Self-Guided Algorithm for Fast Image Reconstruction in Photo-Magnetic Imaging: Artificial Intelligence-Assisted Approach
Abstract
1. Introduction
2. Materials and Methods
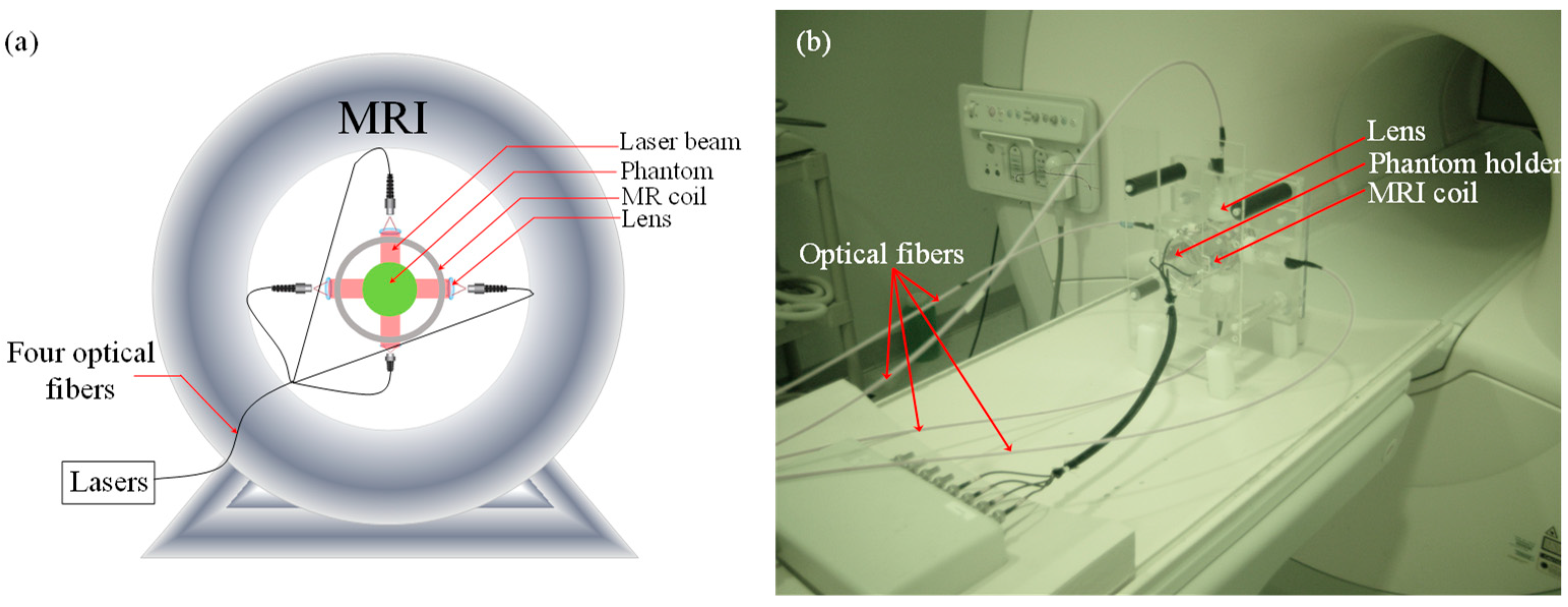
2.1. PMI Methodology
2.2. PMI Image Reconstruction Algorithm
2.2.1. PMI Forward Problem
2.2.2. PMI Inverse Problem
2.3. AI-Based A Priori Information Generation
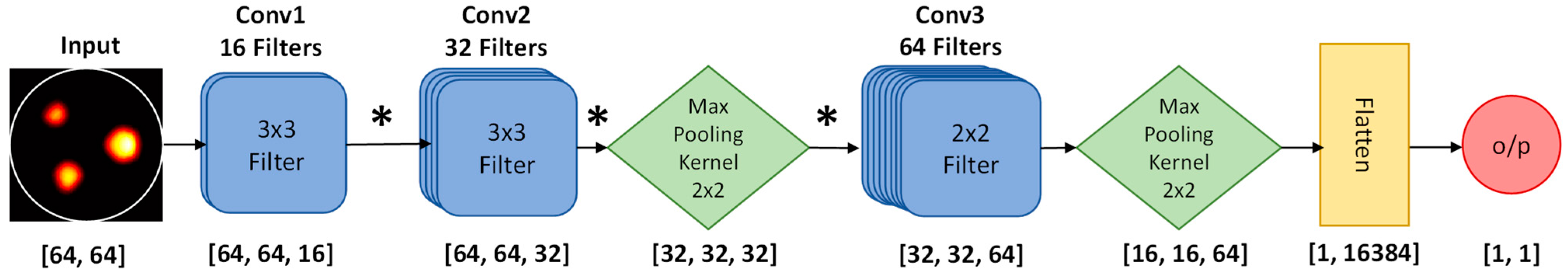
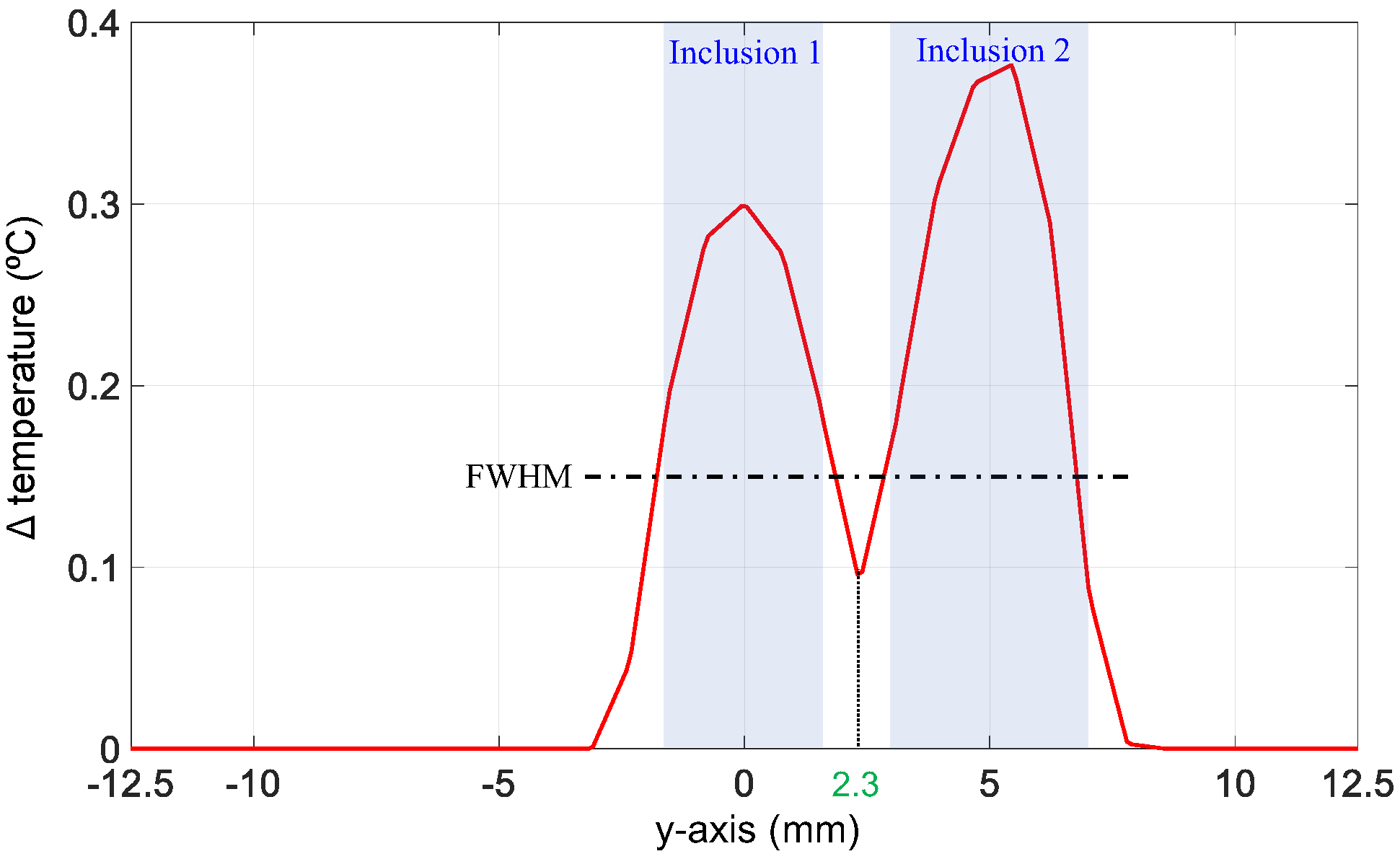
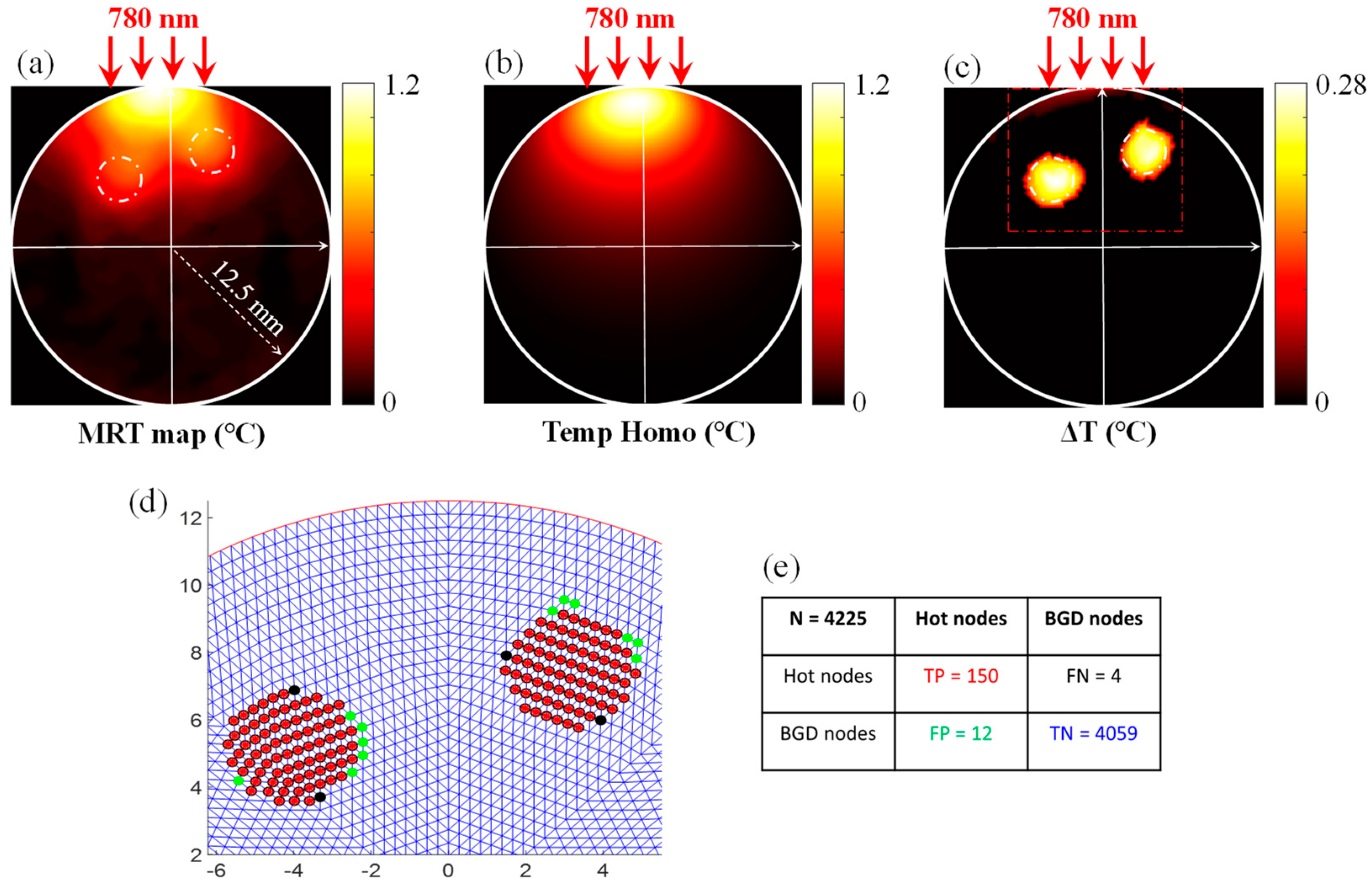
2.3.1. ML-Based Delineation of Region of Interest
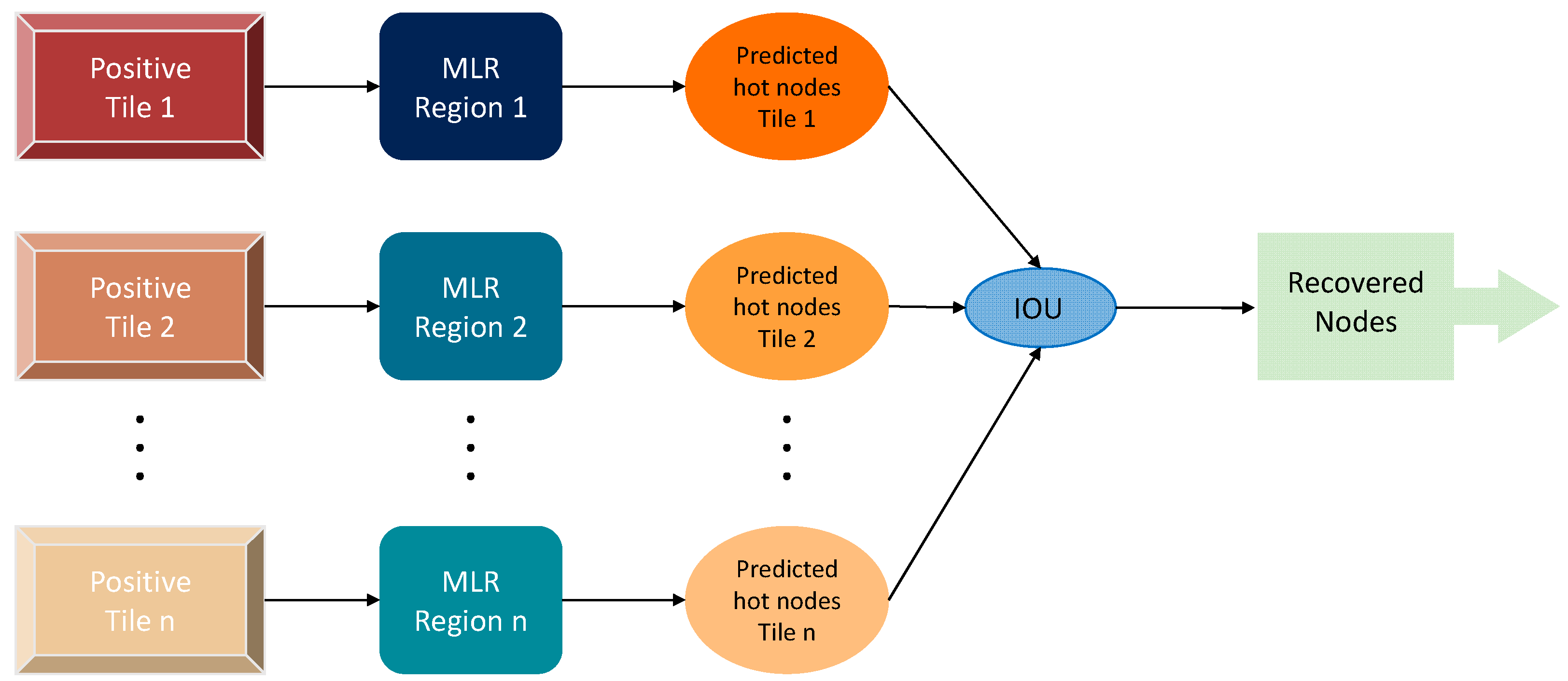
2.3.2. Prediction of Hot Nodes
3. Results
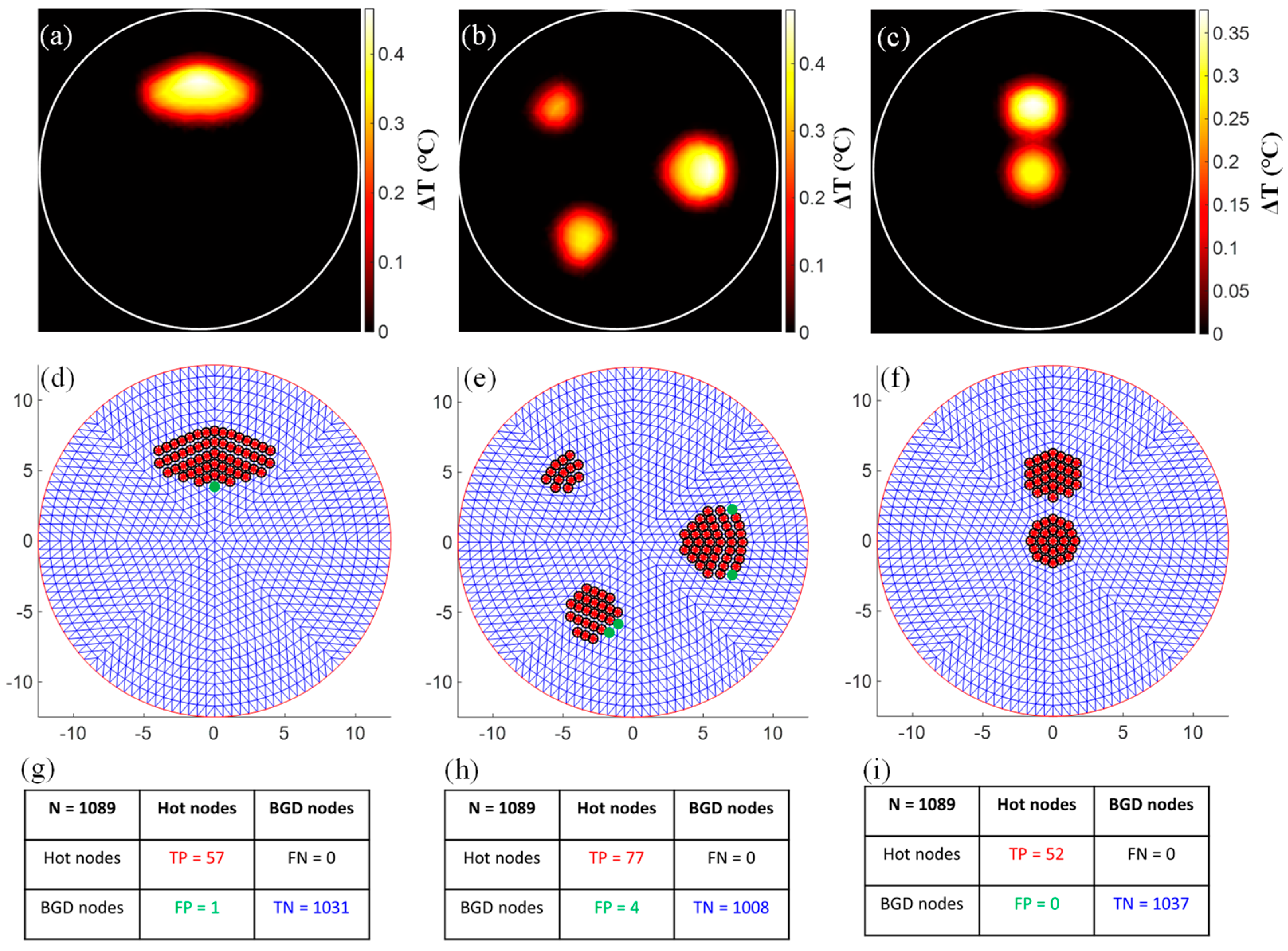
3.1. Evaluation of the Functional A Priori Mask Recovery
3.2. PMI Reconstruction with vs. without A Priori Information
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwong, J.; Nouizi, F.; Cho, J.; Zheng, J.; Li, Y.; Chen, J.; Su, M.; Gulsen, G. Diffuse optical imaging of the breast using structured-light. In Proceedings of the SPIE BiOS, San Francisco, CA, USA, 7 February 2015; pp. 93190K–93195K. [Google Scholar]
- Uddin, K.S.; Zhang, M.; Anastasio, M.; Zhu, Q. Optimal breast cancer diagnostic strategy using combined ultrasound and diffuse optical tomography. Biomed. Opt. Express 2020, 11, 2722–2737. [Google Scholar] [CrossRef]
- Cochran, J.M.; Busch, D.R.; Lin, L.; Minkoff, D.L.; Schweiger, M.; Arridge, S.; Yodh, A.G. Hybrid time-domain and continuous-wave diffuse optical tomography instrument with concurrent, clinical magnetic resonance imaging for breast cancer imaging. J. Biomed. Opt. 2019, 24, 051409. [Google Scholar] [CrossRef]
- Cheong, W.F.; Prahl, S.A.; Welch, A.J. A review of the optical properties of biological tissues. IEEE J. Quantum Electron. 1990, 26, 2166–2185. [Google Scholar] [CrossRef]
- Dehghani, H.; Srinivasan, S.; Pogue, B.W.; Gibson, A. Numerical modelling and image reconstruction in diffuse optical tomography. Philos. Trans. A Math. Phys. Eng. Sci. 2009, 367, 3073–3093. [Google Scholar] [CrossRef]
- Applegate, M.; Istfan, R.; Spink, S.; Tank, A.; Roblyer, D. Recent advances in high speed diffuse optical imaging in biomedicine. APL Photonics 2020, 5, 040802. [Google Scholar] [CrossRef]
- Carp, S.A.; Sajjadi, A.Y.; Wanyo, C.M.; Fang, Q.; Specht, M.C.; Schapira, L.; Moy, B.; Bardia, A.; Boas, D.A.; Isakoff, S.J. Hemodynamic signature of breast cancer under fractional mammographic compression using a dynamic diffuse optical tomography system. Biomed. Opt. Express 2013, 4, 2911–2924. [Google Scholar] [CrossRef]
- Zhu, Q.; Ademuyiwa, F.O.; Young, C.; Appleton, C.; Covington, M.F.; Ma, C.; Sanati, S.; Hagemann, I.S.; Mostafa, A.; Uddin, K.S. Early assessment window for predicting breast cancer neoadjuvant therapy using biomarkers, ultrasound, and diffuse optical tomography. Breast Cancer Res. Treat. 2021, 188, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Chae, E.Y.; Kim, H.H.; Sabir, S.; Kim, Y.; Kim, H.; Yoon, S.; Ye, J.C.; Cho, S.; Heo, D.; Kim, K.H. Development of digital breast tomosynthesis and diffuse optical tomography fusion imaging for breast cancer detection. Sci. Rep. 2020, 10, 13127. [Google Scholar] [CrossRef] [PubMed]
- Nouizi, F.; Diaz-Ayil, G.; Blé, F.-X.; Dubois, B.; Uhring, W.; Poulet, P. Time-gated near-infrared spectroscopic imaging of brain activation: A simulation proof of concept. In Proceedings of the SPIE BIOS, San Francisco, CA, USA, 17 February 2011; pp. 78960L–78968L. [Google Scholar]
- Yücel, M.A.; Evans, K.C.; Selb, J.; Huppert, T.J.; Boas, D.A.; Gagnon, L. Validation of the hypercapnic calibrated fMRI method using DOT-fMRI fusion imaging. Neuroimage 2014, 102 Pt 2, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Dehghani, H.; Eggebrecht, A.T. Investigation of effect of modulation frequency on high-density diffuse optical tomography image quality. Neurophotonics 2021, 8, 045002. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Frijia, E.M.; Vidal Rosas, E.; Collins-Jones, L.; Smith, G.; Nixon-Hill, R.; Powell, S.; Everdell, N.L.; Cooper, R.J. Design and validation of a mechanically flexible and ultra-lightweight high-density diffuse optical tomography system for functional neuroimaging of newborns. Neurophotonics 2021, 8, 015011. [Google Scholar] [CrossRef] [PubMed]
- Arridge, S.R.; Lionheart, W.R.B. Non-uniqueness in diffusion-based optical tomography. Opt. Lett. 1998, 23, 882. [Google Scholar] [CrossRef] [PubMed]
- Brooksby, B.; Jiang, S.; Dehghani, H.; Pogue, B.W.; Paulsen, K.D.; Kogel, C.; Doyley, M.; Weaver, J.B.; Poplack, S.P. Magnetic resonance-guided near-infrared tomography of the breast. Rev. Sci. Instrum. 2004, 75, 5262–5270. [Google Scholar] [CrossRef]
- Xue, M.; Zhang, M.; Li, S.; Zou, Y.; Zhu, Q. Automated pipeline for breast cancer diagnosis using US assisted diffuse optical tomography. Biomed. Opt. Express 2023, 14, 6072–6087. [Google Scholar] [CrossRef] [PubMed]
- Poplack, S.P.; Park, E.-Y.; Ferrara, K.W. Optical breast imaging: A review of physical principles, technologies, and clinical applications. J. Breast Imaging 2023, 5, 520–537. [Google Scholar] [CrossRef] [PubMed]
- Nouizi, F.; Luk, A.; Thayer, D.; Lin, Y.; Ha, S.; Gulsen, G. Experimental validation of a high-resolution diffuse optical imaging modality: Photomagnetic imaging. J. Biomed. Opt. 2016, 21, 16009. [Google Scholar] [CrossRef] [PubMed]
- Luk, A.T.; Ha, S.; Nouizi, F.; Thayer, D.; Lin, Y.; Gulsen, G. A true multi-modality approach for high resolution optical imaging: Photo-magnetic imaging. In Proceedings of the SPIE BiOS, San Francisco, CA, USA, 3 March 2014; pp. 89370G–89377G. [Google Scholar]
- Luk, A.T.; Thayer, D.; Lin, Y.; Nouizi, F.; Gao, H.; Gulsen, G. A novel high-resolution optical imaging modality: Photo-magnetic imaging. In Proceedings of the SPIE BiOS, San Francisco, CA, USA, 13 March 2013; pp. 857404–857410. [Google Scholar]
- Algarawi, M.; Erkol, H.; Luk, A.; Ha, S.; Burcin Unlu, M.; Gulsen, G.; Nouizi, F. Multi-Wavelength Photo-Magnetic Imaging System for Photothermal Therapy Guidance. Lasers Surg. Med. 2020, 53, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Algarawi, M.; Erkol, H.; Luk, A.; Ha, S.; Ünlü, M.B.; Gulsen, G.; Nouizi, F. Resolving tissue chromophore concentration at MRI resolution using multi-wavelength photo-magnetic imaging. Biomed. Opt. Express 2020, 11, 4244–4254. [Google Scholar] [CrossRef]
- Algarawi, M.; Nouizi, F.; Luk, A.; Mehrabi, M.; Erkol, H.; Ünlü, M.B.; Gulsen, G.; Ha, S. High-resolution chromophore concentration recovery using multi-wavelength photo-magnetic imaging. In Proceedings of the Multimodal Biomedical Imaging XIV, San Francisco, CA, USA, 27 February 2019; p. 108710F. [Google Scholar]
- Nouizi, F.; Algarawi, M.; Erkol, H.; Luk, A.; Gulsen, G. Photo-magnetic imaging: A new functional imaging modality for more accurate photothermal therapy planning. In Proceedings of the Optical Methods for Tumor Treatment and Detection: Mechanisms and Techniques in Photodynamic and Photobiomodulation Therapy XXX, San Francisco, CA, USA, 4 March 2022; pp. 1194006–1194013. [Google Scholar]
- Nouizi, F.; Algarawi, M.; Erkol, H.; Luk, A.; Gulsen, G. Multiwavelength photo-magnetic imaging algorithm improved for direct chromophore concentration recovery using spectral constraints. Appl. Opt. 2021, 60, 10855–10861. [Google Scholar] [CrossRef]
- Nouizi, F.; Erkol, H.; Luk, A.; Marks, M.; Unlu, M.B.; Gulsen, G. An accelerated photo-magnetic imaging reconstruction algorithm based on an analytical forward solution and a fast Jacobian assembly method. Phys. Med. Biol. 2016, 61, 7448–7465. [Google Scholar] [CrossRef]
- Nouizi, F.; Erkol, H.; Luk, A.; Lin, Y.; Gulsen, G. Analytical Photo Magnetic Imaging. In Proceedings of the Optical Tomography and Spectroscopy, Fort Lauderdale, FL, USA, 28 April 2016; p. OW4D. 7. [Google Scholar]
- Nouizi, F.; Erkol, H.; Luk, A.; Unlu, M.B.; Gulsen, G. Real-time photo-magnetic imaging. Biomed. Opt. Express 2016, 7, 3899–3904. [Google Scholar] [CrossRef] [PubMed]
- Erkol, H.; Nouizi, F.; Unlu, M.B.; Gulsen, G. An extended analytical approach for diffuse optical imaging. Phys. Med. Biol. 2015, 60, 5103. [Google Scholar] [CrossRef]
- Erkol, H.; Nouizi, F.; Luk, A.; Unlu, M.B.; Gulsen, G. Comprehensive analytical model for CW laser induced heat in turbid media. Opt. Express 2015, 23, 31069–31084. [Google Scholar] [CrossRef]
- Ishihara, Y.; Calderon, A.; Watanabe, H.; Okamoto, K.; Suzuki, Y.; Kuroda, K.; Suzuki, Y. A precise and fast temperature mapping using water proton chemical shift. Magn. Reson. Med. 1995, 34, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, M.; Arridge, S.R.; Hiraoka, M.; Delpy, D.T. The finite element method for the propagation of light in scattering media: Boundary and source conditions. Med. Phys. 1995, 22, 1779–1792. [Google Scholar] [CrossRef]
- Nouizi, F.; Torregrossa, M.; Chabrier, R.; Poulet, P. Improvement of absorption and scattering discrimination by selection of sensitive points on temporal profile in diffuse optical tomography. Opt. Express 2011, 19, 12843–12854. [Google Scholar] [CrossRef]
- Wissler, E.H. Pennes’ 1948 paper revisited. J. Appl. Physiol. 1998, 85, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Diaz, S.H.; Aguilar, G.; Lavernia, E.J.; Wong, B.J.F. Modeling the thermal response of porcine cartilage to laser irradiation. IEEE Sel. Top. Quantum Electron. 2001, 7, 944–951. [Google Scholar] [CrossRef]
- Lin, Y.; Gao, H.; Thayer, D.; Luk, A.L.; Gulsen, G. Photo-magnetic imaging: Resolving optical contrast at MRI resolution. Phys Med. Biol. 2013, 58, 3551–3562. [Google Scholar] [CrossRef]
- Arridge, S. Optical tomography in medical imaging. Inverse Probl. 1999, 15, R41–R93. [Google Scholar] [CrossRef]
- Nouizi, F.; Kwong, T.C.; Cho, J.; Lin, Y.; Sampathkumaran, U.; Gulsen, G. Implementation of a new scanning method for high-resolution fluorescence tomography using thermo-sensitive fluorescent agents. Opt. Lett. 2015, 40, 4991–4994. [Google Scholar] [CrossRef] [PubMed]
- Yalavarthy, P.K.; Pogue, B.W.; Dehghani, H.; Carpenter, C.M.; Jiang, S.; Paulsen, K.D. Structural information within regularization matrices improves near infrared diffuse optical tomography. Opt. Express 2007, 15, 8043–8058. [Google Scholar] [CrossRef] [PubMed]
- Luk, A.; Nouizi, F.; Erkol, H.; Unlu, M.B.; Gulsen, G. Ex vivo validation of photo-magnetic imaging. Opt. Lett. 2017, 42, 4171–4174. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Hegde, P.U.; Ricci Jr, A.; Kane, M.; Cronin, E.B.; Ardeshirpour, Y.; Xu, C.; Aguirre, A.; Kurtzman, S.H.; Deckers, P.J. Early-stage invasive breast cancers: Potential role of optical tomography with US localization in assisting diagnosis. Radiology 2010, 256, 367–378. [Google Scholar] [CrossRef]
- Nouizi, F.; Brooks, J.; Zuro, D.M.; Hui, S.K.; Gulsen, G. Development of a theranostic preclinical fluorescence molecular tomography/cone beam CT-guided irradiator platform. Biomed. Opt. Express 2022, 13, 6100–6112. [Google Scholar] [CrossRef]


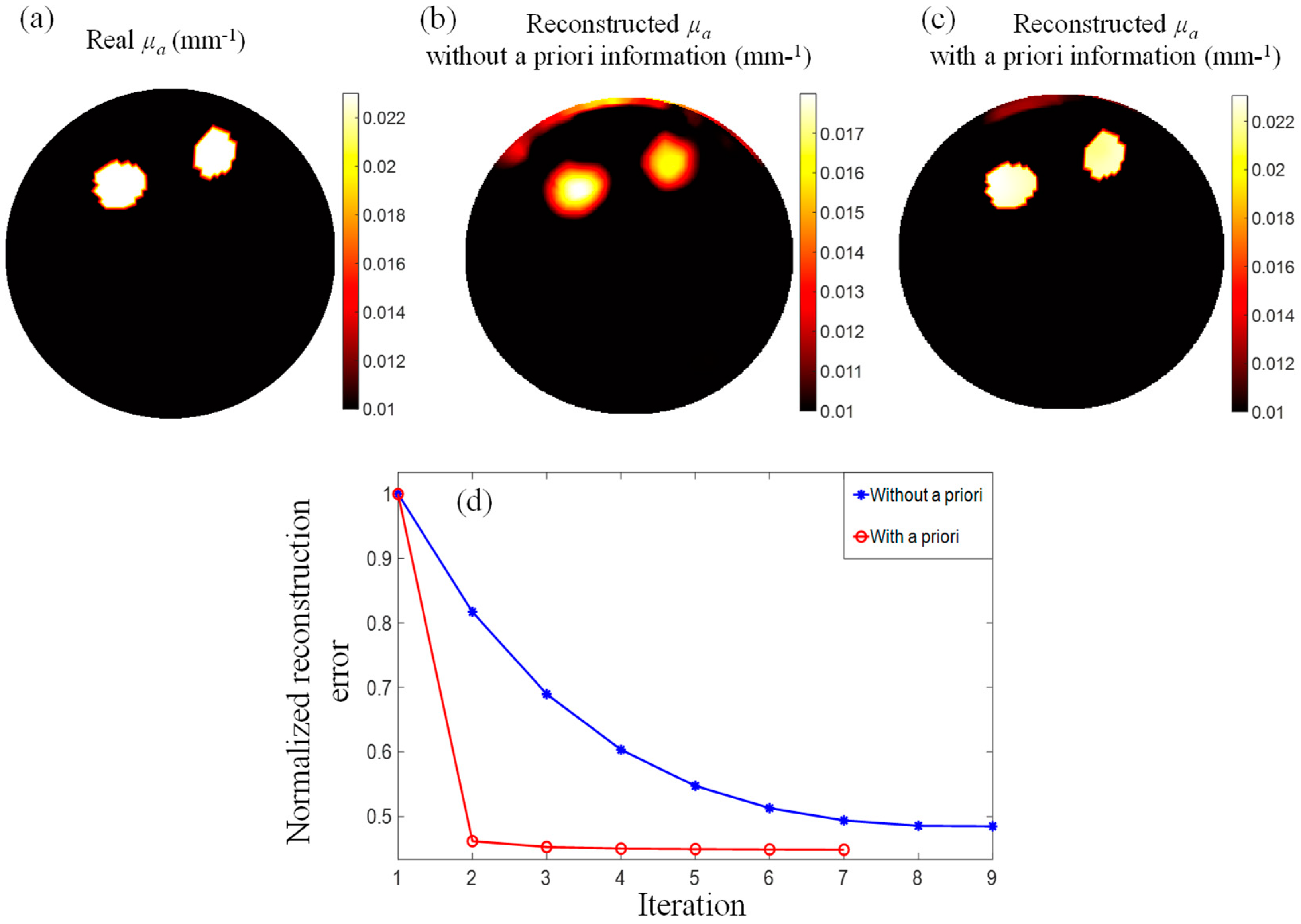
| Real | Reconstructed without A Priori Information | Reconstructed with A Priori Information | |
|---|---|---|---|
| Inclusion 1 | 0.023 | 0.0157 ± 0.00140 | 0.0226 ± 0.00030 |
| Inclusion 2 | 0.023 | 0.0148 ± 0.00130 | 0.0220 ± 0.00026 |
| Background | 0.010 | 0.0094 ± 0.00089 | 0.0100 ± 0.00040 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Algarawi, M.; Saraswatula, J.S.; Pathare, R.R.; Zhang, Y.; Shah, G.A.; Eresen, A.; Gulsen, G.; Nouizi, F. Self-Guided Algorithm for Fast Image Reconstruction in Photo-Magnetic Imaging: Artificial Intelligence-Assisted Approach. Bioengineering 2024, 11, 126. https://doi.org/10.3390/bioengineering11020126
Algarawi M, Saraswatula JS, Pathare RR, Zhang Y, Shah GA, Eresen A, Gulsen G, Nouizi F. Self-Guided Algorithm for Fast Image Reconstruction in Photo-Magnetic Imaging: Artificial Intelligence-Assisted Approach. Bioengineering. 2024; 11(2):126. https://doi.org/10.3390/bioengineering11020126
Chicago/Turabian StyleAlgarawi, Maha, Janaki S. Saraswatula, Rajas R. Pathare, Yang Zhang, Gyanesh A. Shah, Aydin Eresen, Gultekin Gulsen, and Farouk Nouizi. 2024. "Self-Guided Algorithm for Fast Image Reconstruction in Photo-Magnetic Imaging: Artificial Intelligence-Assisted Approach" Bioengineering 11, no. 2: 126. https://doi.org/10.3390/bioengineering11020126
APA StyleAlgarawi, M., Saraswatula, J. S., Pathare, R. R., Zhang, Y., Shah, G. A., Eresen, A., Gulsen, G., & Nouizi, F. (2024). Self-Guided Algorithm for Fast Image Reconstruction in Photo-Magnetic Imaging: Artificial Intelligence-Assisted Approach. Bioengineering, 11(2), 126. https://doi.org/10.3390/bioengineering11020126






