Establishment of a Human Immunocompetent 3D Tissue Model to Enable the Long-Term Examination of Biofilm–Tissue Interactions
Abstract
1. Introduction
| 3D Model System | 2D Model System |
|---|---|
| Improved proliferation and differentiation | Loss of cellular phenotype |
| Bacteria can surpass into deeper tissues | Change in morphology and functionality |
| Interaction with fibroblasts | Loss of cell signalling |
| Cell–cell and cell–ECM interactions | Standardizable |
2. Materials and Methods
2.1. Cell Culture
2.1.1. THP-1 Differentiation
2.1.2. Primary Fibroblasts
2.1.3. SIS-muc
2.2. Bacteria Culture
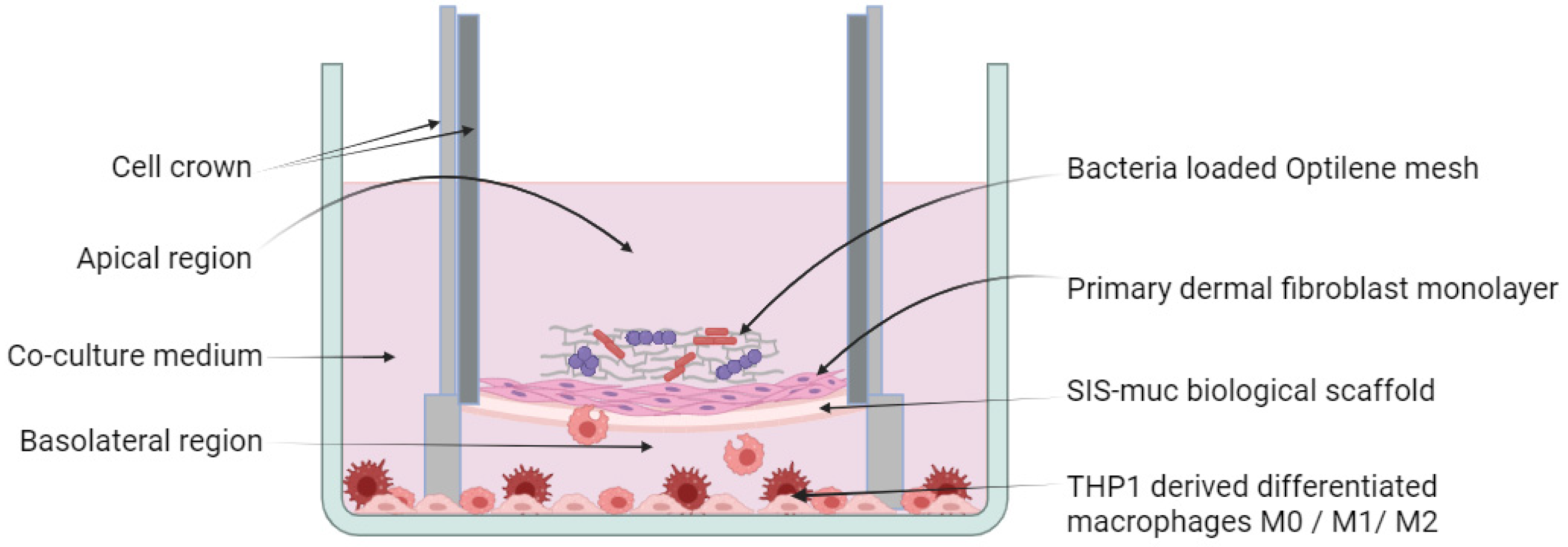
2.3. Establishment of a 3D Model
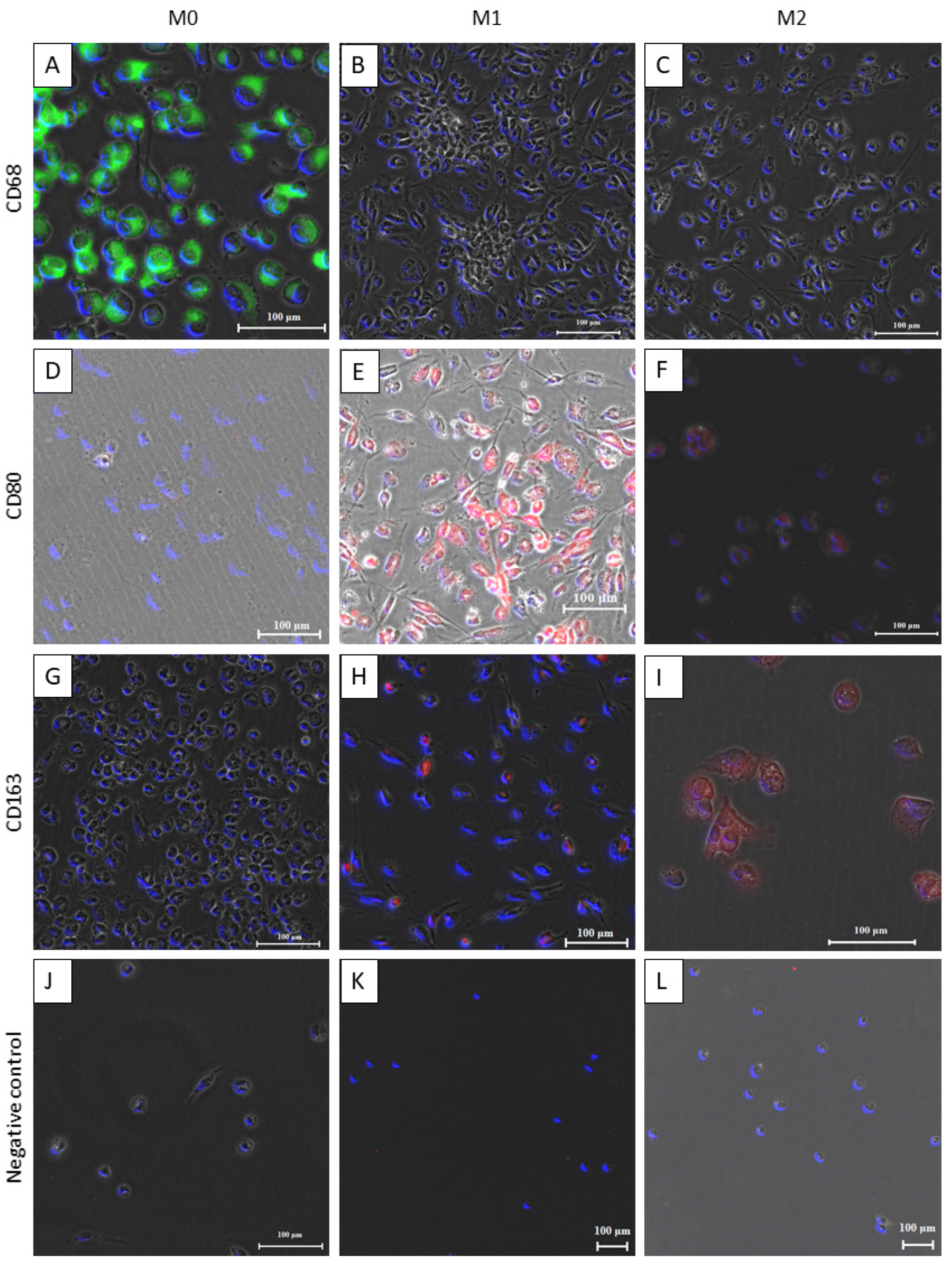
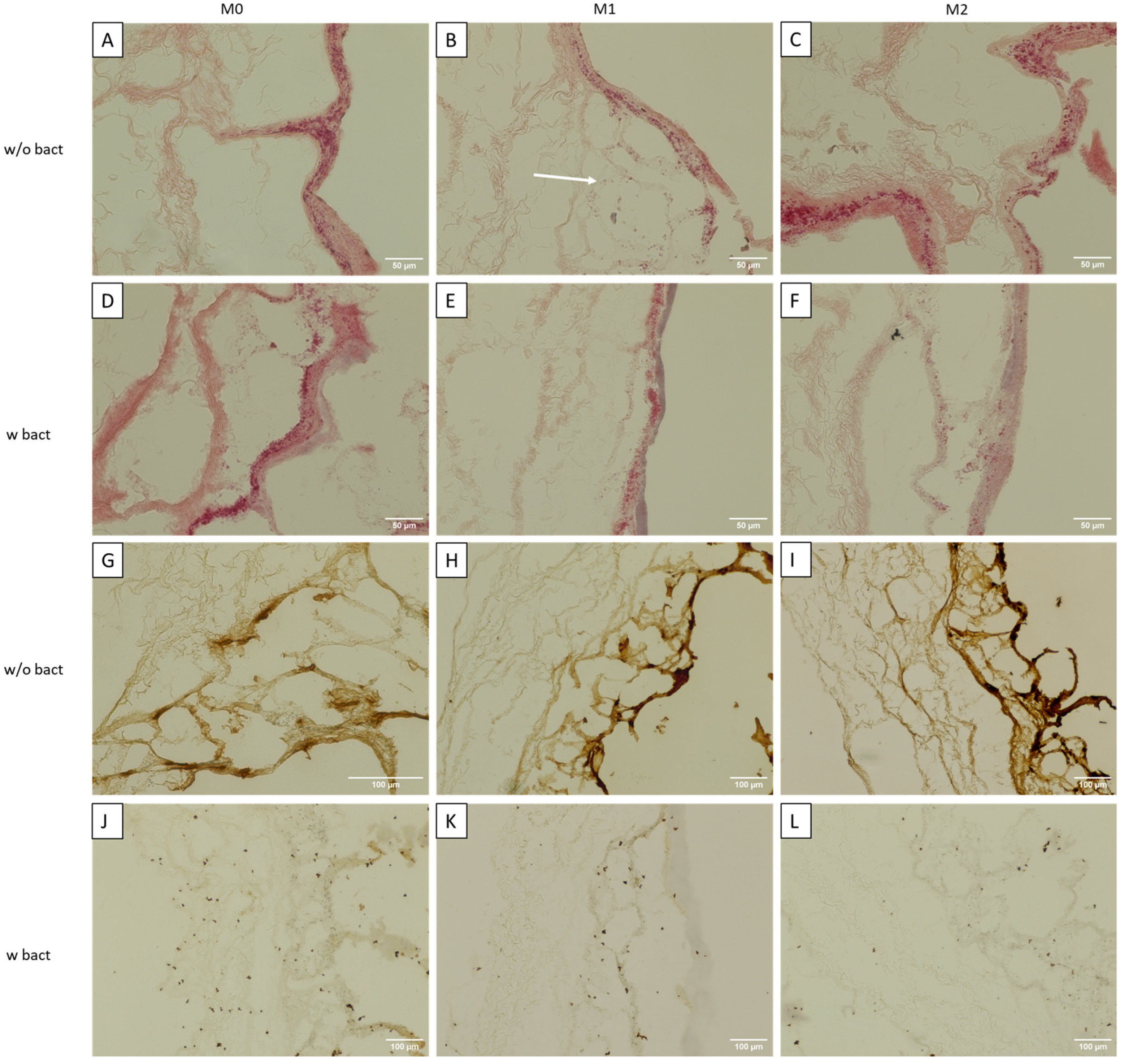
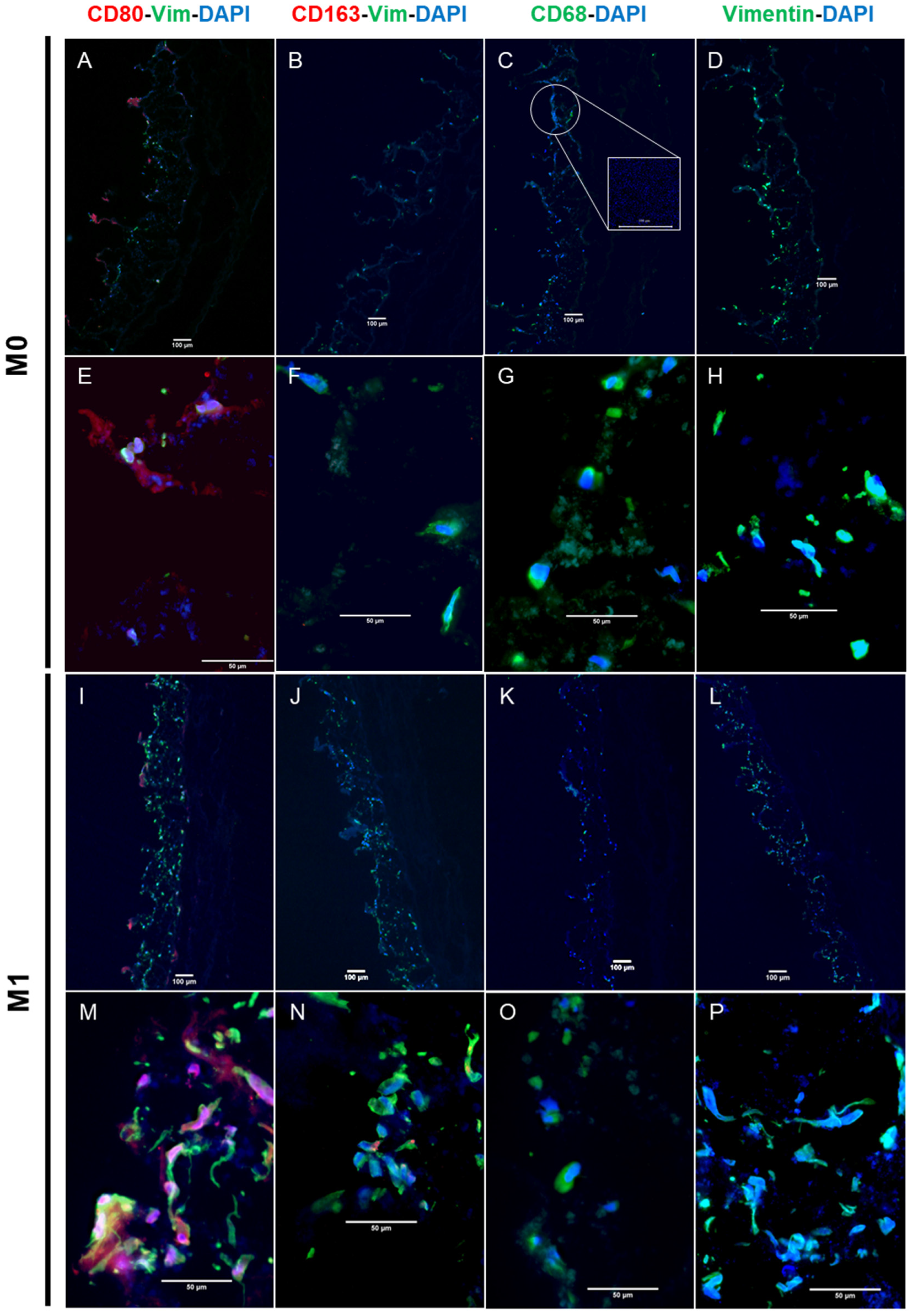
2.4. Qualitative Analysis
2.4.1. Immunohistochemical Analysis
2.4.2. Immunofluorescence Staining
2.5. Quantitative Analysis
2.5.1. OD Measurement
2.5.2. Agar Plates
2.5.3. ELISA
2.5.4. Image Processing and Quantification of IHC Images
2.6. Statistics
3. Results
3.1. Differentiation of THP-1 Monocyte-Like Cells into Macrophages
3.2. Immunocompetent Tissue Model (Bacteria)
3.3. Cytokine Secretion
4. Discussion
4.1. 3D Immunocompetent Model System
4.2. Immunocompetence
4.3. M0 Macrophage Models
4.4. M1 Macrophage Models
4.5. M2 Macrophage Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choi, E.; Murray, B.; Choi, S. Biofilm and Cancer: Interactions and Future Directions for Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 12836. [Google Scholar] [CrossRef]
- Radaic, A.; Ganther, S.; Kamarajan, P.; Grandis, J.; Yom, S.S.; Kapila, Y.L. Paradigm shift in the pathogenesis and treatment of oral cancer and other cancers focused on the oralome and antimicrobial-based therapeutics. Periodontol 2000 2021, 87, 76–93. [Google Scholar] [CrossRef]
- Karpinski, T.M. Role of Oral Microbiota in Cancer Development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Kakabadze, M.Z.; Paresishvili, T.; Karalashvili, L.; Chakhunashvili, D.; Kakabadze, Z. Oral microbiota and oral cancer: Review. Oncol. Rev. 2020, 14, 476. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef]
- Vestby, L.K.; Gronseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Maharjan, S.; Cecen, B.; Zhang, Y.S. 3D Immunocompetent Organ-on-a-Chip Models. Small Methods 2020, 4, 2000235. [Google Scholar] [CrossRef]
- Tran, F.; Klein, C.; Arlt, A.; Imm, S.; Knappe, E.; Simmons, A.; Rosenstiel, P.; Seibler, P. Stem Cells and Organoid Technology in Precision Medicine in Inflammation: Are We There Yet? Front. Immunol. 2020, 11, 573562. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zheng, J.; Hu, Q.; Liang, L.; Yang, D.; Cheng, Y.; Li, S.S.; Chen, L.J.; Yang, Y. Smart acoustic 3D cell construct assembly with high-resolution. Biofabrication 2022, 14, 045003. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Hu, X.; Yang, D.; Hu, Q.; Zheng, J.; Zhao, S.; Zhu, C.; Xiao, X.; Yang, Y. Acoustic quasi-periodic bioassembly based diverse stem cell arrangements for differentiation guidance. Lab Chip 2023, 23, 4413–4421. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Linke, K.; Schanz, J.; Hansmann, J.; Walles, T.; Brunner, H.; Mertsching, H. Engineered liver-like tissue on a capillarized matrix for applied research. Tissue Eng. 2007, 13, 2699–2707. [Google Scholar] [CrossRef] [PubMed]
- Mertsching, H.; Walles, T.; Hofmann, M.; Schanz, J.; Knapp, W.H. Engineering of a vascularized scaffold for artificial tissue and organ generation. Biomaterials 2005, 26, 6610–6617. [Google Scholar] [CrossRef]
- Franklin, R.A. Fibroblasts and macrophages: Collaborators in tissue homeostasis. Immunol. Rev. 2021, 302, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Van Linthout, S.; Miteva, K.; Tschope, C. Crosstalk between fibroblasts and inflammatory cells. Cardiovasc. Res. 2014, 102, 258–269. [Google Scholar] [CrossRef]
- Benoit, M.; Desnues, B.; Mege, J.L. Macrophage polarization in bacterial infections. J. Immunol. 2008, 181, 3733–3739. [Google Scholar] [CrossRef] [PubMed]
- Pidwill, G.R.; Gibson, J.F.; Cole, J.; Renshaw, S.A.; Foster, S.J. The Role of Macrophages in Staphylococcus aureus Infection. Front. Immunol. 2020, 11, 620339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zou, J.; Chen, R. An M0 macrophage-related prognostic model for hepatocellular carcinoma. BMC Cancer 2022, 22, 791. [Google Scholar] [CrossRef] [PubMed]
- Nietzer, S.; Baur, F.; Sieber, S.; Hansmann, J.; Schwarz, T.; Stoffer, C.; Hafner, H.; Gasser, M.; Waaga-Gasser, A.M.; Walles, H.; et al. Mimicking Metastases Including Tumor Stroma: A New Technique to Generate a Three-Dimensional Colorectal Cancer Model Based on a Biological Decellularized Intestinal Scaffold. Tissue Eng. Part C Methods 2016, 22, 621–635. [Google Scholar] [CrossRef]
- Wiese-Rischke, C.; Murkar, R.S.; Walles, H. Biological Models of the Lower Human Airways-Challenges and Special Requirements of Human 3D Barrier Models for Biomedical Research. Pharmaceutics 2021, 13, 2115. [Google Scholar] [CrossRef]
- Lee, J.; Cuddihy, M.J.; Kotov, N.A. Three-dimensional cell culture matrices: State of the art. Tissue Eng. Part B Rev. 2008, 14, 61–86. [Google Scholar] [CrossRef]
- Heydarian, M.; Schweinlin, M.; Schwarz, T.; Rawal, R.; Walles, H.; Metzger, M.; Rudel, T.; Kozjak-Pavlovic, V. Triple co-culture and perfusion bioreactor for studying the interaction between Neisseria gonorrhoeae and neutrophils: A novel 3D tissue model for bacterial infection and immunity. J. Tissue Eng. 2021, 12, 2041731420988802. [Google Scholar] [CrossRef]
- Kapalczynska, M.; Kolenda, T.; Przybyla, W.; Zajaczkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Blizniak, R.; Luczewski, L.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.P.; Young, H.; Hurlstone, A.; Wellbrock, C. Differentiation of THP1 Cells into Macrophages for Transwell Co-culture Assay with Melanoma Cells. Bio-Protocol 2015, 5, e1638. [Google Scholar] [CrossRef]
- Shang, L.; Yan, Y.; Zhan, Y.; Ke, X.; Shao, Y.; Liu, Y.; Yang, H.; Wang, S.; Dai, S.; Lu, J.; et al. A regulatory network involving Rpo, Gac and Rsm for nitrogen-fixing biofilm formation by Pseudomonas stutzeri. npj Biofilms Microbiomes 2021, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Gajewska, J.; Chajecka-Wierzchowska, W. Biofilm Formation Ability and Presence of Adhesion Genes among Coagulase-Negative and Coagulase-Positive Staphylococci Isolates from Raw Cow’s Milk. Pathogens 2020, 9, 654. [Google Scholar] [CrossRef]
- Parekh, R. Fundamentals of Image, Audio, and Video Processing Using MATLAB; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Jannasch, M.; Gaetzner, S.; Weigel, T.; Walles, H.; Schmitz, T.; Hansmann, J. A comparative multi-parametric in vitro model identifies the power of test conditions to predict the fibrotic tendency of a biomaterial. Sci. Rep. 2017, 7, 1689. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.; Gresnigt, M.S.; Last, A.; Wollny, T.; Berlinghof, F.; Pospich, R.; Cseresnyes, Z.; Medyukhina, A.; Graf, K.; Groger, M.; et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 2019, 220, 119396. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Stout, R.D.; Suttles, J. Functional plasticity of macrophages: Reversible adaptation to changing microenvironments. J. Leukoc. Biol. 2004, 76, 509–513. [Google Scholar] [CrossRef]
- Stout, R.D.; Suttles, J. T cell signaling of macrophage function in inflammatory disease. Front. Biosci. 1997, 2, d197–d206. [Google Scholar] [CrossRef]
- D’Andrea, A.; Ma, X.; Aste-Amezaga, M.; Paganin, C.; Trinchieri, G. Stimulatory and inhibitory effects of interleukin (IL)-4 and IL-13 on the production of cytokines by human peripheral blood mononuclear cells: Priming for IL-12 and tumor necrosis factor alpha production. J. Exp. Med. 1995, 181, 537–546. [Google Scholar] [CrossRef]
- Mohd Yasin, Z.N.; Mohd Idrus, F.N.; Hoe, C.H.; Yvonne-Tee, G.B. Macrophage polarization in THP-1 cell line and primary monocytes: A systematic review. Differentiation 2022, 128, 67–82. [Google Scholar] [CrossRef]
- Rao Muvva, J.; Parasa, V.R.; Lerm, M.; Svensson, M.; Brighenti, S. Polarization of Human Monocyte-Derived Cells with Vitamin D Promotes Control of Mycobacterium tuberculosis Infection. Front. Immunol. 2019, 10, 3157. [Google Scholar] [CrossRef]
- Chen, S.; Saeed, A.; Liu, Q.; Jiang, Q.; Xu, H.; Xiao, G.G.; Rao, L.; Duo, Y. Macrophages in immunoregulation and therapeutics. Signal Transduct. Target. Ther. 2023, 8, 207. [Google Scholar] [CrossRef]
- Singh, S.; Anshita, D.; Ravichandiran, V. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol. 2021, 101, 107598. [Google Scholar] [CrossRef] [PubMed]
- Meniailo, M.E.; Malashchenko, V.V.; Shmarov, V.A.; Gazatova, N.D.; Melashchenko, O.B.; Goncharov, A.G.; Seledtsova, G.V.; Seledtsov, V.I. Interleukin-8 favors pro-inflammatory activity of human monocytes/macrophages. Int. Immunopharmacol. 2018, 56, 217–221. [Google Scholar] [CrossRef]
- Wang, L.X.; Zhang, S.X.; Wu, H.J.; Rong, X.L.; Guo, J. M2b macrophage polarization and its roles in diseases. J. Leukoc. Biol. 2019, 106, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yuan, X.; Wang, C. The clinical value of IL-3, IL-4, IL-12p70, IL17A, IFN-gamma, MIP-1beta, NLR, P-selectin, and TNF-alpha in differentiating bloodstream infections caused by gram-negative, gram-positive bacteria and fungi in hospitalized patients: An Observational Study. Medicine 2019, 98, e17315. [Google Scholar] [CrossRef]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Kaya, E.; Batoni, G.; Di Luca, M.; Apolloni, E.; Mazzoni, A.; Maisetta, G.; Esin, S. Planktonic and Biofilm-Associated Pseudomonas aeruginosa and Staphylococcus epidermidis Elicit Differential Human Peripheral Blood Cell Responses. Microorganisms 2021, 9, 1846. [Google Scholar] [CrossRef] [PubMed]
- Le, K.Y.; Park, M.D.; Otto, M. Immune Evasion Mechanisms of Staphylococcus epidermidis Biofilm Infection. Front. Microbiol. 2018, 9, 359. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Nakamura, K.; Cornforth, M.; Suzuki, F. Role of M2b macrophages in the acceleration of bacterial translocation and subsequent sepsis in mice exposed to whole body [137Cs] gamma-irradiation. J. Immunol. 2012, 189, 296–303. [Google Scholar] [CrossRef] [PubMed]



| Antibody | Manufacturer | Incubation Time/ Temperature | Dilution IHC | Dilution IF | Cells Positive |
|---|---|---|---|---|---|
| mCD68 | Invitrogen, Darmstadt, Germany, 14-0688-82 | Overnight, 4 °C | 1:500 | 5 µg/mL | M0 |
| rCD80 | Invitrogen, Darmstadt, Germany, PA585913 | Overnight, 4 °C | 1:500 | 1:100 | M1 |
| rCD163 | BIOSUSA, Massachusetts U.S.A. bsm-54015R | Overnight, 4 °C | 1:100 | 1:100 | M2 |
| Vimentin | Sigma-Aldrich, Taufkirchen, Germany, V2258 | 1 h, room temperature | 1:400 | 1:400 | Fibroblasts |
| Anti-mouse IgG FITC | Sigma Aldrich, Taufkirchen, Germany, F0257 | 1 h, room temperature | -- | 1:50 | -- |
| Anti-rabbit IgG (H+L) | Sigma Aldrich, SAB4600084 | 1 h, room temperature | -- | 10 µg/mL | -- |
| DAPI (Fluoromount-G) | Invitrogen, Darmstadt, Germany, E139612 | -- | -- | 1 drop/slide | -- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murkar, R.; von Heckel, C.; Walles, H.; Moch, T.B.; Arens, C.; Davaris, N.; Weber, A.; Zuschratter, W.; Baumann, S.; Reinhardt, J.; et al. Establishment of a Human Immunocompetent 3D Tissue Model to Enable the Long-Term Examination of Biofilm–Tissue Interactions. Bioengineering 2024, 11, 187. https://doi.org/10.3390/bioengineering11020187
Murkar R, von Heckel C, Walles H, Moch TB, Arens C, Davaris N, Weber A, Zuschratter W, Baumann S, Reinhardt J, et al. Establishment of a Human Immunocompetent 3D Tissue Model to Enable the Long-Term Examination of Biofilm–Tissue Interactions. Bioengineering. 2024; 11(2):187. https://doi.org/10.3390/bioengineering11020187
Chicago/Turabian StyleMurkar, Rasika, Charlotte von Heckel, Heike Walles, Theresia Barbara Moch, Christoph Arens, Nikolaos Davaris, André Weber, Werner Zuschratter, Sönke Baumann, Jörg Reinhardt, and et al. 2024. "Establishment of a Human Immunocompetent 3D Tissue Model to Enable the Long-Term Examination of Biofilm–Tissue Interactions" Bioengineering 11, no. 2: 187. https://doi.org/10.3390/bioengineering11020187
APA StyleMurkar, R., von Heckel, C., Walles, H., Moch, T. B., Arens, C., Davaris, N., Weber, A., Zuschratter, W., Baumann, S., Reinhardt, J., & Kopp, S. (2024). Establishment of a Human Immunocompetent 3D Tissue Model to Enable the Long-Term Examination of Biofilm–Tissue Interactions. Bioengineering, 11(2), 187. https://doi.org/10.3390/bioengineering11020187







