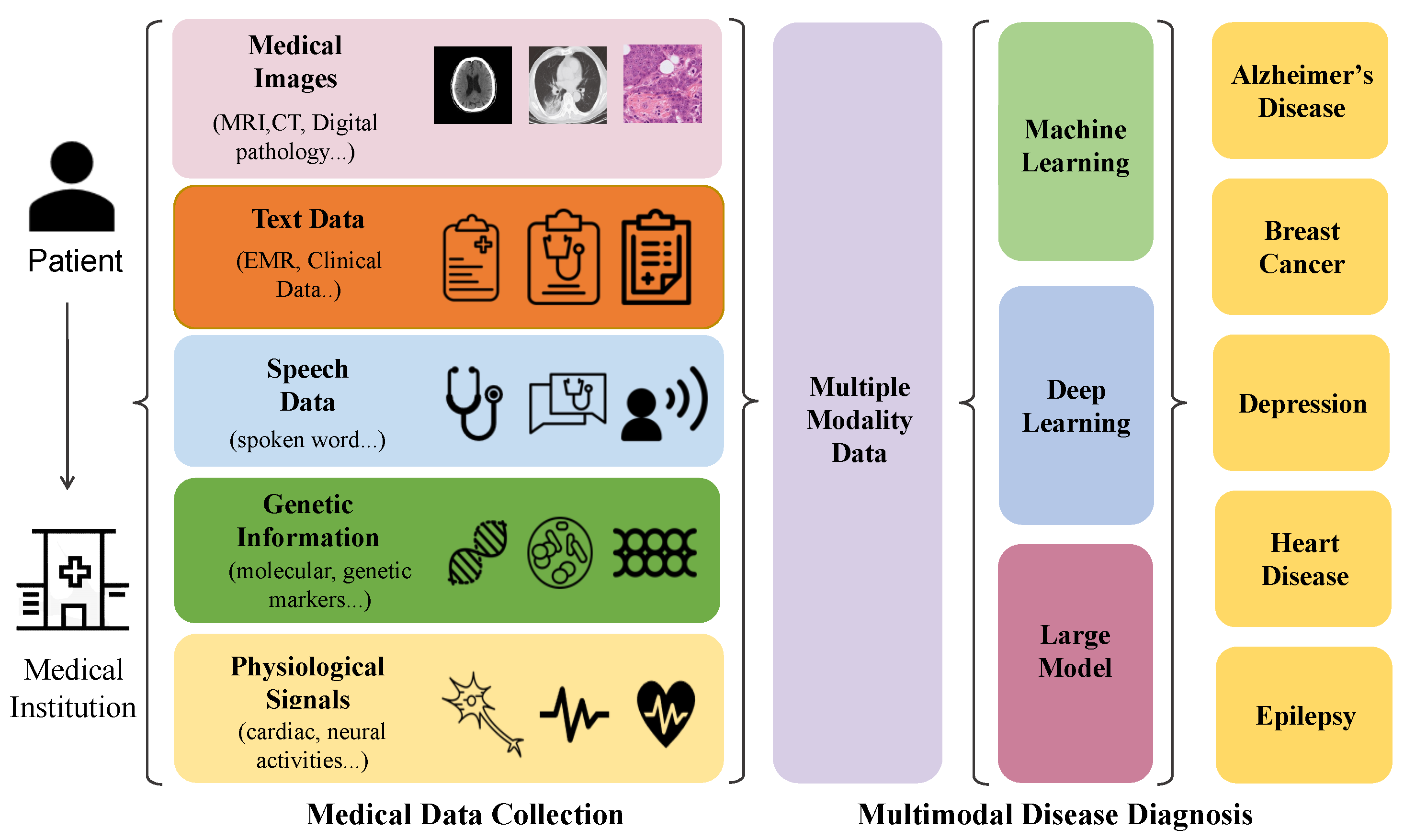
3.1. Diagnosis of Alzheimer’s Disease
Alzheimer’s disease constitutes a progressive neurodegenerative disorder, characterized by cognitive decline, memory impairment, and compromised communicative abilities. In the realm of AI-driven diagnostic investigations for Alzheimer’s disease, medical imaging modalities such as MRI and PET are universally recognized as indispensable tools. They offer profound insights into the alterations of brain structure and functionality, thus furnishing critical information for diagnosis. Concurrently, the analysis of speech patterns has also surfaced as a promising domain. Changes in language and communication frequently serve as precursors to cognitive deterioration, making them significant markers for early detection. This section delves into and evaluates the pertinent literature on automated Alzheimer’s disease diagnosis, leveraging MRI, PET, speech, and other multi-modal strategies. A consolidated synopsis of the model and its attributes is presented herein, with detailed elaborations provided in
Table 3.
Magnetic resonance imaging (MRI). MRI is pivotal in Alzheimer’s disease (AD) diagnostics, offering a non-invasive modality that provides intricate images capturing the brain’s structural and tissue details. There has been a substantial focus on harnessing morphological attributes from MRI scans as the central criterion for facilitating automated AD diagnosis. To illustrate, Li et al. [
52] initiate the process by pinpointing the hippocampal regions in structural MRI (sMRI) images that are productive for diagnosis, drawing on prior knowledge. Subsequently, they deploy a deep learning architecture to distill distinctive patterns pertinent to AD diagnosis. Building upon this, Lian et al. [
70] amalgamate a discriminative localization phase for brain atrophy with the subsequent stages of feature extraction and classification framework development. They introduce a Hierarchical Fully Convolutional Network (H-FCN) designed to autonomously and systematically discern patch-level and region-level indicative sites within the entire brain MRI scan. This model embraces a data-driven strategy that concurrently learns and amalgamates feature representations spanning multiple scales—from patch to region to subject level—to formulate a comprehensive AD diagnostic model. Addressing the nuances of brain atrophy, which pose significant diagnostic challenges in MRI imaging, Zhu et al. [
59] unveil DA-MIDL, a novel deep learning framework endowed with a dual attention mechanism. This mechanism is adept at singling out the most salient pathological locales for AD diagnosis. DA-MIDL is composed of a patch network replete with spatial attention blocks, an attention Multiple Instance Learning (MIL) pooling module, and an attention-aware global classifier. The patch network is engineered to extract salient structural features from myriad local sMRI patches disseminated throughout the brain. The attention MIL pooling phase is adept at assigning variable weights to patch-level features, orchestrating them into a holistic representation of the entire brain’s architecture. This global representation forms the foundation for the subsequent AD diagnostic classifier.
Furthermore, the quantification of hippocampal volume attrition has been recognized as a seminal marker for AD diagnosis. Uysal et al. leverage semi-automatic segmentation software ITK-SNAP to calculate hippocampal volume metrics. They construct a dataset incorporating parameters such as age, gender, diagnostic status, and volumetric data for left and right hippocampal regions. Utilizing this dataset, they apply machine learning algorithms to effectively differentiate between Alzheimer’s disease (AD), Mild Cognitive Impairment (MCI), and cognitively normal (CN) cohorts.
Positron emission tomography (PET). While MRI images primarily yield extensive data on brain structure, they fall short of providing insights at the molecular level. This is where Positron Emission Tomography (PET) imaging gains its prominence. As a molecular imaging technique, PET scrutinizes specific biological processes such as protein aggregation, metabolic rates, or receptor concentrations using radiolabeled tracers. PET imaging thus offers an intricate depiction of biological and metabolic dynamics within the brain and is routinely employed in diagnosing and monitoring Alzheimer’s disease (AD). In the study by Chen et al. [
60], a novel contrastive learning paradigm is introduced, utilizing brain 18F-FDG PET images to surmount the challenges associated with the paucity of data and the low signal-to-noise ratio, which are typical in PET images pertinent to AD prediction. They implement a data augmentation strategy to amplify the volume of training data, and they apply the adversarial loss to expand the distances between features of different classes while consolidating the similarities within the same class.
Furthermore, they develop a dual convolutional mixed attention module, fine-tuning the network’s proficiency in discerning diverse perceptual fields. By aligning the predictive outcomes of individual PET slices with clinical neuropsychological evaluations, they advance a diagnostic methodology conducive to refining AD diagnoses. Baydargil et al. [
71] deliver an unsupervised adversarial parallel model tailored for the anomaly analysis in AD, sharply delineating AD, mild cognitive impairment (MCI), and normal control groups. The model exhibits robust classification with rates and area under the curve (AUC) scores reaching 96.03% and 75.21%, respectively, underscoring its effective discriminative performance. Lu et al. lay the groundwork for a cutting-edge deep learning infrastructure, utilizing FDG-PET metabolic imaging to pinpoint subjects with symptomatic pre-AD in the MCI phase, setting them apart from other MCI cohorts (non-AD/non-progressive). They pioneer a multi-scale deep neural network that reports a classification precision of 82.51%, relying solely on a single-modal metric (FDG-PET metabolic data). Cheng et al. [
53] present an innovative classification scheme that amalgamates a two-dimensional Convolutional Neural Network (CNN) with a Recurrent Neural Network (RNN). Their strategy is oriented towards deconstructing 3D images into a succession of 2D slices to capture the features inherent to 3D PET imagery. Within this framework, they architect a hierarchical 2D cellular neural network tasked with the extraction of intra-slice features, while the Gated Recurrent Unit (GRU) within the RNN is deployed to elucidate inter-slice features that contribute to the final classification outcome.
Speech. The manifestation of Alzheimer’s disease (AD) in speech signals offers a distinctive avenue for diagnosis, as individuals with AD exhibit notable speech pattern alterations compared to those without the condition. Employing speech recognition technology for AD diagnostics is not only non-invasive and safe but also cost efficient, making it an appealing methodology for widespread application. Before the infusion of deep learning into the field, traditional approaches to speech analysis for AD diagnosis relied heavily on manual feature extraction. Techniques such as analysis of static features, utilization of feature sets like ComParE 2016 and eGeMAPS, as well as Mel-Frequency Cepstral Coefficients (MFCC), were common practices. These extracted features were then analyzed using machine learning classifiers, including logistic regression, random forests, and support vector machines, to distinguish between affected and healthy individuals. Studies by Hason et al. [
72], Hernández et al. [
73], and Yu et al. [
74] are examples of such research efforts.
With the advent of deep learning, there has been a paradigm shift in research methodologies for AD diagnosis. Deep learning techniques have taken precedence, given their ability to automatically extract complex patterns from raw data without the need for manual feature selection. In this context, Lopez et al. [
55] have made strides in early AD detection by implementing classical Multilayer Perceptrons (MLPs) and Convolutional Neural Networks (CNNs), illustrating the potential of deep learning in enhancing diagnostic accuracy. Further advancing the field, Liu et al. [
75] leveraged an Automatic Speech Recognition (ASR) model to derive speaker-independent bottleneck features, which are highly discriminative and robust. They coupled this with a CNN for modeling local context and an RNN for capturing the global context within speech. An attention mechanism was integrated to selectively focus on the most salient features for AD detection, improving the model’s interpretability and effectiveness. Additionally, Bertini et al. [
76] introduced an end-to-end model for AD detection, innovatively applying SpecAugment [
77] for data augmentation to enhance the robustness and generalizability of the model against variability in speech data. They then utilized the auDeep [
78] autoencoder, followed by fully connected layers for feature learning and classification, streamlining the process from raw speech input to the diagnostic output. This end-to-end approach simplifies the pipeline and potentially improves the model’s accuracy and applicability in clinical settings.
MRI-PET image fusion. The integration of MRI and PET imaging modalities has yielded a synergistic approach in medical diagnostics, particularly for disorders such as Alzheimer’s disease (AD). This technique of image fusion leverages the unique strengths of each imaging method to offer a more holistic representation of the brain’s structure and function. The pioneering work of Shi et al. [
79] introduced the multi-modal Stacked Denoising Predictive Network (MM-SDPN). This algorithm is structured in two phases specifically tailored to merge and learn from the feature representations of multi-modal neuroimaging data. This integration enhances the diagnostic process for Alzheimer’s disease, offering a deepened insight into the complex interactions between different types of brain changes associated with the disease. Sharma et al. [
80] took a different approach, utilizing wavelet packet transform as their method of fusing MRI and PET images. Their methodology involves an eight-layer Convolutional Neural Network (CNN) that meticulously extracts features across multiple layers. The extracted features are then processed through an ensemble of non-iterative Random Vector Functional Link (RVFL) networks. This ensemble strategy aims to robustly capture the intricate patterns from the fused data for accurate AD diagnosis.
Further advancing the field, Zhou et al. [
81] proposed a unique method for latent representation learning that encompasses data from various modalities, including MRI, PET, and genetic information. Their approach focuses on deducing latent representations and then projects these representations into the label space for diagnostic purposes. This technique underscores the potential of combining structural, functional, and biological data to enhance the accuracy of Alzheimer’s disease diagnostics. Addressing the potential issue of overfitting when dealing with the fusion of high-dimensional data, Ning et al. [
72] developed a relation-induced multi-modal shared representation learning approach. Their model is an integrative framework that combines the processes of representation learning, dimensionality reduction, and classifier design. It operates by learning bidirectional mappings between the original feature space and a shared representation space, thereby distilling the essence of multi-modal inputs into a cohesive, shared format that is conducive to diagnostic analysis. These studies illustrate a growing trend in leveraging sophisticated computational models and algorithms to enhance the accuracy and reliability of Alzheimer’s disease diagnostics by capitalizing on complementary information from multiple imaging modalities.
Speech–Text fusion. The nuanced extraction of acoustic features from speech datasets, coupled with the semantic analysis of textual data, fosters an enriched comprehension of Alzheimer’s disease (AD). By amalgamating speech and text data, a more extensive spectrum of AD-related features is captured, bolstering the diagnostic accuracy for this condition. Historically, the nascent stages of AD research leveraged machine learning techniques for analytical purposes. Shah et al. [
42] focused on the extraction of word-level duration features, datasets on pause rates, and measures of speech clarity. They explored a variety of models, such as logistic regression, random forest, support vector machine (SVM), extreme gradient boosting, and neural networks in isolation and in combination, targeting both classification and regression tasks. Martinc et al. [
43] commenced with spectrum subtraction for noise abatement, progressing to the use of a bag-of-n-grams approach for textual feature extraction. Concurrently, they extracted eGeMAPS features from speech data. A suite of classifiers, including XGBoost, SVM, random forest, logistic regression, and linear discriminant classifiers, was then deployed for classification tasks.
In the landscape of recent advancements, deep learning techniques have increasingly been harnessed for the automated diagnosis of Alzheimer’s disease. Cai et al. [
82] applied Graph Neural Networks (GNNs) for the extraction of textual features and introduced audio data by utilizing the WavLM model to extract salient audio features. They then integrated these features with text features via various methodologies. Mei et al. [
83] extracted a plethora of features comprising static acoustic features, the ComParE 2016 feature set, the eGeMAPS feature set, along with feature vectors from the wav2vec2 pre-trained model, and the Hubert pre-trained model for AD detection. They meticulously fine-tuned the wav2vec2.0 model on speech from assorted frequency bands, culminating in a remarkable accuracy of 87% and an RSME of 3.727. Agbavor et al. [
84] procured deep representation features through data2vec and wav2vec2, subsequently refining an end-to-end model with fully connected layers for enhanced AD detection efficacy.
Other models. A diverse array of molecular and multi-omics approaches, including RNA-seq, single nucleotide polymorphisms (SNPs), protein sequences, and integrated omics data, have been employed to unravel the complexities of Alzheimer’s disease diagnosis. For instance, groundbreaking work by Li et al. [
84], Taeho et al. [
85], Xu et al. [
86], Javier et al. [
87], and Park et al. [
88] has significantly contributed to the field by leveraging these techniques. Further, Park et al. [
88] have pioneered a deep learning approach tailored for AD prediction that synergistically utilizes multiple heterogeneous omics data. In a similar vein, Golovanevsky et al. [
89] have devised a multi-modal Alzheimer’s Disease Diagnostic framework (MADDi), ingeniously combining neural networks with attention mechanisms to harness the power of imaging, genetic, and clinical data for enhanced AD diagnostic precision. In addition to these genomic and proteomic strategies, electrophysiological methods such as EEG have been instrumental in AD diagnosis. Notable research by Djemili et al. [
90], Pandya et al. [
91], Kim et al. [
92], along with studies cited as [
93], have demonstrated the utility of EEG in capturing the neurophysiological hallmarks of Alzheimer’s disease, adding a valuable dimension to the diagnostic toolkit.
Table 3.
Summary of different medical features for Alzheimer’s disease diagnosis.
Table 3.
Summary of different medical features for Alzheimer’s disease diagnosis.
| Literature | Feature Name | Modality | Dateset | Results |
|---|
| Li et al. [52] | Hippocampal morphology feature | MRI | ADNI | 0.939 (AUC) |
| Lian et al. [70] | Original MRI scan feature | MRI | ADNI | 0.9 (ACC); 0.95 (AUC:AD vs. NC) |
| Zhu et al. [59] | Patch proposals selected from the MRI scans | MRI | ADNI, AIBL | 0.9193 (ACC: AD vs. NC vs. MCI) 0.9287 (AUC) |
| Chen et al. [60] | optimized anchor data from brain 18F-FDG PET slices | PET | ADNI | 0.9193 (ACC: AD vs. NC vs. MCI) 0.9287 (AUC) |
| Baydargil et al. [71] | Original PET slices | PET | ADNI | 0.9603 (ACC: AD vs. NC vs. MCI) 0.7521 (AUC) |
| Cheng et al. [53] | a sequence of 2D slice groups from 3D PET | PET | ADNI | 0.9528 (AUC: AD vs. NC) |
| Shi et al. [79] | high-level features of MRI and PET | MRI, PET | ADNI | 0.9713 ± 0.0444 (ACC: AD vs. NC) |
| Sharma et al. [80] | Fused image by wavelet packet transform (WPT) | MRI, PET | ADNI | 0.9603 (ACC: AD vs. NC vs. MCI) 0.7521 (AUC) |
| Zhou et al. [81] | magnetic resonance imaging (MRI), positron emission tomography (PET), and genetic data | MRI, PET,
Gene | ADNI | - |
| Ning et al. [72] | magnetic resonance imaging (MRI) and positron emission tomography (PET) | MRI, PET | ADNI | 0.976 (AUC: AD vs. NC) 0.969 (ACC: AD vs. NC) |
| Li et al. [84] | RNA-seq | Gene-based | GEO | 0.859 (AUC), 0.781 (ACC) |
| Taeho et al. [85] | SNP | Gene-based | ADNI | 0.82 (AAUC) |
| Xu et al. [86] | protein | sequence Gene-based | UniProt | 0.857 (ACC) |
| Javier et al. [87] | genetic variation data | Gene-based | ADNI | 0.719 (ACC) |
| Park et al. [88] | Multi-omics data | Gene-based | GEO | 0.823 (ACC) |
| Golovanevsky et al. [89] | imaging, genetic, and clinical data | Gene-based | GEO | 0.9688 (ACC) |
| Djemili et al. [90] | statistical characteristics (1. Maximum value in each IMF. 2. Minimum value in each IMF. Mean of the absolute values in each IMF. 4. Standard deviation in each IMF.) | EEG | Bonn dataset | The classification accuracy for normal and abrupt cessation electroencephalogram (EEG) signals is 1, while the classification accuracy for intermittent and abrupt cessation EEG signals reaches 0.977 |
| Pandya et al. [91] | Amplitude, period and waveform offset of K-Complex | EEG | Private dataset | - |
| Kim et al. [92] | EEG segment with respect to RP(Absolute power of EEG signals in three different frequency bands) | EEG | Private dataset | 0.75 (ACC) |
| Deepthi et al. [93] | Frequency domain features extracted by Fast Fourier Transform (FFT) | EEG | ADNI | - |
| Hason et al. [72] | MFCC | speech | ADReSS | Accuracy: 0.822 |
| Hernández et al. [73] | Speech duration, descriptive statistical variables | specch | private dataset | Accuracy: 0.8 |
| Yu et al. [74] | Based on phoneme characteristics, pronunciation coordination characteristics, and pitch variance | speech | private dataset | Accuracy: 0.93 |
| Lopez et al. [55] | Linear features include spectral domain features and time domain features, such as harmonicity, spectrum centroid, formants, etc. Nonlinear characteristics include fractal dimension, permutation entropy, multi-scale permutation entropy, etc. | speech | private dataset | Accuracy: 0.89 |
| Liu et al. [75] | Bottleneck feature vector (depth representation feature) | speech | Dementia-
Bank Pitt | F1: 0.7802 |
| Bertini et al. [76] | spectrogram | specch | Dementia-
Bank Pitt | Accuracy is 0.933, F1 score is 0.885 |
| Shah et al. [42] | Word-level duration feature set, pause rate data set, speech intelligibility feature set | speech, text | ADReSS-M | Accuracy: 0.696, RMSE: 4.8 |
| Martinc et al. [43] | bag-of-n-grams features (text)
eGeMAPS feature set (voice) | speech, text | Dementia-
Bank Pit | Accuracy: 0.9167 |
| Cai et al. [82] | GNN (text features)
WavLM (voice features) | Speech, text | Dementia-
Bank Pit | Accuracy: 0.8484 ± 0.0544 |
| Mei et al. [83] | Silent characteristics
ComParE 2016 feature set, eGeMAPS feature set
wav2vec2 pre-trained model feature vector
Hubert pre-trained model feature vector | Speech, text | AADReSS-M | Accuracy: 0.87, RMSE: 3.727 |
| Agbavor et al. [84] | data2vec, wav2vec2 | Speech, text | ADReSSo | F1: 0.728, RMSE: 3.493 |
3.2. Diagnosis of Breast Cancer
Breast cancer, originating in the breast cell tissue, stands as a pivotal health challenge for individuals across the globe. The key to enhancing survival and ensuring a better quality of life for those impacted by this disease lies in early detection and an integrated approach to treatment, involving a diverse team of medical professionals. The conventional diagnostic toolkit for breast cancer includes mammography, which is instrumental in visualizing breast tissue and identifying any irregularities that may indicate the presence of cancerous cells. Clinical breast exams conducted by healthcare professionals also play a significant role in early detection, as they involve a thorough palpation of the breast tissue to detect lumps or other changes. Additionally, gene screening is becoming increasingly important in breast cancer diagnosis, particularly for women with a family history of the disease, as it can identify inherited genetic mutations that may elevate the risk of breast cancer, such as mutations in the BRCA1 and BRCA2 genes. In this section, the diagnostic methodologies driven by the aforementioned modalities are rigorously explored and demonstrated. To provide a clear and concise representation of the various models and their attributes, reference is made to the details encapsulated in the accompanying tables, labeled as
Table 4. These tables present a summarized outlook of the models, delineating their features, performance metrics, and other pertinent details that contribute to the overarching domain of breast cancer diagnosis.
X-ray mammography. Breast Lesion Classification is a critical facet of breast cancer diagnosis, as it aims to accurately differentiate between benign and malignant lesions discovered during screenings. X-ray mammography remains the cornerstone of early breast cancer detection, enabling physicians to spot minuscule masses or calcifications that could indicate the presence of cancer cells within the breast tissue. To augment the diagnostic efficiency for breast lesions, Al-antari et al. [
94] have presented a comprehensive Computer-Aided Diagnosis (CAD) system that harnesses the power of deep learning, leveraging data from the DDSM and INbreast databases, which are prominent digital mammography datasets. The innovation began with the utilization of a You Only Look Once (YOLO) [
95] deep learning detector specifically calibrated for the identification of breast lesions across whole mammograms. Subsequently, Al-antari et al. assessed and fine-tuned three deep learning classifiers—the standard feedforward CNN, ResNet-50, and InceptionResNet-V2—for the nuanced task of breast lesion classification.
Furthering the advancement in this domain, Yeman et al. [
96] introduced an inventive approach employing a parallel deep Convolutional Neural Network (CNN) designed to analyze and learn from the symmetrical deep features extracted from the bilateral views of breast X-ray images. They innovatively computed the probability of pixels being part of a lesion by examining the local line and gradient direction features distribution, which then pinpointed the centers of suspected lesions. A global threshold was applied to these likelihood images to discern potential lesion-bearing regions. Ensuring symmetry, right and left breast X-ray images were horizontally flipped for congruent orientation, and the analysis proceeded with patched images fed into two mirrored deep CNN structures. The concatenated deep features from this twin-CNN setup were introduced into a Neural Network (NN) classifier, which achieved a remarkable prediction accuracy rate of 93.33%. In another groundbreaking work, Riyadh et al. [
97] conceived a novel mixed deep learning Computer-Aided Diagnosis system for breast lesions, which combined a backbone residual deep learning network to generate profound features with a transformer that incorporates self-attention mechanisms for the classification of cancer. This innovative model achieved a perfect 100% accuracy rate for binary classification and an impressive 95.80% for multi-class prediction tasks, a testament to the potential of mixed AI models in discerning between benign and malignant breast tissues with high precision.
Magnetic resonance imaging. Breast MRI is a powerful diagnostic tool that excels in providing detailed insights into breast cancer lesions, surpassing other imaging modalities in delivering precise evaluations of lesion size, location, and type. The robust magnetic field and non-ionizing radiation technique of MRI make it a choice modality for comprehensive breast cancer assessment. Abunasser et al. [
98] have made significant strides in the realm of breast MRI by training six advanced deep learning models, each with the capability to classify eight specific types of breast cancer, encompassing both benign and malignant forms. Their study incorporated a diverse set of models including their own proposed Breast Cancer Neural Network (BCNN), as well as Xception, InceptionV3, VGG16, MobileNet, and ResNet50, all fine-tuned to analyze MRI images for this purpose. These models demonstrated remarkable accuracy in their classification tasks, with rates of 97.54%, 95.33%, 98.14%, 97.67%, 93.98%, and 98.28% respectively, showcasing their potential to serve as reliable diagnostic aides. Complementing these efforts, Huang et al. [
99] embarked on a comprehensive study involving the extraction of an extensive array of 4198 radiomic features from pre-biopsy multiparametric MRI datasets, which included dynamic contrast-enhanced T1-weighted images, fat-suppressed T2-weighted images, and apparent diffusion coefficient maps. In their pursuit of optimal feature selection, they employed a suite of methodologies such as the Least Absolute Shrinkage and Selection Operator (LASSO), Recursive Feature Elimination (RFE), Maximum Relevance Minimum Redundancy (mRMR), Boruta, and Pearson correlation analysis. Leveraging these strategically chosen features, Huang et al. proceeded to construct 120 diagnostic models that varied by classification algorithms, MRI sequence-segmented feature sets, and the employed selection strategies. These models were adeptly designed to not just categorize breast cancer lesions but also to predict cancer molecular subtypes and androgen receptor expression, potentially offering a nuanced approach to personalized cancer care.
Ultrasound images. The field of medical imaging for breast cancer diagnosis has been greatly enhanced by the incorporation of artificial intelligence, with ultrasound imaging being a key focus due to its safety and non-invasive nature. Jabeen et al. [
100] introduced a cutting-edge classification framework specifically designed for ultrasound images, which effectively combines the prowess of deep learning with optimal feature selection techniques. This framework is composed of a structured five-step process: (i) Data augmentation is applied to expand the dataset, thereby providing a more robust foundation for training Convolutional Neural Network (CNN) models. (ii) The pre-trained DarkNet-53 model is adapted by modifying its output layer to align with the categories of the augmented dataset. (iii) Transfer learning is employed to train this modified model, with feature extraction carried out from the global average pooling layer. (iv) Two enhanced optimization algorithms, the Improved Differential Evaluation (RDE) and Improved Grey Wolf (RGW), are utilized for the selection of the most discriminative features. (v) A novel, probability-based sequential method is used to combine these optimally selected features, followed by the application of machine learning algorithms for the final classification task. The implementation of this framework on the Augmented Breast Ultrasound Images (BUSI) dataset resulted in an impressive highest accuracy of 99.1%, demonstrating its potential to significantly improve diagnostic processes.
Building on the momentum of innovation in the field, Ragab et al. [
101] spearheaded the development of an Integrated Deep Learning Clinical Decision Support System for Breast Cancer Diagnosis and Classification (EDLCDS-BCDC). This innovative technology is engineered to detect the presence of cancer through the analysis of ultrasound images. The process involves an initial preprocessing stage using Wiener filtering and contrast enhancement to prepare the images. Image segmentation is then carried out using the Chaos Krill Herd Algorithm (CKHA) and Kapur Entropy (KE). The feature extraction is performed through an ensemble of three sophisticated deep-learning models, namely VGG-16, VGG-19, and SqueezeNet. The final stage of the classification process employs the Cat Swarm Optimization (CSO) algorithm to optimize a Multi-Layer Perceptron (MLP) model, ensuring precise categorization of the cancer images. Both these studies showcase the innovative intersection of deep learning and optimization algorithms in improving the accuracy and efficiency of breast cancer classification using ultrasound imaging.
Medical text data. The use of advanced natural language processing (NLP) techniques to analyze and classify medical data, including patient self-reports and medical records, has become increasingly prevalent in breast cancer research. Leveraging the power of these techniques can provide valuable insights and assist in the early detection and treatment of breast cancer. Kumar et al. [
102] tailored a BERT-based model to specifically address the classification of breast cancer-related posts on Twitter, as described in Shared Task 8 of SMM4H-2021. Their approach was to employ BlueBERT [
103], which is pre-trained on a comprehensive biomedical corpus acquired from PubMed, enhancing the model’s understanding of medical terminology and context. To bolster the model’s resilience against adversarial inputs, they incorporated gradient-based adversarial training, which ultimately resulted in the model achieving F1 scores of 0.8625 on the development set and 0.8501 on the test set, reflecting high accuracy in the automatic classification of breast cancer mentions in social media posts.
Further innovations in NLP, as seen in the works of Chen et al. [
104] and Zhou et al. [
105], push the boundaries of model interpretability and domain-specific accuracy. Chen et al. [
104] took the capabilities of BERT further by integrating semantic trees into the model, thus constructing an interpretable neural network. They harnessed a capsule network with multiple attention heads to refine the semantic representations, while backpropagation and dynamic routing algorithms were implemented to provide local interpretability. This level of interpretability is particularly important in medical applications where understanding the reasoning behind a model’s prediction is as crucial as the prediction itself. Zhou et al. [
105] explored the benefits of pre-training BERT on a cancer-specific dataset, which aimed to enhance the model’s ability to extract breast cancer phenotypes from pathology reports and clinical records. Their findings underscore the significance of domain-specific pre-training, as it substantially improved the performance of the model, making it more attuned to the nuances of cancer-related data. Addtionally, Deng et al. [
106] investigated the potential assistance provided by advanced language models like GPT-4 in the context of breast cancer diagnosis. The authors emphasized GPT-4’s capability to rapidly mine crucial information from extensive medical records, which could potentially influence the diagnosis of breast cancer. By automating the extraction of key data points, GPT-4 could enhance the accuracy and efficiency of diagnostic procedures, supporting healthcare professionals in making informed decisions. These studies collectively highlight the transformative impact that state-of-the-art NLP models can have on the medical field, particularly in the realm of breast cancer diagnosis and classification.
Genetic data. Human cancer is a heterogeneous disease caused by stochastic cellular mutations and driven by various genomic alterations [
107,
108]. Currently, numerous research efforts are focused on utilizing genetic data and artificial intelligence algorithms to develop diagnostic models to enhance the clinical efficiency and accuracy of breast cancer diagnosis [
109,
110,
111]. Presently, artificial intelligence techniques in breast cancer diagnosis research based on genomics primarily focus on RNA-seq data, single nucleotide polymorphisms (SNPs), protein sequences, and the integration of multi-omics data. (1) RNA-seq. Xu et al. [
112] proposed a multi-granularity cascade forest (gcForest) for predicting four subtypes of breast cancer (Basal, Her2, Luminal A, and Luminal B). They compared the gcForest classifier with three different machine learning methods (KNN, SVM, and MLP). The results showed that gcForest showed a higher accuracy score of 92%. (2) MicroRNA. Sherafatian et al. [
50] employed three tree-based algorithms (Random Forest, Rpart, and tree bag) to classify breast cancer subtypes (Luminal, HER2-enriched, basal) using miRNA data from TCGA. The results showed that Rpart achieved the best classification performance. For the Luminal subtype, the accuracy, sensitivity, and specificity were 88.9%, 82.4%, and 95.4%, respectively. For the HER2-enriched subtype, the accuracy, sensitivity, and specificity were 90.2%, 93.9%, and 86.4%, respectively. For the basal subtype, the accuracy, sensitivity, and specificity were 84.5%, 75%, and 94%, respectively. (3) Multi-omics data. Mohaiminul et al. [
58] proposed a comprehensive deep-learning framework for classifying molecular subtypes of breast cancer. The framework utilized copy number alteration and gene expression data from the METABRIC. The results achieved an accuracy of 76.7% and an AUC of 83.8%.
Table 4.
Summary of different medical features for breast cancer diagnosis.
Table 4.
Summary of different medical features for breast cancer diagnosis.
| Literature | Feature Name | Modality | Dateset | Results |
|---|
| Al-Antari et al. [94] | Original X-ray mammographic data | X-ray | CBIS-DDSM and DDSM | 0.985 (ACC) |
| Yeman et al. [96] | Breast lesion detection from entire mammograms by object detection model | X-ray | DDSM and INbreast | ACC of three models: 94.50%, 95.83%, and 97.50% |
| Riyadh et al. [97] | Extracted patches centered on the points from the original X-ray | X-ray | General Electric, Siemens, and Hologic | 0.933 (AUC) |
| Abunasser et al. [98] | Original MRI data | MRI | Kaggle depository | 98.28 (F1-score) |
| Huang et al. [99] | multi-parametric MRI | MRI | Private dataset | Multilayer Perceptron (MLP): 0.907 (AUC) and 85.8% (ACC) |
| Jabeen et al. [100] | Original ultrasound images data | Ultrasound Images | BUSI dataset | 99.1% (ACC) |
| Ragab et al. [101] | Segmented regions from original | ultrasound images Ultrasound Images | - | 96.92% (ACC) |
| Kumar et al. [102], Peng et al. [103] | Word embedding | Text | witter self-report | F1: 0.8501 |
| Chen et al. [104] | Word embedding, syntactic structure | Text | Shanghai Ruijin Hospital Molybdenum Mammography X-ray Report | Mi-P(%) = 91.58 Mi-R(%) = 91.58 Mi-F1(%) = 91.58 Ma-P(%) = 75.95 Ma-R(%) = 79.73 Ma-F1(%) = 77.14 |
| Zhou et al. [105] | mutil feature | Text | private dataset | exact match and lenient
match, macro-F1: 0.876, 0.904 |
| Xu et al. [112] | RNA-seq | Gene-based | Medical Records | - |
| Sherafatian et al. [50] | miRNA | Gene-based | TCGA | 92% (ACC) |
| Mohaiminul Islam M et al. [58] | Copy number alteration (CNA), RNA-seq | Gene-based | METABRIC | 76.7% (ACC), 83.8% (AUC) |
| Sun et al. [108] | Clinical, CNV, RNA-seq | Gene-based | METABRIC | 82% (AUC) |
3.3. Diagnosis of Depression
Depression is a common mental health disorder characterized by persistent feelings of sadness, hopelessness, and a lack of interest or pleasure in daily activities. It can affect a person’s thoughts, emotions, and physical well-being, often leading to challenges in daily functioning. Depression varies in severity, and its impact on individuals can range from mild to severe. In the realm of diagnosis, text, speech, and EEG analysis have emerged as crucial tools for assessing and understanding depression. These modalities offer valuable insights into an individual’s mental state, providing a nuanced understanding of their emotional well-being. This section aims to delve into various approaches and methodologies related to the diagnosis of depression using these modalities. This section provides a summarized overview of the model and its features, as detailed in the accompanying
Table 5.
Medical text data. Aragon et al. [
58] introduced a sophisticated deep emotional attention model tailored for the detection of anorexia and depression. This model integrates nuanced sub-emotion embeddings with the advanced architectures of Convolutional Neural Networks (CNNs), Gated Recurrent Units (GRUs), and attention mechanisms to attain high predictive accuracy. Verma et al. [
113] explored depression detection through the analysis of tweet data, utilizing four established machine learning models: Naive Bayes, Support Vector Machines (SVMs), K-Nearest Neighbors (KNNs), and Random Forest. Of these, the Random Forest model demonstrated superior performance, achieving an impressive accuracy peak of 78%.
Furthering the field, Ghosh et al. [
114] adopted a novel deep multi-task learning strategy that simultaneously addresses emotion recognition and depression detection. Their findings suggest that the multi-tasking framework significantly boosts the efficacy of both tasks when learned concurrently. Xu et al. [
115] ventured into the domain of psychological health with the introduction of their Linguistic Landscape Model (LLM). This model was rigorously tested across a spectrum of tasks, including psychological stress classification, depression severity assessment, suicide ideation detection, and suicide risk evaluation. The empirical results underscored the LLM’s robust performance, placing it on par with the leading task-specific models in the field. Lastly, Qi et al. [
116] presented an all-encompassing benchmark that capitalizes on supervised learning techniques alongside the LLM framework, with a specific emphasis on the capabilities of the GPT series. Their research offers an in-depth analysis of these advanced LLMs, particularly in their application to cognitive distortion diagnosis and suicide risk stratification. This study not only highlights the models’ proficiency in capturing and interpreting complex emotional states but also provides a critical examination of their inherent potential and current limitations within the psychological domain.
Speech. From the initial forays into the realm of machine learning for depression diagnosis, a vast array of approaches has emerged. Liu et al. [
117] introduced a multi-task ensemble learning technique that utilizes speaker embeddings to facilitate depression classification. Long et al. [
118] devised an innovative multi-classifier system dedicated to depression recognition, distinguished by its synthesis of various speech types and emotional nuances. Jiang et al. [
119] developed the Ensemble Logistic Regression Model for Depression Detection (ELRDD), representing a significant stride in predictive modeling. Complementing this, Liu et al. [
120] proposed an inventive decision tree-based method for the fusion of speech segments, aimed at bolstering the accuracy of depression recognition.
As deep learning forges ahead, its methodologies are increasingly being adopted for diagnosing depression. Yin et al. [
121] presented a deep learning model that harnesses the strengths of parallel Convolutional Neural Networks (CNNs) and Transformers, balancing effective information extraction with computational tractability for depression detection. Adding to this body of work, Tasnim et al. [
122] examined the predictive utility of two acoustic feature sets—conventional handcrafted features and those derived from deep representations—in assessing depression severity through speech analysis. He et al. [
123] proposed a hybrid approach combining handcrafted elements with deep learning features to precisely gauge depression severity from speech. Dubagunta et al. [
124] conducted an exploration into methods for modeling speech source-related information in the context of depression, mindful of the potential neural physiological changes impacting vocal cord function. Zhao et al. [
125] sought to advance depression detection by tapping into inherent speech information, advocating for a Long Short-Term Memory (LSTM) model augmented with multi-head temporal attention. In a similar vein, Dong et al. [
126] recommended the application of pre-trained models for the extraction of deep Speaker Recognition (SR) and Speech Emotion Recognition (SER) features. Their approach synergizes these two profound speech features to capture the complementary data embedded within speaker voice characteristics and emotional variances.
EEG. The field of depression diagnosis has witnessed the burgeoning integration of electroencephalogram (EEG) and machine learning techniques, marking a pivotal research trajectory. In the reported literature [
127], a novel deep learning method named the Asymmetry Matrix Image (AMI) is introduced, which constructs spatial distribution maps from EEG signals by assessing the asymmetry between cerebral hemispheres. AMI has been shown to outperform traditional methods, delivering superior classification accuracy and enhancing the distinction between depression patients and healthy controls. Additional research [
128] delves into the utilization of nonlinear EEG signal features, such as Higuchi’s fractal dimension (HFD) and sample entropy (SampEn), which serve as indicators of signal complexity and irregularity. These nonlinear metrics have proven efficacious in segregating depression patients from healthy individuals, with high accuracy figures reported across a range of machine learning classifiers. In a different approach, literature [
129] focuses on power spectral features and asymmetry measures within the alpha, beta, delta, and theta frequency bands. Notably, findings suggest that asymmetries in the alpha2 and theta bands, particularly when analyzed with a Support Vector Machine (SVM), lead to higher diagnostic precision, with an accuracy rate of 88.33%. Explorations into the use of EEG data for depression diagnosis have also extended to single-channel and multi-channel formats [
130]. By refining feature selection and classification models via genetic algorithms, it has been discovered that single-channel analysis can effectively differentiate depression patients, underscoring the potential for employing portable EEG devices in preliminary depression screening despite a noted limitation in clinical generalizability due to small sample sizes. The literature [
131] investigates four feature selection techniques and five classification algorithms for processing EEG data. Through rigorous data preprocessing and feature extraction—identifying noise types and harnessing both linear and nonlinear features—the critical role of the data preparation phase is emphasized for achieving optimal classification accuracy.
A novel article [
47] presents a multi-modal feature fusion method that integrates EEG with eye movement (EM) signals, aiming to refine the identification of mild depression. The application of deep learning to fuse these multi-modal data sets enables real-time monitoring and detection of mild depression, with the fusion approach in the hidden layers yielding improved recognition accuracy over single-feature methods, and showcasing the benefits of combining diverse physiological signals. The melding of EEG and machine learning has advanced the diagnostic and treatment prediction capabilities for depression. Although challenges such as limited sample sizes and variability in feature extraction persist, forthcoming research endeavors are expected to tackle these issues, thereby enhancing the precision and utility of predictive models. Importantly, these advancements lay the groundwork for tailored treatment modalities, contributing to the delivery of more accurate and efficacious interventions for those suffering from depression.
Multi-modal. The landscape of depression diagnosis is rapidly evolving with the advent of multi-modal approaches, harnessing the rich data from speech, text, and video to create more nuanced and comprehensive diagnostic tools. Ehghaghi et al. [
132] embarked on an interpretable analysis to discern the distinct characteristics between dementia and depression. They pinpointed a spectrum of differentiators such as auditory anomalies, repetitive speech patterns, word retrieval struggles, coherence degradation, and variance in lexical density and richness—all of which are pivotal in distinguishing these disorders. Diep et al. [
133] ventured further by proposing a model that synthesizes deep learning features from both audio and text modalities, enriched with manually curated attributes deriving from domain expertise. Mao et al. [
134] introduced a novel approach using an attention-based multi-modal framework to generate a joint speech and text representation, specifically for the prediction of depression. Exploring the intersection of speech and video modalities, Jan et al. [
135] investigated the capability of cognitive machines and robots to autonomously recognize psychological states. By analyzing gestures and facial expressions, these intelligent systems aim to play a role in monitoring depressive states. Uddin et al. [
136] optimized the data processing workflow by segmenting audio and video into fixed-length units for input into a spatiotemporal network. This network is tailored to extract both spatial and temporal characteristics, with the introduction of dynamic feature descriptors like the Volume Local Directional Structure Pattern (VLDSP) to capture the nuances of facial dynamics.
Not content with dual-modal analyses, some studies have ambitiously integrated all three modalities—speech, text, and video—to push the boundaries of depression detection. Yang et al. [
137] contributed to this growing body of work by discussing a multi-modal depression analysis framework comprising deep convolutional neural networks (DCNNs) and deep neural networks (DNNs). This composite approach leverages the strengths of each modality, offering a more robust and potentially accurate detection system. The convergence of such diverse modalities represents a significant step forward in the field of mental health diagnostics. By combining distinct but complementary data sources, these integrated approaches aim to mirror the complex nature of depression more closely, offering promising directions for future research and potential clinical applications. The ultimate goal is to refine these tools for enhancing early detection and personalizing treatment strategies, thus providing a beacon of hope for individuals grappling with depression.
3.4. Diagnosis of Heart Disease
Heart diseases, particularly Cardiovascular Diseases (CVD), stand as the leading cause of death worldwide. Hypertrophic Cardiomyopathy (HCM) poses significant challenges due to the thickening of the left ventricular walls of the heart. The modern era has seen a paradigm shift in heart disease diagnosis, leveraging advanced technologies across various modalities. This chapter will diagnostic methods for heart disease using hypertrophic cardiomyopathy (HCM) as an example. We will gain a deeper understanding of HCM-assisted diagnostic techniques based on echocardiography, medical text data, and electrocardiograms (ECG) and explore other heart disease diagnostic methods based on genetic data. The comprehensive application of these diagnostic tools provides support for the early identification and treatment of heart disease and is of great significance for improving patient prognosis and quality of life. This section provides a summarized overview of the model and its features, as detailed in the accompanying
Table 6.
Echocardiography. Deep learning frameworks have shown remarkable promise in enhancing the accuracy and efficiency of heart disease detection and classification. Among these advancements, the work of Almadani et al. [
138] stands out with the introduction of the HCM Dynamic Echo, an end-to-end deep learning framework designed for the binary classification of echocardiography videos into hypertrophic cardiomyopathy (HCM) or normal categories. This system includes two analytical components: Branch 1, dubbed the Slow Path, which focuses on extracting spatial features, and Branch 2, known as the Fast Path, which is dedicated to capturing temporal structure information, thereby improving the accuracy of video recognition. They applied transfer learning and pre-trained HCM Dynamic Echo on the large Stanford EchoNet Dynamic Echocardiography dataset, enabling HCM detection in smaller echocardiography video datasets. In rigorous evaluations, HCM Dynamic Echo outperformed state-of-the-art baselines, with an accuracy of 93.13%, an F1 score of 92.98%, a Positive Predictive Value (PPV) of 94.64%, a specificity of 94.87%, and an Area Under the Curve (AUC) of 93.13%.
Parallel to these developments, other researchers have also made significant contributions to the field. For instance, Madani et al. [
139] developed a high-efficiency deep learning classifier for binary Left Ventricular Hypertrophy (LVH) diagnosis using echocardiography images. The core framework of their model included a U-Net for eliminating auxiliary information from image and a series of convolutional neural networks, resulting in an accuracy of 91.2%. To counter data scarcity, they proposed data augmentation using semi-supervised Generative Adversarial Networks (GANs). GANs demonstrated superior performance than traditional CNNs with limited data, attaining a test accuracy of 92.3%. Nasimova et al. [
140] introduced a deep convolutional neural network for classifying echocardiography videos as Dilated Cardiomyopathy or Hypertrophic Cardiomyopathy. Their study initially generated an Echo dataset from internet-sourced Echo videos and EchoNet database videos. The team trimmed the collected videos to 2–5 s to remove unnecessary echo information and redundant frames before segmenting them into 112 × 112 × 3 images for manual feature extraction. These images and extracted features were input into a six-layer CNN for classification, achieving a test accuracy of 98.2%.
Moreover, some studies have contributed to the field by applying deep learning models to diagnose various cardiac conditions from echocardiography. Zhang et al. [
141] utilized the VGG-16 model to automatically detect three diseases from echocardiography: Hypertrophic Cardiomyopathy, Pulmonary Arterial Hypertension, and Cardiac Amyloidosis. They trained separate networks for each disease, using three random images per video. The images were processed through the VGG-16 model with a fully connected layer featuring two output units, achieving an AUC of 93% and
p-value of 0.23 for HCM detection. Ghorbani et al. [
142] analyzed 3312 consecutive comprehensive non-stress echocardiography studies collected from June to December 2018. The process started with the first frame of each video, sampling 20 frames at intervals of 100 milliseconds. The Inception-Resnet-v1 network processed each frame individually, and the final prediction was determined by averaging the predictions from all individual frames. This method achieved an AUC-ROC of 0.75 and an F1 score of 0.57.
Medical text data. Sundaram et al. [
143] developed a Random Forest (RF) model to automatically identify patients with Hypertrophic Cardiomyopathy (HCM) using features extracted from Cardiac Magnetic Resonance (CMR) imaging reports. The Random Forest (RF) model attained an accuracy of 86% using 608 features and achieved 85% accuracy with 30 features. Mishra et al. [
144] introduced an innovative application within the medical Internet of Things (IoMT) domain. They utilized a Recurrent convolutional neural network (Rec-CONVnet) to accurately estimate the risk of heart disease. The system design compiles various data points such as age, gender, symptoms of chest discomfort, blood sugar levels, blood pressure (BP), and other relevant clinical factors. Through comprehensive simulations and evaluations, the Rec-CONVnet demonstrated remarkable performance, achieving an impressive F1 score of 97%. Jayasudha et al. [
145] designed a Social Water Cycle Driving Training Optimization (SWCDTO) ensemble classifier for heart disease detection. The classifier showed outstanding performance across specificity, accuracy, and sensitivity, reaching 95.84%, 94.80 and 95.36% in each metric. Levine et al. [
146] investigated the performance of a large model (GPT-3) in diagnosing and triaging diseases like heart disease. The findings indicated that GPT-3’s performance nearly approached that of professional medical practitioners.
Genetic data. Peng et al. [
147] employed a Support Vector Machine (SVM), Random Forest (RF), and Logistic Regression (LR) to develop a classification model for coronary atherosclerosis heart disease (CAD). This model utilized datasets GSE12288, GSE7638, and GSE66360 from the GEO database. The ROC curve analysis revealed for SVM, RF, and LR in validation to be 75.58%, 63.57%, and 63.95%, respectively. Their respective areas under the curve were 81.3% (95% CI 0.761–0.866,
p < 0.0001), 72.7% (95% CI 0.665–0.788,
p < 0.0001), and 78.3% (95% CI 0.725–0.841,
p < 0.0001). Liu et al. [
148] created a classification model for Coronary Artery Disease (CAD) using LASSO logistic regression, random forest, and SVM. They used data from the GEO dataset GSE113079, achieving an AUC of 97.1% in the training set and 98.9% in the testing set. Zhang et al. [
44] introduced the Integration Machine Learning (IML) algorithm, incorporating a SVM, neural network (NN), RF, gradient boosting machine (GBM), decision trees (DT), and LASSO. This algorithm was applied to classify patients with Acute Myocardial Infarction (AMI) and stable coronary artery disease (SCAD), using GEO datasets GSE60993, GSE62646, GSE48060, and GSE59867, achieving an AUC over 90%. Hou et al. [
149] utilized SVM for classifying CAD without heart failure (CAD-non HF), CAD complicated with heart failure (CAD-HF), and healthy controls, using GEO datasets GSE20681 and GSE59867. The study achieved an AUC of 0.944. Finally, Samadishadlou et al. [
150] applied SVM for classifying myocardial infarction (MI), stable CAD, and healthy individuals, using datasets GSE59867, GSE56609, and GSE54475 from GEO. Their model demonstrated an AUC-ROC of 96% and an accuracy of 94%.
Electrocardiogram. The integration of Convolutional Neural Networks (CNN) into the analysis of Electrocardiogram (ECG) data has marked a significant leap forward in detecting Hypertrophic Cardiomyopathy (HCM) and other cardiovascular diseases (CVDs) [
151]. Among the notable contributions, Tison et al. [
152] developed an automated and highly interpretable method for analyzing patient ECG features. This method processed and analyzed 36,186 ECG datum from the University of California, San Francisco (UCSF) database. Researchers utilized Hidden Markov Models (HMM) to extract ECG vector representations containing 725 features, which were then trained using CNNs to estimate cardiac structural and functional indices and classify diseases. Compared to traditional neural network models, this vectorized processing approach better retained meaningful features in ECGs, thus enhancing the interpretability and accuracy of diagnostic results. Similarly, Dai et al. [
151] used a deep CNN to classify five cardiovascular diseases (CVDs) using standard 12-lead ECG signals. The study utilized the public Physiobank (PTB) ECG database. The researchers have segmented ECG signals into different intervals—1 s, 2 s, and 3 s—without detecting individual waves, thus forming three distinct datasets. They applied ten-fold cross-validation on one-second-long ECG signals and tested on the other two datasets (two and three seconds long). The proposed CNN model achieved an accuracy, sensitivity, and specificity of 99.59%, 99.04%, and 99.87%, respectively, for one-second signals, demonstrating superior performance. For two-second signals using pre-trained models, the system achieved an overall accuracy, sensitivity, and specificity of 99.80%, 99.48%, and 99.93%. For three-second signal detection, the accuracies were 99.84%, sensitivity 99.52%, and specificity 99.95%. These results indicate that the proposed system achieved high performance while maintaining simplicity and flexibility, suggesting its potential for real-time application in medical settings.
Furthermore, Tison et al. [
153] highlighted the application value of AI-enhanced ECG (AI-ECG) in assessing disease states and treatment responses for obstructive HCM. The study noted that AI-ECG could extract more physiologically and pathophysiologically relevant information related to obstructive HCM from ECGs, surpassing traditional manual interpretation methods. Moreover, the study mentioned the potential of AI-ECG for remote monitoring through smartphone electrodes to assess disease states and treatment responses. The authors also foresaw the future application of this technology in medication adjustment and enhancing treatment safety.
Another impressive study is conducted by the Mayo Clinic [
154]: they used digital 12-lead ECGs from 2448 diagnosed HCM patients and 51,153 age and gender-matched non-HCM controls to train and validate a CNN. The algorithm performed impressively in adult HCM patient ECG detection, with an AUC of 0.96, sensitivity of 87%, and specificity of 90%. The algorithm’s performance in a test of 300 children and over 18,000 age and gender-matched controls was equally impressive: the HCM detection model achieved an AUC of 0.98, sensitivity of 92%, specificity of 95%, Positive Predictive Value (PPV) of 22%, and Negative Predictive Value (NPV) of 99%. The study found that the algorithm generally performed better in the adolescent group than in the pediatric group.
Table 6.
Summary of different medical features for heart disease diagnosis.
Table 6.
Summary of different medical features for heart disease diagnosis.
| Literature | Feature Name | Modality | Dataset | Results |
|---|
| Almadani et al. [138] | Echocardiography | echocar-
diogram videos | Stanford EchoNet- Dynamic echocardiogram dataset | ACC: 93.13%,
F1-score: 92.98%,
Positive Predictive Value (PPV): 94.64%,
specificity: 94.87%,
AUC: 93.13% |
| Madani et al. [139] | echocardiography | Original echocardiograms | Private dataset | 92.3% accuracy: binary left ventricular hypertrophy classification |
| Nasimova et al. [140] | Echocardiography | Clipped echocardiogram video frames | (1) EchoNet database; (2) Echo videos from the Internet | ACC: 98.2% (dilated cardiomyopathy vs. hyper-trophic cardiomy-opathy (HCM)) |
| Zhang et al. [141] | Echocardiography | Original echocardiograms | Private dataset | AUC: 0.93 |
| Ghorbani et al. [142] | Echocardiography | Cropped echocardiogram regions (inside of the scanning sector) | Private dataset | AUC: 0.75 |
| Sundaram et al. [143] | Word Embedding, Part of Speech (POS) | Text | CMR | 86% (ACC) for 608
features and 85% (ACC) for 30 features |
| Mishra et al. [144] | Word Embedding | Text | Real clinical records in hospital databases | 97% F1 score, FPR of 64.6%, accuracy of 96.4%, and accuracy of 76.2% |
| Levine et al. [146] | Multivariate Features | Text | Recruited participants | Brier score = 0.18 for disease, Brier score = 0.22 for triage |
| Peng et al. [147] | Gene-based | RNA-seq | GEO | SVM: 81.3% (ACC); RF: 72.7% (ACC); LR: 78.3% (ACC) |
| Liu et al. [148] | Gene-based | RNA-seq | GEO | Training: 97.1% (AUC), test: 98.9% (AUC) |
| Zhang et al. [44] | Gene-based | RNA-seq | GEO | 90% (AUC) |
| Hou et al. [149] | Gene-based | RNA-seq | GEO | 94.4% (AUC) |
| Samadishadlou et al. [150] | Gene-based | MicroRNA | GEO | 96% (AUC), 94% (ACC) |
| Dai et al. [151] | End-to-end Auto-learned Features | ECG | Physiobank (PTB) Public Dataset | Accuracy: 99.84%, Sensitivity: 99.52%, Specificity: 99.95% |
| Tison et al. [152] | 725 Features Extracted using Hidden Markov Models | ECG | UCSF Database | AUR: Range 0.94 to 0.77 |
| Tison et al. [153] | End-to-end Auto-learned Features | ECG | UCSF Database | - |
| Ko et al. [154] | End-to-end Auto-learned Features | ECG | Public Mayo Clinic Developed Database | AUC: 0.96, Sensitivity: 87%, Specificity: 90% |
3.5. Diagnosis of Epilepsy
Epilepsy, a prevalent neurological disorder affecting approximately 60 million people worldwide [
155], poses significant diagnostic challenges. A range of symptoms characterizes it, and an effective diagnosis requires a multidisciplinary approach. This article explores various diagnostic methods employed in epilepsy detection, utilizing advanced technology and medical imaging. This chapter will explore auxiliary diagnostic techniques for epilepsy based on images, medical text data, and electroencephalography (EEG). These methods play a crucial role in improving the accuracy and efficiency of epilepsy diagnosis, providing us with a new perspective to understand this complex disease and bringing better medical services to patients. This section provides a summarized overview of the model and its features, as detailed in the accompanying
Table 7.
Medical video. Using video data for computer-assisted diagnosis has become essential for the timely detection of epilepsy. Karácsony et al. [
156] employed clinical Motion Capture (MoCap) to quantitatively analyze seizure-related symptoms such as ictal head turning and upper limb automatisms, marking a pioneering discovery in differentiating epilepsy syndromes, providing clinical localization and lateralization information. Maia et al. [
157] applied a threshold-based approach to first detect regions of interest (beds) in video data, aligning them vertically for consistency, then utilized Convolutional Neural Networks and Multilayer Perceptrons to classify epileptic seizures, achieving 65% AUC. Achilles et al. [
158] recorded 52 seizures at 15 frames per second using infrared and depth imaging sensors, training distinct Deep Convolutional Neural Network architectures (CNNs) on video frames (one CNN for infrared frames, another for depth frames). Combining outputs from both networks, they achieved the prediction of ictal or interictal epilepsy phases, with their method demonstrating high sensitivity (87%) and specificity (81%) for generalized tonic-clonic seizures.
Building upon these advancements, Ahmedt-Aristizabal [
159] unveiled an innovative network approach that integrates 3D facial reconstruction with deep learning. The design of this approach aims to detect and measure orofacial semiotics in a collection of 20 seizure videos, featuring recordings from patients with temporal and extra-temporal lobe epilepsy. The developed network demonstrated its capability to differentiate between two types of epileptic seizures, achieving an average classification accuracy of 89%. It marks a significant advancement in computer vision and deep learning within non-contact systems, particularly for identifying common semiotics in real-world clinical environments. Significantly, this method departs from earlier epilepsy monitoring techniques by moving beyond the reliance on single-angle image information. In contrast, Kunekar et al. [
160] proposed improving accuracy by utilizing information from multiple modalities instead of relying solely on features from a single viewpoint. Ahmedt-Aristizabal et al. [
161] proposed a new modular, hierarchical, multi-modal system aimed at detecting and quantifying semiotic signs recorded in 2D monitoring videos. This method combines computer vision with deep learning architectures to learn semiotic features from facial, body, and hand movements.
MRI. MRI-generated 2D or 3D images enable a better understanding of the brain’s internal structure, pinpointing brain issues associated with epileptic seizures. fMRI has become indispensable tools in the detection and understanding of epileptic seizures by providing detailed images of the brain’s internal structure. Garner et al. [
162] applied a machine learning approach using a Random Forest classifier, trained with resting-state functional MRI (fMRI) data, to predict epilepsy outcomes. The model achieved a 69% accuracy rate in predicting epilepsy outcomes on the test set after 100 stratified cross-validation rounds, using 70% of resting-state fMRI scans for training and 30% for testing. Similarly, Sahebzamani et al. [
163] employed the Gram-Schmidt orthogonalization method alongside a unified tissue segmentation approach for segmenting brain tissues in MRI images. They calculated first-order statistical and Gray Level Co-occurrence Matrix (GLCM) texture features and trained SVM classifiers using features from either the entire brain or the hippocampus to diagnose epilepsy. This comprehensive segmentation and whole-brain analysis methodology yielded a 94% accuracy rate.
In the quest for early and accurate diagnosis, researchers like Si et al. [
164] have turned to diffusion MRI techniques to detect subtle brain changes in conditions such as Juvenile Myoclonic Epilepsy. They emphasized the importance of early diagnosis in Juvenile Myoclonic Epilepsy (JME), a disorder that predominantly affects adolescents and poses significant developmental challenges. They utilized two advanced diffusion MRI techniques—High Angular Resolution Diffusion Imaging (HARDI) and Neurite Orientation Dispersion and Density Imaging (NODDI)—to create connectivity matrices that capture subtle white matter changes. By adopting transfer learning, they trained sophisticated Convolutional Neural Network (CNN)-based models for JME detection. Pominova et al. [
165] explored various deep 3D neural architecture building blocks for epilepsy detection, using both structural and functional MRI data. They experimented with 12 different architectural variants of 3D convolution and 3D recurrent neural networks. Santoso et al. [
166] proposed a novel integrated Convolutional Neural Network approach for classifying brain abnormalities (epilepsy vs. non-epilepsy) using axial multi-sequence MR images. The model comprised base learners with distinct architectures and lower parameter counts. By aggregating the outputs and predictions of these base models (through methods like majority voting, weighted majority voting, and weighted averaging) and feeding them into a meta-learning process with a SVM, they significantly enhanced the final classification performance.
Medical text data. Hamid et al. [
167] showcased the potential to differentiate epileptic patients from those with psychogenic non-epileptic seizures (PNES). They developed an NLP tool based on an annotator modular pipeline to analyze electronic medical records, identifying grammatical structures and named entities. This algorithm was proficient in detecting concepts indicative of PNES and those negating its presence. Taking a different approach, Pevy and colleagues [
168] utilized written records of conversations between patients and doctors to distinguish between epileptic seizures and PNES. They employed an NLP toolkit to extract specific features of speech formulation efforts, such as hesitations, reformulations, and grammatical repairs, from these transcripts. The algorithm then trained machine learning classifiers with these features, enabling it to distinguish patients based on their verbal expression patterns. Connolly et al. [
169] further affirmed the effectiveness of NLP in differentiating among various epilepsy types, including partial epilepsy, generalized epilepsy, and unclassified epilepsy. By analyzing text features extracted from electronic medical records, their algorithm successfully classified different subtypes of epilepsy with remarkable accuracy.
EEG. Researchers frequently use CNN (Convolutional Neural Network) architectures, which can extract features automatically, unlike traditional machine learning classifiers that require manual extraction of features for detecting and classifying epileptic seizures effectively. Clarke et al. [
170] developed a deep Convolutional Neural Network (CNN) for detecting epileptic seizure discharges, trained using a dataset comprising over 6000 marked events from a group of 103 patients diagnosed with Idiopathic Generalized Epilepsy (IGE). This newly proposed automatic detection algorithm showcased exceptional performance in identifying epileptic seizures from clinical EEGs. The system achieved an impressive average sensitivity of 95% and kept the average false positive rate to just one per minute. These results indicate that AI-powered computer-assisted EEG analysis could significantly improve the speed and precision of EEG assessments, thereby potentially enhancing treatment outcomes for epilepsy patients. Fürbass et al. [
171] employed the Fast R-CNN method for object detection, using deep regression for localization estimation of EDs (negative peaks) and the UDA training process to handle noise and artefacts in EEG. The authors used EEG data from 590,000 epochs of 289 patients for unsupervised training and tested it against 100 proprietary datasets. The experimental results indicated that the DeepSpike algorithm attained a sensitivity of 89%, a specificity of 70%, and an overall accuracy rate of 80%, showcasing its high effectiveness in identifying EEG discharges. Thara et al. [
172] used a two-layer stacked bidirectional Long Short-Term Memory (LSTM) technique for detecting epileptic seizures. The researchers built a model with two LSTM layers, dropout and dense layers, and trained and optimized it using activation functions such as sigmoid and softmax, achieving good results with an accuracy of 99.89% on the training set and 99.08% on the test set. Yao et al. [
173] experimented with ten different and independently improved RNN (IndRNN) architectures, achieving the best accuracy with a 31-layer Dense IndRNN with attention (DIndRNN).
Multi-modality. Torres-Velázquez et al. [
174] evaluated the performance of multi-channel deep neural networks in Temporal Lobe Epilepsy (TLE) classification tasks under single and combined datasets. They trained, validated, and tested several multi-channel deep neural network models using brain structural indices from structural MRI, MRI-based region of interest correlation features, and personal demographic and cognitive data (PDC). Results indicated that PDC alone provided the most accurate TLE classification, followed by the combination of PDC with MRI-based brain structural indices. These findings affirm the potential of deep learning methods, like mDNN models, in TLE classification when combined with multiple datasets.
Table 7.
Summary of different medical features for epilepsy diagnosis.
Table 7.
Summary of different medical features for epilepsy diagnosis.
| Literature | Feature Name | Modality | Dataset | Results |
|---|
| Karácsony et al. [156] | Medical video | 2D + 3D video feature | Neuro-
Kinect | - |
| Maia et al. [157] | Medical video | Original Infrared video feature | Private data | 0.65 (AUC) |
| Achilles et al. [158] | Medical video | infrared and depth video frames | ADNI, AIBL | sensitivity (87%) specificity (81%) |
| Ahmedt-Aristizabal et al. [159] | Medical video | Regions of interest by 3D face reconstruction from the original video sequences | Private dataset | 0.89 (ACC) |
| Ahmedt-Aristizabal [161] | Medical video | 2D monitoring videos | Private dataset | 83.4 % (ACC: face); 80.1% (ACC: body) body; 69.3% (ACC:hand)e |
| Garner et al. [162] | MRI | functional magnetic resonance imaging (fMRI) data | REDCap | 0.69 (ACC) |
| Sahebzamani et al. [163] | MRI | first-order statistical and volumetric gray-level co-occurrence matrix (GLCM) texture features from structural MRI data | Private dataset | 0.94 (ACC) |
| Si et al. [164] | MRI | the connectivity matrix which can describe tiny changes in white matter | Private dataset | 75.2% (ACC) and the 0.839 (AUC) |
| Pominova et al. [165] | MRI | 3D + 4D MRI data | Private dataset | 0.73 (AUC) |
| Santos et al. [166] | MRI | axial multi-sequences of MRI | Private dataset | 86.3% (ACC)
90.75% (F1-score) |
| Hamid et al. [167] | stemming features, POS, bag of concepts | Text | VA national clinical
database | The accuracy, sensitivity, and F-score are 93%, 99%, and 96% |
| Pevy et al. [168] | Word embedding | Text | Recording, transcribing, and writing records of
interview corpora | 71% (ACC) |
| Connolly et al. [169] | N-gram | Text | DrWare-
house (DrWH) | 0.708 (F1) for partial epilepsy (PE), generalized epilepsy (GE), and unclassified epilepsy (UE), 0.899 (F1) for PE and GE |
| Clarke et al. [170] | End-to-end Auto-learned | EEG | Public Ad-hoc | Average Sensitivity:
95% |
| Fürbass et al. [171] | End-to-end Auto-learned | EEG | Private Dataset (Test); 590,000
Epochs from 289 Patients in Temple University’s Public EEG Corpus (Training) | Sensitivity: 89%, Specificity: 70%, Overall Accuracy: 80% |
| Thara et al. [172] | End-to-end Auto-learned | EEG | Private Dataset | Accuracy: 99.89% |
| Yao et al. [173] | End-to-end Auto-learned | EEG | CHB-MIT Dataset | Average Sensitivity: 88.80%, Specificity: 88.60%, Precision: 88.69% |
| Torres-Velázquez et al. [174] | Multi-modality | brain structure metrics from structural MRI, MRI-based region of interest correlation features, and personal demographic and cognitive data (PDC) | Private Dataset | Acc = 69.46% ± 20.82%, AUC = 70.00% ± 26.00% |
3.6. Discussion
Modality distinction. In our comprehensive review, we examine the different methods used to automatically diagnose five specific diseases: Alzheimer’s disease (AD), breast cancer, depression, heart disease, and epilepsy. The medical data produced from different disease diagnosis processes has commonalities, mainly encompassing image, text, genetic, signal, and voice modalities. Distinctive preferences for specific modalities exist across different diseases. Even within the realm of single medical imaging, nuanced differences become apparent. For Alzheimer’s disease diagnosis, Magnetic Resonance Imaging (MRI) and Positron Emission Tomography (PET) images emerge as the predominant modalities, supplemented by the inclusion of voice data. The widespread use of MRI and PET stems from their effectiveness in capturing the structural and functional brain changes associated with Alzheimer’s disease (AD). The unique characteristics of neurodegenerative alterations make these imaging modalities particularly suitable for early detection and monitoring of disease progression.
Contrastingly, in breast cancer diagnostics, a multifaceted approach involves genetic data, X-ray imaging, ultrasound, and a notable amount of textual information. The rationale behind this approach lies in the heterogeneity of breast cancer itself, necessitating a comprehensive analysis of genetic predispositions, coupled with various imaging techniques and textual data to enhance diagnostic accuracy. Each modality contributes valuable insights into different aspects of breast cancer pathology, collectively enhancing the overall diagnostic efficacy. In the context of depression diagnosis, the emphasis shifts toward textual data and Electroencephalogram (EEG). The reliance on text data could be attributed to the subjective nature of depression symptoms, requiring a nuanced analysis of linguistic patterns and sentiment. EEG captures brain wave activity and complements textual data by providing physiological markers that indicate depression.
For heart disease diagnosis, the prevalent modalities include echocardiography, electrocardiography, and medical texts. The dominance of ultrasound-based echocardiography comes from its ability to provide real-time images of the heart’s structure and function, which is essential for assessing cardiac health. Electrocardiography contributes information on the heart’s electrical activity, while medical texts further contextualize the diagnostic process. For epilepsy diagnostics, a comprehensive strategy incorporates Magnetic Resonance Imaging (MRI), video data capturing patient movements, Electroencephalogram (EEG), and relevant textual information. The utilization of these diverse modalities is driven by the intricate nature of epilepsy itself, demanding a thorough examination of various aspects. MRI provides structural insights, video data offers observations of seizures and associated movements, EEG captures electrical activity in the brain, while textual information contributes contextual details.
In conclusion, the selection of modalities for automated diagnosis is intricately tied to the unique characteristics and pathological features of each disease. Understanding the rationale behind the prevalence of specific modalities facilitates a targeted and effective approach to automated disease diagnosis.
Modality fusion. Contemporary diagnostic methodologies increasingly favour the integration of multi-modal approaches. The advantages of the multi-modal paradigm lie in its ability to provide a more comprehensive and accurate understanding of complex phenomena by integrating diverse data modalities. This approach enhances robustness, improves interpretability, and allows for personalized and optimized solutions across various domains.
In diagnosing Alzheimer’s Disease (AD), where subtle but significant changes in language patterns and cognitive function are markers, combining speech and text analysis is extremely valuable. This multi-modal approach adeptly captures the intricate linguistic nuances and potential confusion in communication exhibited by AD patients. Integrating genetic data and electroencephalogram (EEG) as supplementary information enriches the diagnostic process, addressing the multifaceted nature of AD symptoms and facilitating a more accurate and holistic understanding. In cancer research, there is a significant emphasis on combining imaging and genetic data. Since genetic mutations play a pivotal role in the development and progression of various types of cancer, identifying specific genetic alterations associated with different types of cancer can provide insights into their molecular mechanisms and potential therapeutic targets.
Besides, specific genetic mutations may present as unique visual patterns. For example, specific genetic alterations in breast cancer, such as those in the BRCA genes, may result in characteristic radiographic features observable in mammograms or other imaging modalities. Therefore, combining genetic data with medical imaging enhances our molecular-level understanding of cancer and supports the creation of tailored, accurate methods for its diagnosis and treatment. Depression diagnosis predominantly relies on speech modalities, with supplementary integration of text or video data. This emphasis on speech is justified by the distinct changes in vocal patterns and tone often exhibited by individuals with depression. Adding text or video data enhances the diagnostic process by providing extra information on the patient’s emotional and behavioural conditions.
For diagnosing heart disease, it’s common to combine ultrasound imaging with medical texts. The rationale behind this lies in the need to comprehensively assess both structural and functional aspects of the heart. Ultrasound provides real-time visualizations of cardiac anatomy, while medical texts offer additional clinical context, creating a synergistic diagnostic approach. Epilepsy diagnosis currently benefits from the mutual utilization of various imaging modalities, such as Magnetic Resonance Imaging (MRI) and Positron Emission Tomography (PET) images. This approach acknowledges the diverse epileptic manifestations and leverages the strengths of multiple imaging techniques to achieve a more comprehensive and accurate diagnosis. In essence, the choice of modalities for fusion explicitly correlates with the diverse manifestations of patients’ conditions. The reasonable multi-modal fusion approach can capture the intricacies of symptoms, ensuring a more nuanced and effective diagnostic outcome tailored to the specificities of each medical condition.
Performance improvement. The evolution of research in automated disease diagnosis is accompanied by the continual improvement of performance. This progression has transitioned from machine learning dominance to primary reliance on deep learning, complemented by innovative techniques such as attention mechanisms and transfer learning. Initially, disease diagnosis methods focused on developing feature engineering within machine learning studies, where manually identifying and selecting pertinent features was vital for the model’s performance. However, this process had limitations, often requiring domain expertise and not fully exploiting the richness of complex datasets. In response to these challenges, the subsequent embrace of deep learning has become a transformative force in medical diagnostics. The distinctive advantage of deep learning lies in its capability to automatically extract hierarchical and intricate features from raw data, eliminating the need for explicit feature engineering. This automated feature extraction significantly enhances the diagnostic model’s performance by allowing it to discern intricate patterns and relationships within the data.
Deep learning has improved the accuracy and efficiency of disease detection. Within the domain of deep learning for medical diagnostics, scholars have proposed innovative techniques to elevate model performance. Inspired by how we humans see, attention mechanisms in deep learning models allow a focus on areas within the data for better analysis. It mimics the human ability to prioritize relevant information, improving the model’s ability to capture subtle or critical features. Attention mechanisms have shown effectiveness in different medical imaging tasks, leading to diagnoses that are more precise and aware of the context. Transfer learning has also become a technique to overcome the issue of scarce medical data samples. In transfer learning, a model pre-trained on a large dataset, often from a related domain, is fine-tuned on a smaller target dataset, which is typically scarce in medical applications. This approach leverages the knowledge gained from the source domain to enhance the model’s performance on the target task, even when training samples are limited. Transfer learning has proven effective in scenarios where acquiring a large, labeled medical dataset is impractical, thus facilitating the development of robust diagnostic models. The evolution from traditional machine learning, reliant on explicit feature engineering, to deep learning, with its automated feature extraction capabilities, has significantly improved disease diagnosis models. Combining attention mechanisms with transfer learning highlights scholars’ dedication to enhancing model performance, improving interpretability, and tackling the problem of limited data in medical contexts. These advancements collectively contribute to the ongoing refinement and enhancement of state-of-the-art diagnostic systems.
Large model application. The emergence of large models in AI has revolutionized many industries, particularly in healthcare. These models, often trained on vast datasets, can analyze complex patterns that lead to more accurate and efficient disease diagnosis. With the increasing use of electronic health records and the integration of various data sources, medical institutions now have access to more information. This dataset comprises patient histories, symptomatology, and genetic profiles, among other details, offering a rich reservoir. Large models can analyze this data to discern patterns and correlations. Currently, most large-scale models in healthcare focus on text, analyzing medical records, discharge summaries, and other types of written data. However, there is potential for models to analyze additional forms of medical data, including images, voice recordings, genetic data, and physiological signals.
As technologies improve and datasets grow, we can expect to see more diverse applications of large models in healthcare. For example, image analysis models can process medical images such as X-rays or CT scans to detect diseases or lesions more accurately. The speech analysis model can process the patient’s speech records and extract useful information from them, such as the severity of symptoms or the development trend of the condition. Genetic analysis models can predict a patient’s response to specific drugs or disease risks based on their genomic data. The physiological signal analysis model can track the patient’s vital signs, like heart rate and blood pressure, identify any irregularities swiftly, and take appropriate action. Notably, some challenges need to be solved. One major challenge is data privacy. Training and refining large models necessitates significant data volumes, yet it is essential to safeguard the privacy and security of medical information. Creating strong encryption and access management systems is crucial for patient data. It’s imperative to address ethical considerations when integrating AI into healthcare practices. It is essential to ensure that AI algorithms do not discriminate against any particular group and that their use complies with ethical standards. Overall, the rise of large models in healthcare can contribute to improving patient outcomes and reduce the burden on the healthcare system in the future.