7,
8], we argue that employing synthetic data is most useful in data scarce scenarios. If large datasets are available, the need for synthetic images can certainly be challenged. In this work, we try to test the limits of generative models in a data scarce scenario (
for some classes), especially for downstream classification tasks.
4. Results
In this section, we examine the results of the five generative methods presented in the following categories: generative performance, medical assessment, and classification performance. In generative performance, we look at the performance metrics FID [
58] and MS-SSIM [
59] to quantify the fidelity and diversity of the generated images. In medical assessment, a dedicated thoracic radiologist (C.K.) with 9 years of experience in lung imaging assessed the quality and plausibility of the generated images from a medical perspective. In classification performance, the effect of synthetic data augmentation on a downstream classification task is evaluated.
4.1. Generative Performance
Generated images should be similar to the underlying, real image distribution (fidelity), and ideally show a large variability in possible outcomes (diversity). Fidelity and diversity can be measured by the Fréchet Inception Distance (FID) [
58] and the Multi-Scale Structural Similarity Index (MS-SSIM) [
59], respectively. Both metrics are commonly used in generative image tasks. We calculate the FID based on the final 2048 feature layer of a pre-trained Inception V3 [
60] model, as is standard. The distance is calculated by comparing 50 synthetic images of each class to 50 real images. The MS-SSIM (Gaussian kernel size 11; sigma, 1.5), which is a generalization of the SSIM [
61], is calculated by a pairwise comparison of all combinations of the same 50 synthetic images and taking the average. Lower values of FID and MS-SSIM show larger fidelity and diversity. It should be noted that the metrics used are contingent on the reference samples and implementation, making direct comparisons with other studies challenging [
62].
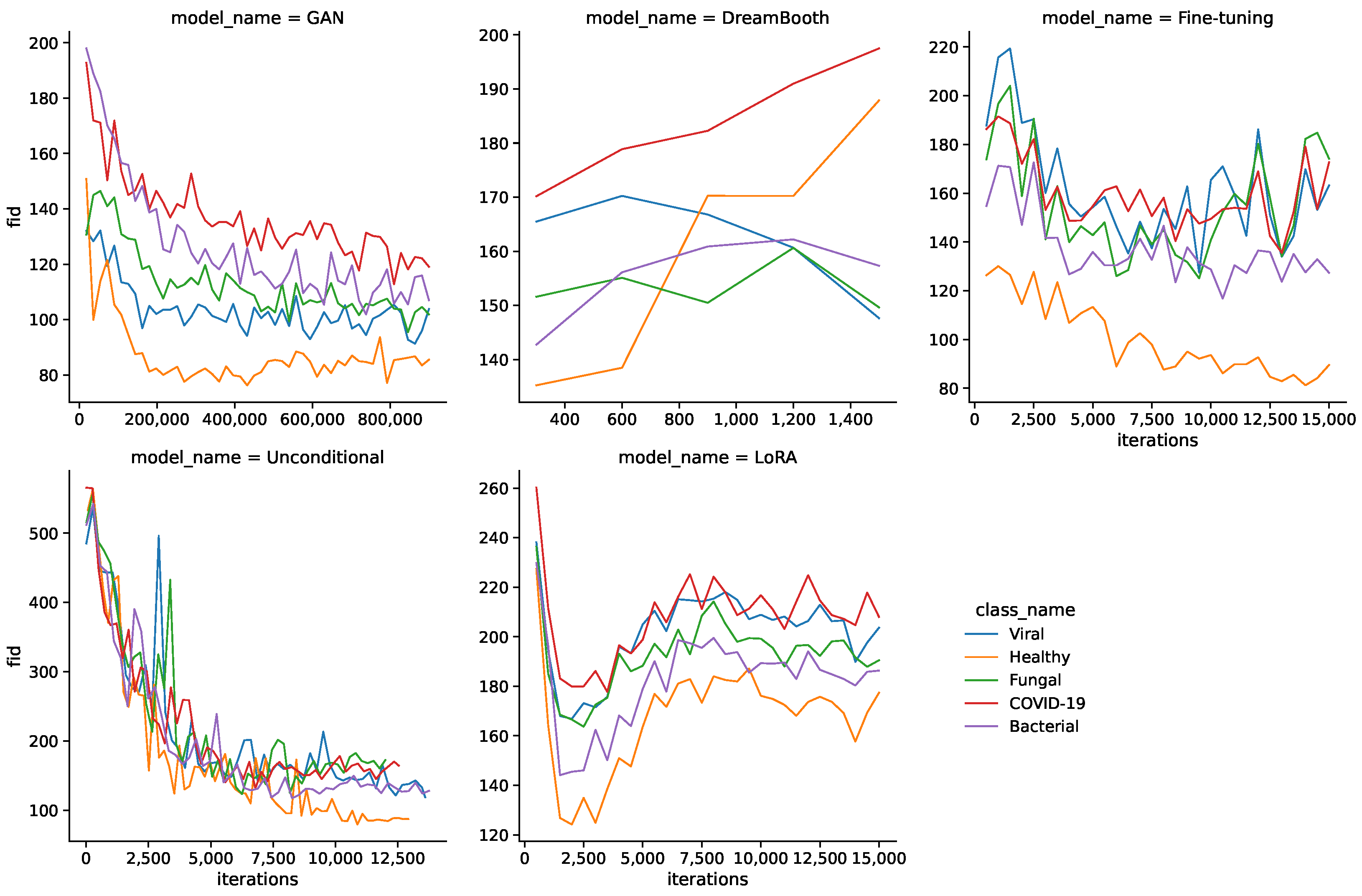
Figure 2 shows the FID values during the training for all models and classes. Interestingly, FID values do not simply decrease during training for all models, but can also increase towards the middle or end of the training process. This is especially apparent for DreamBooth, LoRA, and fine-tuning models, where the FID increases from the start or later during training. The LoRA model sees a sharp drop in FID values in the beginning and shows another decline towards the end of training.
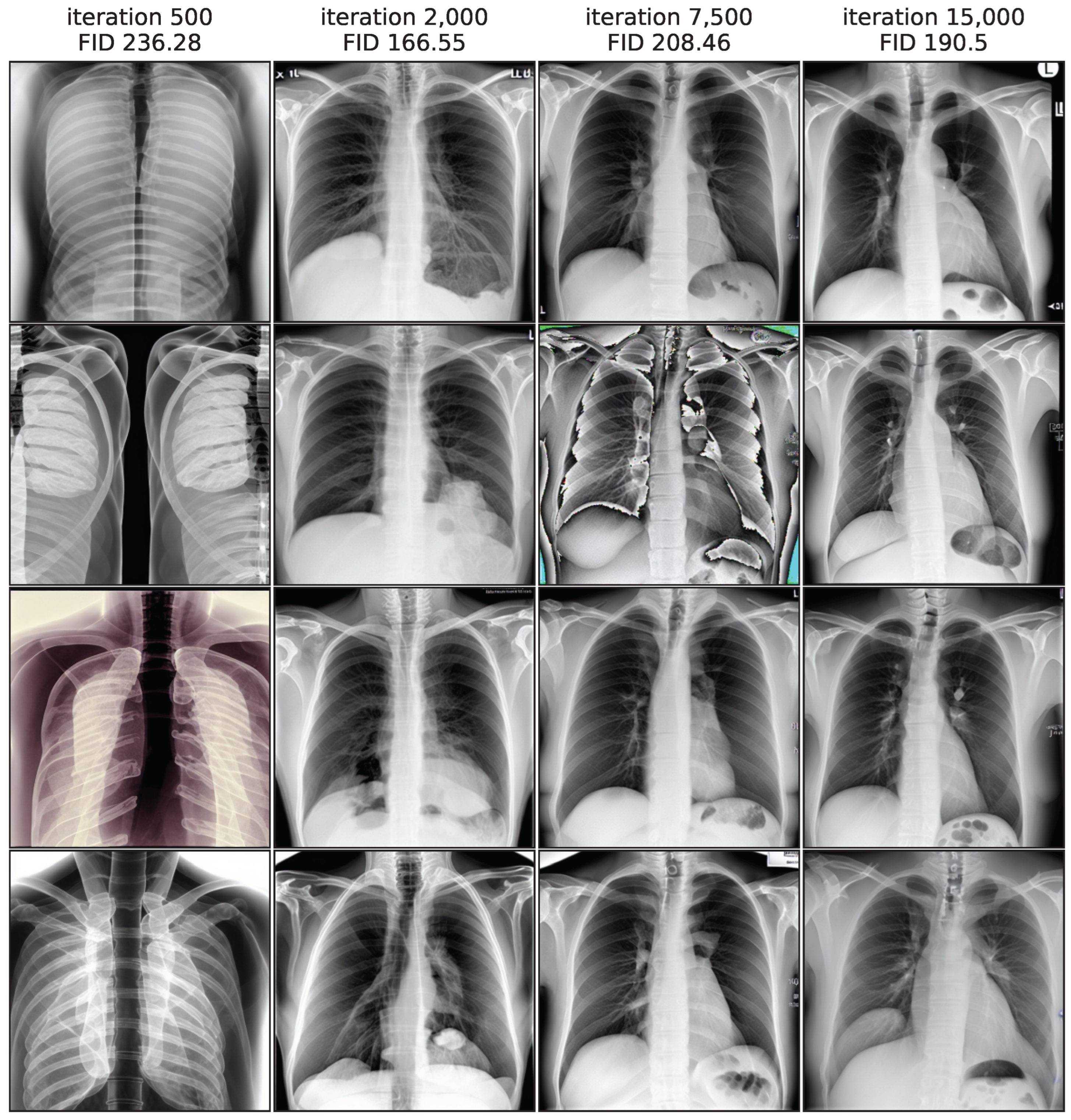
Figure 3 illustrates this by showing image samples of the LoRA model from different training iterations, while images from iteration 500 exhibit the largest FID values and a low quality, the quality improves substantially in iteration 2000. In iteration 7500 the FID rises again and the quality decreases, as shown clearly by a faulty image. In the end of the training process, the FID decreases again slightly and quality seems to increase, while not fully reaching earlier levels. This non-monotonic FID progression confirms the general usefulness of monitoring the FID values during the training process to pick the best model iteration.
Another noteworthy observation is a difference in FID curves between classes. Healthy images generally have lower FID values than the pathological classes, which might stem from a substantial difference in quantity of the underlying real images.
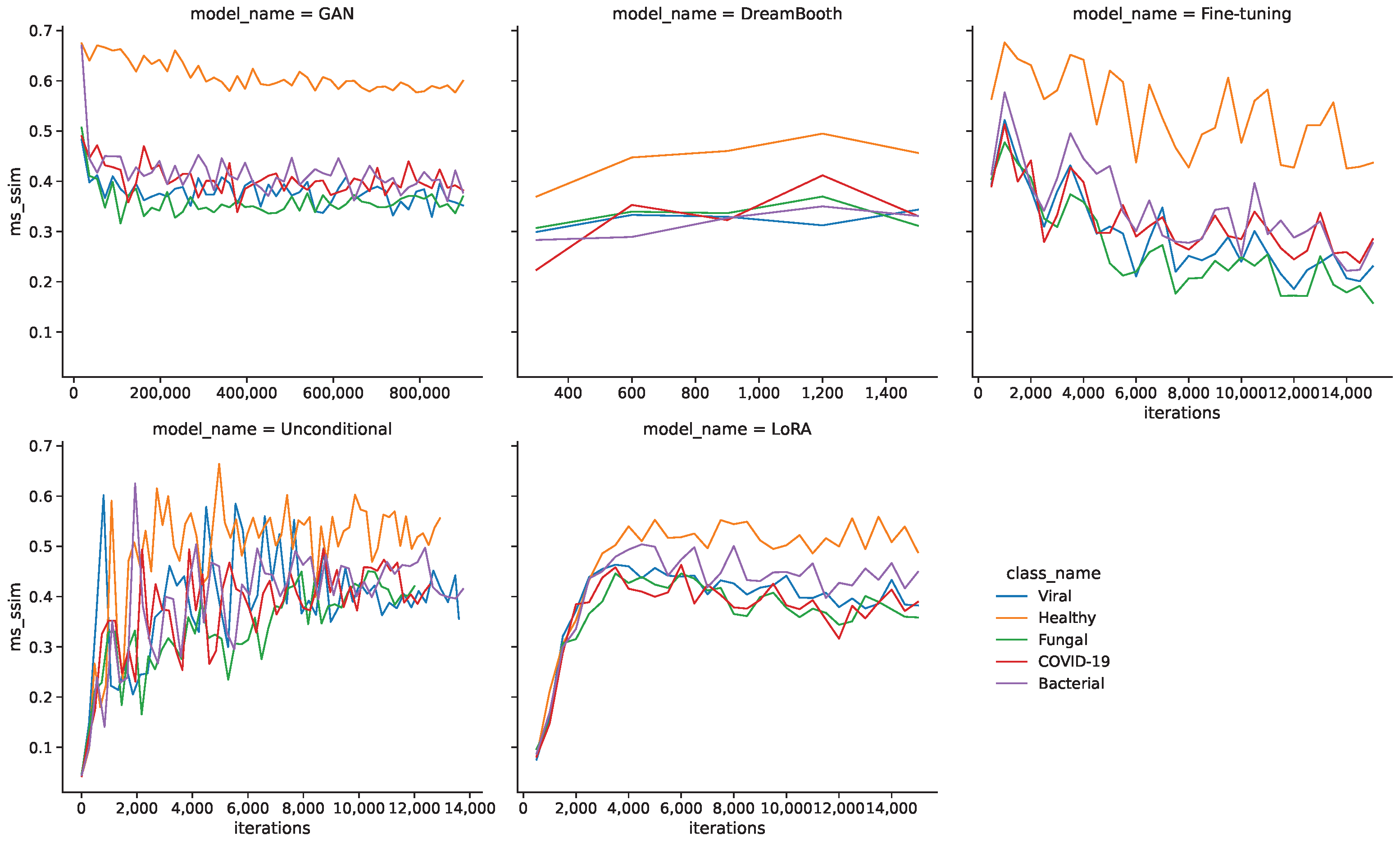
Figure 4 shows the MS-SSIM values during the training for all models and classes. In this case, the larger quantity of healthy images seems to be a disadvantage, as they generally produce higher MS-SSIM values and therefore show a lower diversity.
Table 5 shows the resulting minimum FID and MS-SSIM values for all models and classes. The GAN model shows the lowest FID values for all classes by a sizable margin, followed by unconditional and fine-tuning models. DreamBooth and LoRA models generally show larger FID values and therefore lower fidelity images. The MS-SSIM values are mostly similar for the minimum FID iterations, with unconditional and DreamBooth models having slightly higher values.
4.2. Medical Assessment
Quantitative distance metrics can be a good first indicator of image quality, but they do not provide any medical assessment of image quality or pathological plausibility of the synthetic images. We argue that an evaluation by a human expert is critical in such a sensitive medical setting. A dedicated thoracic radiologist (C.K.) with 9 years of experience in lung imaging has therefore assessed the quality and plausibility of the generated images from a medical perspective.
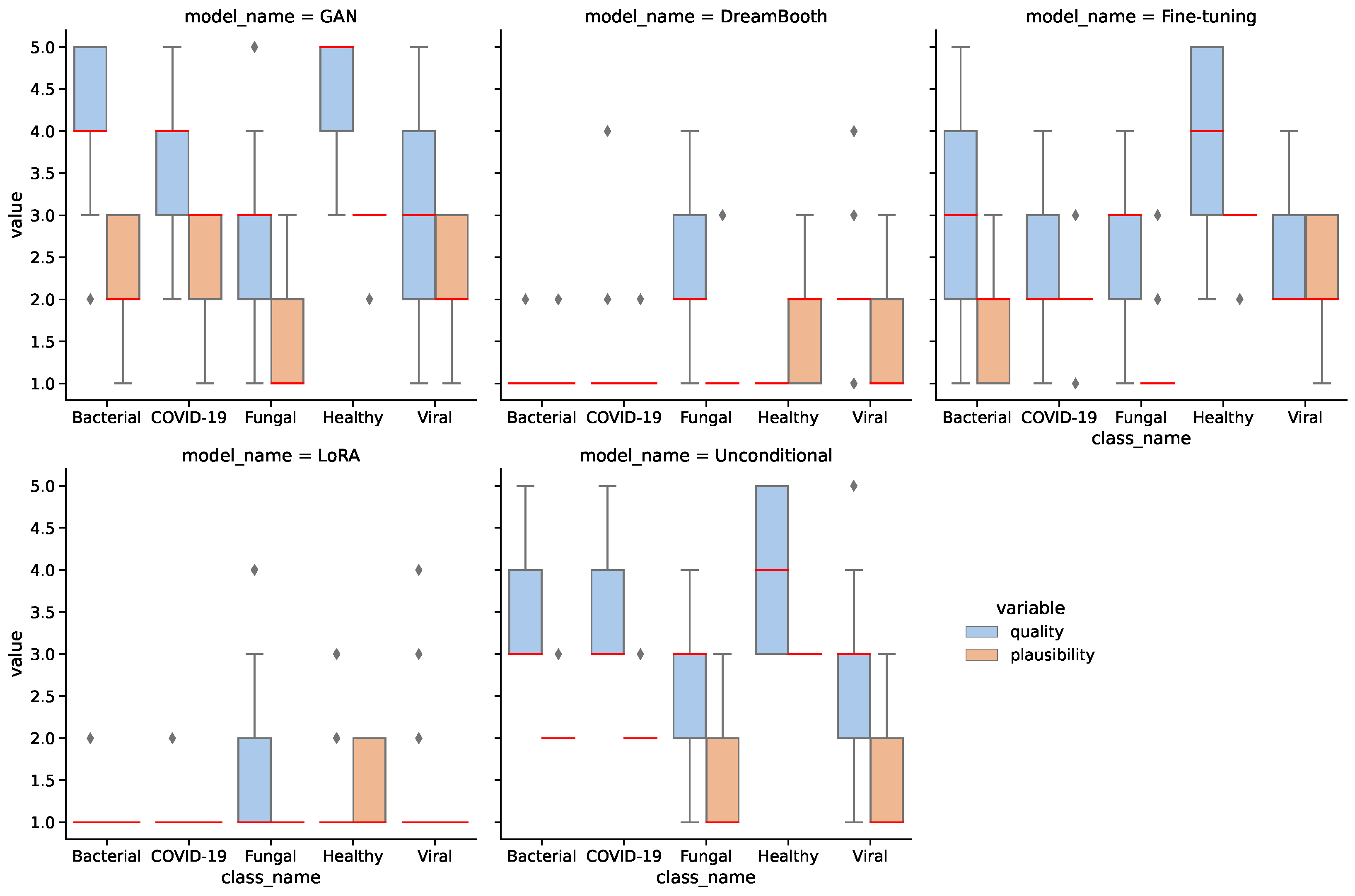
The quality of an image was assessed on a scale of 1 (lowest quality) to 5 (highest quality). Important aspects of the quality assessment are that thorax and lungs are shown as a whole, so that the anatomy is reproduced correctly and to scale. The sharpness and contours must be reproduced correctly. If a pathology was present, it was evaluated according to its characteristic appearance on a scale from 1 to 3 (1 = completely inappropriate, 2 = partly characteristic, 3 = characteristic). Important aspects of the plausibility assessment are to what extent the typical appearance of pneumonia is reproduced. This includes the density, the sharpness compared to the lungs, the relationship to other anatomical structures (heart, pleura) as well as the distribution pattern within both lungs (centrally emphasized, peripherally emphasized, division into the individual lung lobes).
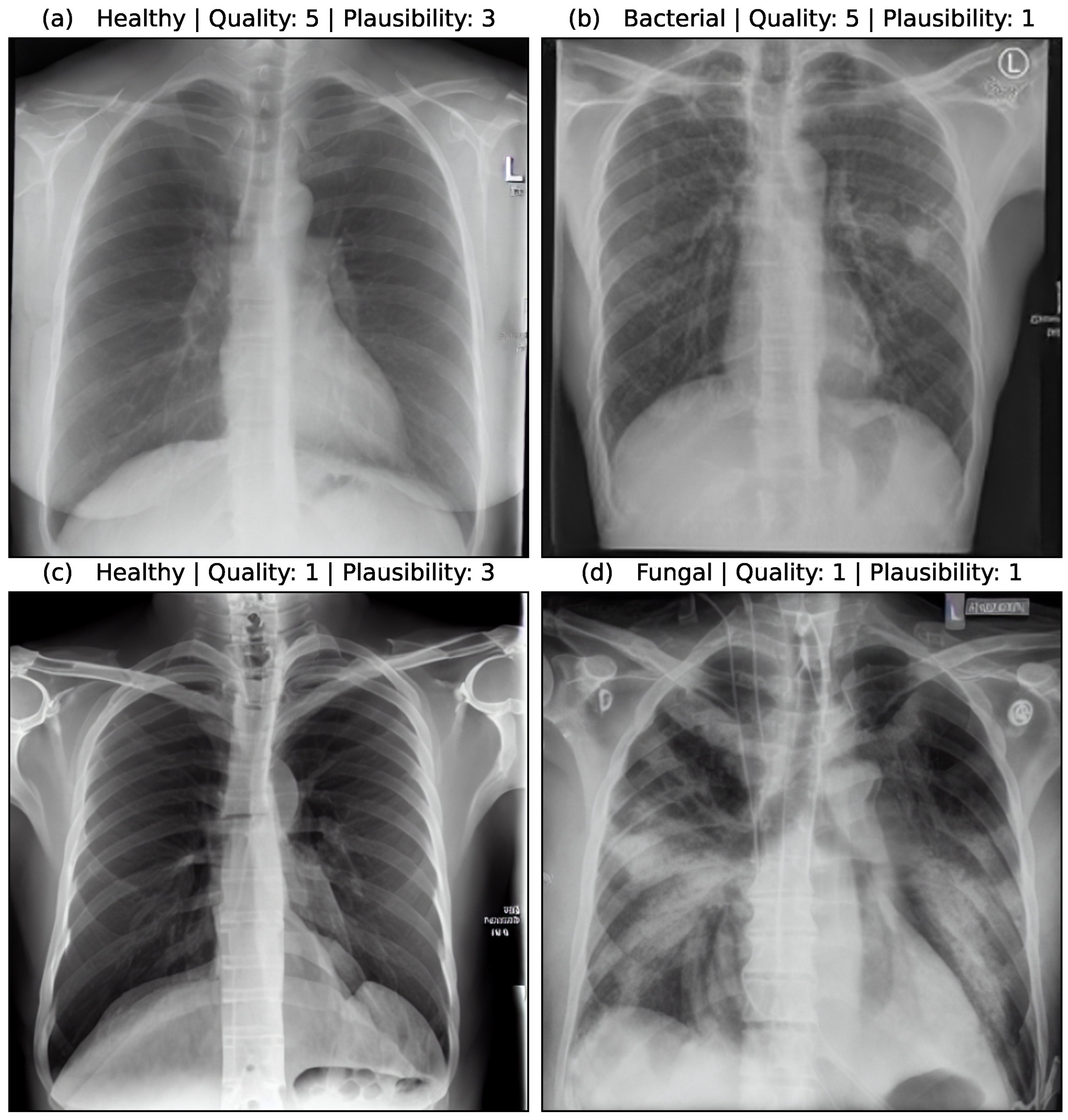
Figure 5 shows the assessment of four synthetic image samples with high/low quality and high/low plausibility scores. In (a, b) the anatomical structures are reproduced realistically and the proportions are accurate. The pleura, diaphragm, heart contour and hilar vessels are reproduced with absolute precision. The breast shadow is also reproduced exactly in (a), which simulates a woman as the gender of the patient. The quality rating is accordingly rated score 5 without any gradations. The healthy state in image (a) is shown regularly with a quality score of 5. The bacterial pneumonia in (b) is rather atypical, a suspect bronchial carcinoma from the simulated image is more realistic. It does not reflect peripheral inflammation in the sense of bronchopneumonia or lobar pneumonia, hence the assessment of plausibility as inappropriate (score 1). Image (c) appears artificial in appearance, the diaphragm contours, the heart silhouette and the bones are not realistically reproduced, the image quality is only rated with score 1. In contrast, no pathology of the lung parenchyma is recognizable, but this is still realistic and therefore rated as score 3 in terms of plausibility. Image (d) also appears artificial in appearance, in particular the hints of foreign material/lines and heart contours are unrealistic (quality score 1). The inflammations of the lungs described also seem unrealistic for any type of pneumonia; a fungal infection is unrealistically shown (plausibility score 1).
We assessed 25 synthetic images of each class for each model in quality and plausibility, totaling 625 images.
Figure 6 shows the results of the assessment for all models and classes. A Kruskal–Wallis test [
63] confirms that the difference between models for both quality and plausibility is significant with
. A Dunn–Bonferroni post hoc test [
64] on quality shows significant differences (
) between most models, except for the GAN-unconditional pair (
), the unconditional-fine-tuning pair (
), and the DreamBooth-LoRA pair (
).
The assessment confirms that the GAN model generates the highest quality and most plausible images, followed by the unconditional and fine-tuning models. DreamBooth and LoRA models seem significantly weaker than the other models. Additionally, the healthy images have higher quality and plausibility on average than the other classes. This is mostly due to larger image quantity and reiterates on the merit of larger data for generative models.
Table 6 aggregates the assessment results for all models. It should be noted that the medical assessment was not used to filter images based on quality for the downstream classification task. Although this might affect the classification task, we deliberately want to omit the integration of further domain knowledge to obtain an unbiased estimation of downstream performance. Furthermore, a comprehensive medical assessment of 70,000 synthetic images would be infeasible.
4.3. Classification Performance
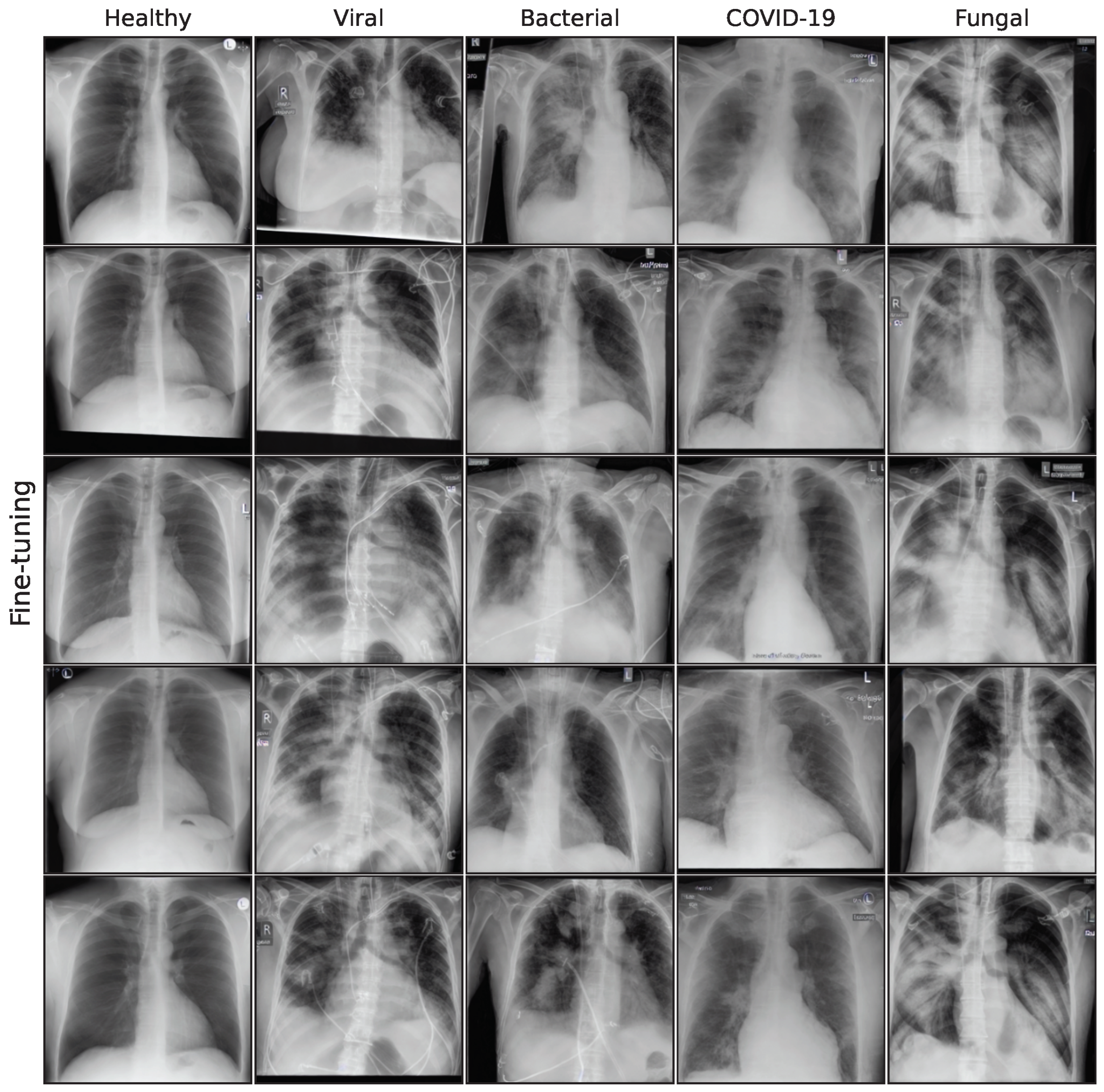
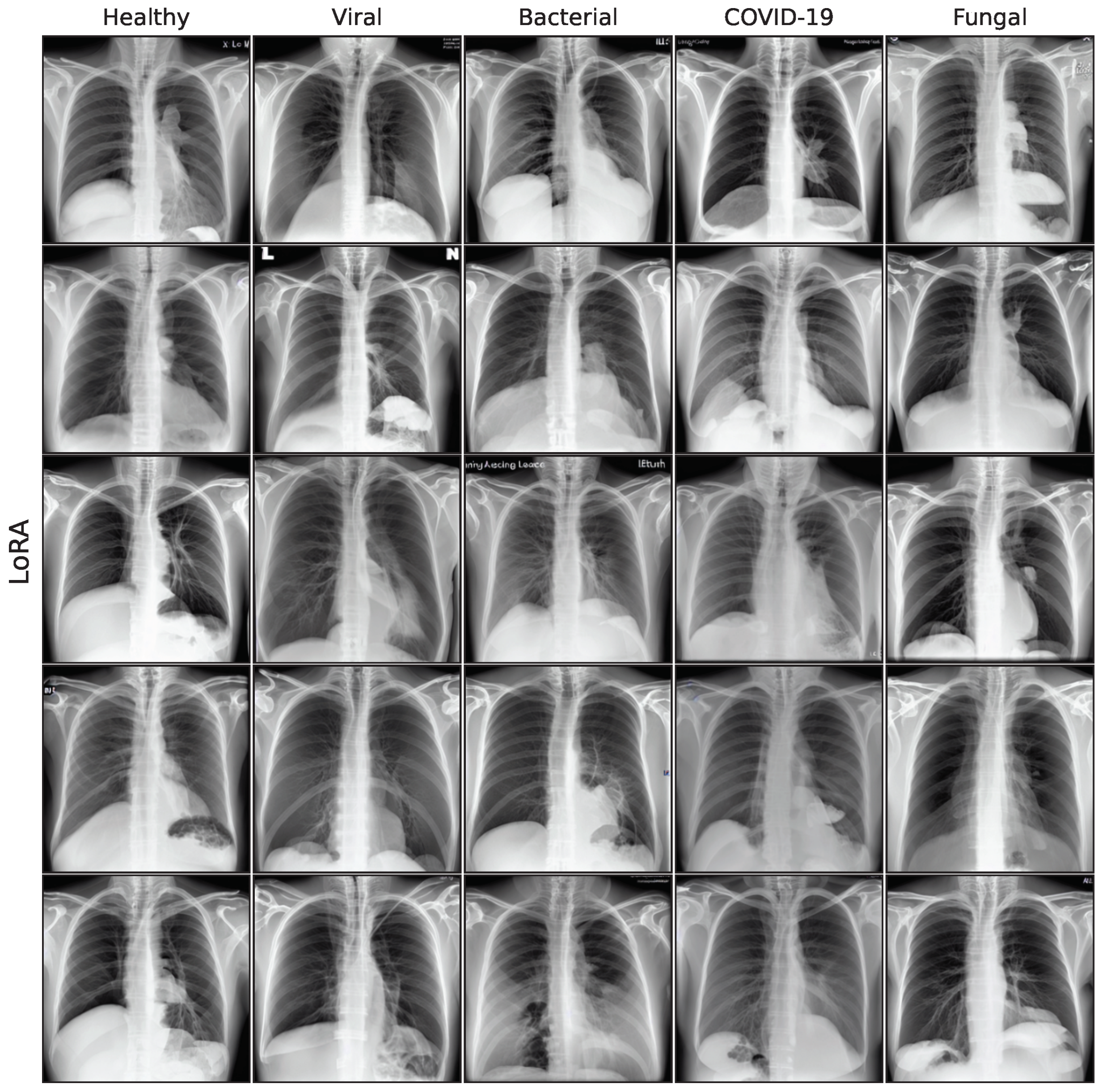
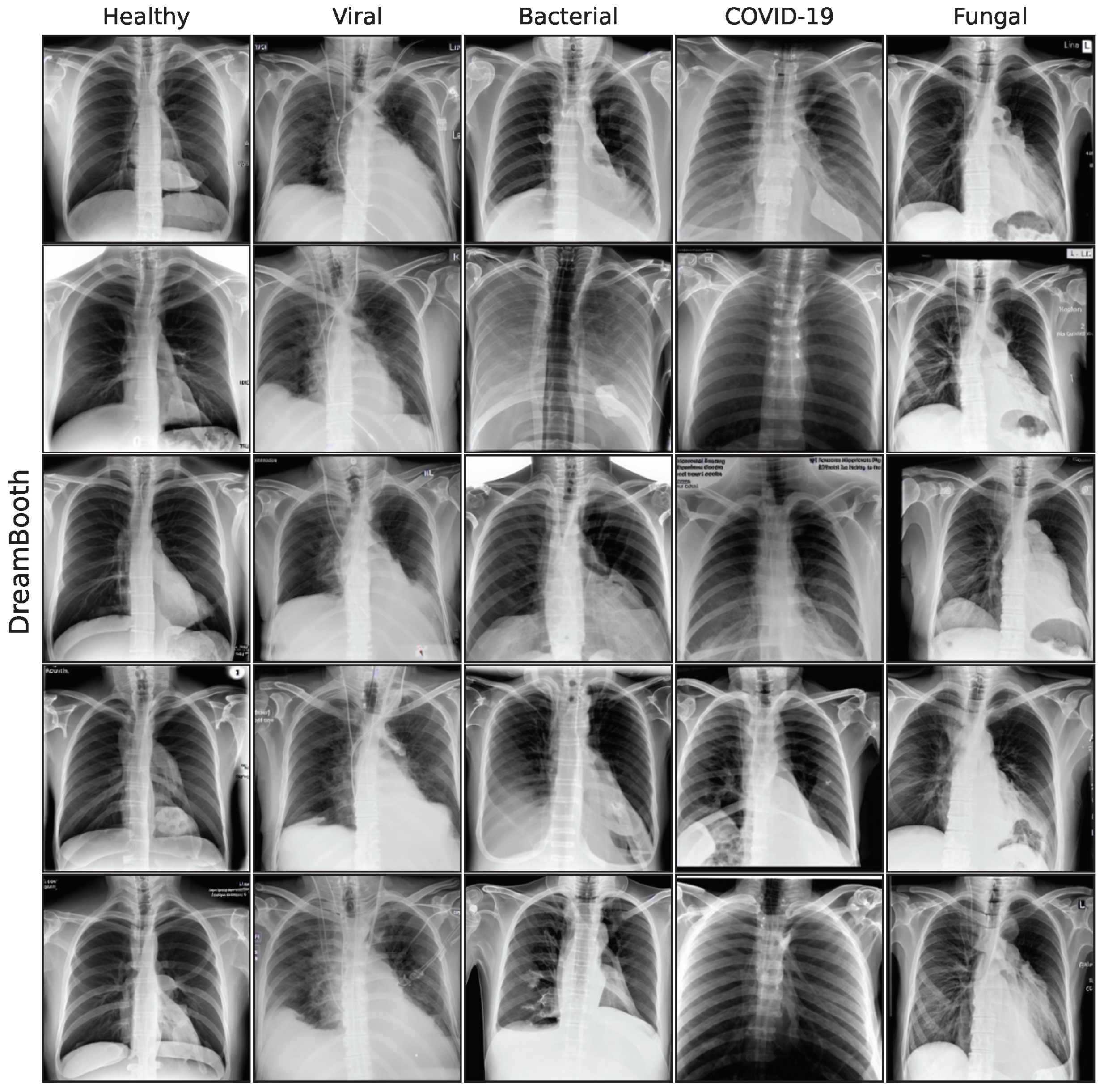
The effect of using the generated images as synthetic data augmentation to improve a classification downstream task is examined, utilizing synthetic images from the lowest FID iterations for all generative models. A selection of these final images is shown in the
Appendix A. The performance is measured on multiple model architectures: ResNet50 [
49], EfficientNet-B0 [
50], EfficientNet-B1 [
50], ConvNeXt-T [
51], and ConvNeXt-S [
51]. The respective baseline models do not use additional synthetic images for training and their performance has been reported in Schaudt et al. [
14]. To gain a better understanding on the synthetic data augmentation methodology, five different generative models with five different classification architectures are benchmarked and also vary the amount of additional synthetic images. All classification model trainings have been repeated 5 times with a different seed to obtain a robust and comprehensive outlook on the expected performance gain. Since the GAN model can generate images much more quickly than the diffusion-based models (in about 1/10th time), we want to examine this advantage by adding even more synthetic images into the training. The +5000 and +10,000 image brackets are therefore only evaluated for the GAN model, since generating such large quantities would be unfeasible for the other models in our setting.
Table 7 reports the accuracy for the presented classification models, trained with additional synthetic images from the presented generative models. The best model is ConvNeXt-S, trained with 250 additional synthetic images from the unconditional model with 81.11% accuracy. This is a notable increase of +2.58 percentage points from the baseline variant, while the best performing models for each architecture all use synthetic images, the results show a large variation in performance across all methods and image brackets. Many configurations even see a decline in classification performance.
Table 8 shows aggregated results as the average change in accuracy in percentage points for all models from baseline over all additional image brackets. The only generative model with a positive impact on classification performance over all classification models is DreamBooth with an average improvement of 0.52. Interestingly, specific generative models can have a substantially higher than average impact on performance for specific classification models. This suggests that some model combinations fit very well together, while others do not. The classification model that shows the highest improvement on average across all generative models is ConvNeXt-S with an increase of 0.12 over baseline.
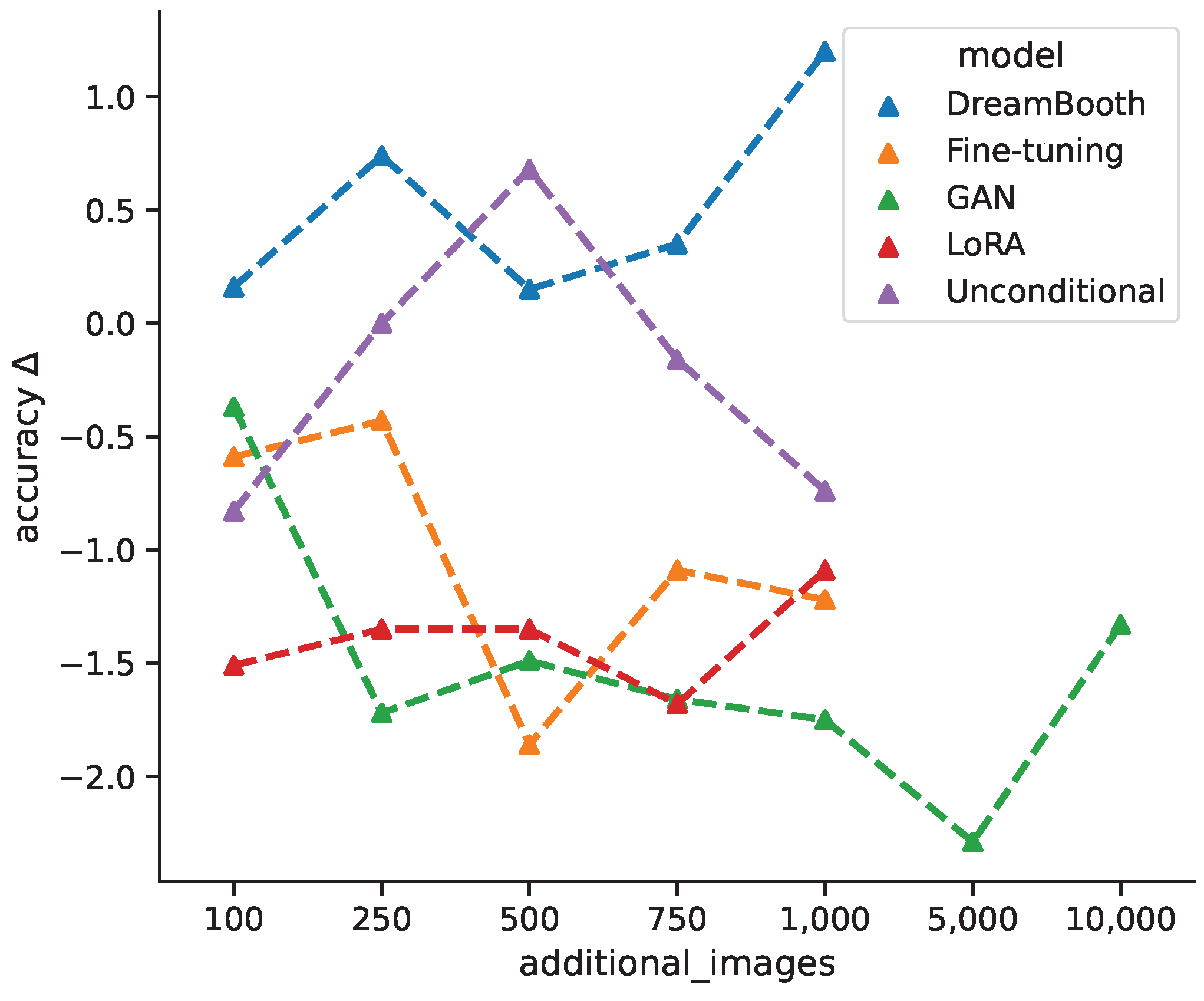
Figure 7 shows the average change in classification accuracy for all generative models from baseline per additional image bracket over all classification models, while DreamBooth favors larger quantities of synthetic images, the unconditional model shows an improvement for intermediate quantities, which decreases again for +750 and +1000 images. Both fine-tuning and GAN models show decreased performances for larger quantities, with a slight incline towards the maximum quantities. This could suggest that even more images might have a positive impact. The LoRA models are largely invariant under different image quantities. Varying the amount of additional synthetic images for training does not follow a clear pattern and seems to depend largely on the specific generative model. For cases where classification accuracy first decreases and then increases could be due to the classification model shifting from learning original features towards generated features. In general, the change in classification accuracy between different models could result from the models ability to generate classifiable features.
To examine class specific performances, we focus on the classification model with the best overall accuracy, which is ConvNeXt-S.
Table 9 shows the precision, recall, and F1-score for all generative models for each class. The metrics have been calculated based on the best performing image bracket for each generative model. We also include a ConvNeXt-S model trained with randomly oversampled classes as a simpler approach to rebalance class distribution during training. Generative approaches show higher recall values for most classes, with sizable differences for some classes (LoRA 0.36 vs. baseline 0.1 for viral cases). All pathological classes show substantially higher recall and F1 values for generative approaches, especially for bacterial, fungal, and viral cases. The baseline model shows the best precision and F1 values for healthy cases, as well as the best precision for COVID-19. This is not very surprising, since these classes are the most frequent and do not benefit as well from the rebalancing effect of synthetic data augmentation. Despite the lower accuracy of the generative approaches, class-specific performances can suggest the use of generative models in imbalanced learning scenarios. This holds true when compared to a simple oversampling approach, which does perform worse for most classes and completely misses the bacterial cases.
5. Discussion
Surprisingly, we found that higher image quality does not translate to better performance on a downstream classification task. Looking at the accuracy, most models performed worse with synthetic data augmentation, which is in line with Müller-Franzes et al. and Packhäuser et al. [
20,
31]. Only the DreamBooth model leads to an overall improvement across all examined classification architectures. We can only guess why image quality does not translate to improved classification models. Images from DreamBooth and LoRA models seem to exhibit higher contrast and unrealistic visibility of the bone structure and appear more cartoon-like in general. This could lead to an indirect regularization effect, leading models to broader areas of the loss function and reduce overfitting. Additionally, those models are pre-trained and already had exposure to limited amounts of chest X-ray images, which could potentially increase image diversity.
It could also be the case that our quality and plausibility assessment does not accurately evaluate whether the generated images hold features that are relevant to classification models. We think that the appearance of typical pathological patterns should, in theory, be the decisive feature used by classification models. Due to the black box nature of these models, it is not possible to finally conclude which features are being used for classification. Even attribution methods like GradCAM [
65] do not map pixel attributions to features in a meaningful way or aggregate the information over many images. Although there might be other metrics for a medical assessment that correlate better with classification performance, we think that the chosen ones are meaningful from the perspective of a human evaluator and present the medical perspective.
Some models benefited more from synthetic data augmentation than others. For example, ConvNeXt-S shows an average improvement of 0.12 percentage points, while EfficientNet-B0 shows a decrease in accuracy of −2.09 percentage points on average. Since EfficientNet-B0 and EfficientNet-B1 show very different improvements (−2.09 pp. vs. 0.02 pp.), model size alone is not a decisive factor to predict model improvement from synthetic data augmentation. More recent architectures with higher capacity show larger improvements on average. We also examined the effect of adding different amounts of synthetic images to the training data. The results are inconclusive with some models benefiting from more images (DreamBooth and LoRA), while others show a performance decline (GAN and fine-tuning). Since the behavior seems to be model-specific, we suggest to experiment with different settings, especially when inference times are short and new images can be generated quickly.
Class-specific metrics like precision, recall, and F1-score, have shown that synthetic data augmentation beats the baseline performance on most classes. This is especially true for classes with low frequency like viral and fungal pneumonia. In cases of imbalanced learning problems, synthetic data augmentation to rebalance class distributions provide a meaningful benefit. This technique performs better than a simple oversampling approach in this study, but comparisons to other sampling methods [
66,
67,
68] might be an interesting direction for further research. Although we could not confirm an overall improvement for a downstream classification task in this study, we did not cherry-pick model configurations where generative models exhibit sizable improvements over baseline models, but opted to give a comprehensive and robust outlook on the expected performance increase over many different scenarios instead. Furthermore, the usefulness of synthetic data exceeds the synthetic data augmentation approach, for example by using synthetic samples from a different institution for pre-training as shown in [
69] for 3D medical images.
Distinguishing between different pneumonia types has advantageous clinical implications, since they require different treatment regimes. Early indications of the cause can help facilitate effective drug treatments, for example in the use of antibiotics or antimicrobial drugs. If the pneumonia is caused by an infectious disease like COVID-19, controlling virus spread becomes an important task and early evidence of such diseases can be very helpful. Early diagnosis and appropriate treatment are essential to prevent complications and improve outcomes. In cases of immunosuppression, e.g., after bone marrow transplantation, a precise differentiation between different infections can be challenging and of crucial importance for the patient. A fast and correct identification is therefore necessary for the survival of the patient. The use of AI can help to break down and identify the correct infection which can often be very similar in appearance [
70].
Our work has limitations. We did not fine-tune the text encoder component of our Stable Diffusion models, which could lead to improved image quality, although recent literature seems to be indecisive on this effect [
23,
30]. We also chose rather simple prompts for text-conditioning of our Stable Diffusion models. Although more detailed prompts could lead to better results, we deliberately wanted to measure the effect of synthetic data augmentation without including further domain knowledge. We have already shown that incorporating domain knowledge can help to improve classification models on this study data [
14]. This also applies to filtering generated images for quality and plausibility before using them on the downstream classification task. Since human evaluation is infeasible for large quantities of images, employing another classification model to filter out bad images could be a promising future approach.
6. Conclusions
In this work, five different generative models for a small pneumonia chest X-ray dataset were evaluated, giving a quantitative and medical assessment of image quality and pathological plausibility. Furthermore, the usefulness of these models as part of a synthetic data augmentation on a downstream classification task was examined. We compare a GAN-based model with diffusion and latent diffusion models on five different classification model architectures to obtain a comprehensive overview of the expected performance gain for synthetic data augmentation. Images generated by the GAN model have shown the best quality in both quantitative and medical assessment and are most plausible as they outperform more recent architectures. Unconditional DDPM and fine-tuning of a Stable Diffusion model follow closely, while the recent fine-tuning approaches LoRA and DreamBooth did not achieve satisfying results in this study.
Interestingly, synthetic healthy findings often exhibit higher quality than pathological findings. Aside from being the most frequent class in this study, we assume that pathological patterns increase image complexity and vary far more than a normal healthy state. We found that the quantitative assessment based on the FID score leads to the same ranking in image quality than our medical assessment. Therefore, we rate the FID score as a solid measure for image quality. Furthermore, image quality and medical plausibility are closely correlated. We have shown that FID scores can fluctuate significantly during training for generative models and therefore advocate to track these metrics closely to decide for the best model checkpoints. In conclusion, this study gives a realistic estimation on the expected performance gain of synthetic data augmentation in a scarce data scenario and encourages many possible directions for further research.