DGNMDA: Dual Heterogeneous Graph Neural Network Encoder for miRNA-Disease Association Prediction
Abstract
1. Introduction
- Combining local structural information and global dependencies: We design a dual heterogeneous graph neural network encoder that integrates a Graph Convolutional Transformer and a Graph Convolutional Attention Network (GCAN). This architecture not only captures the global dependencies of nodes but also effectively learns local structural information, generating more comprehensive node embedding encodings.
- Adaptive fusion of multi-level features: We introduce a fine-grained feature interaction gating mechanism to gradually fuse and refine feature representations from the two encoders at different levels. This adaptive fusion mechanism allows the model to dynamically adjust feature combinations based on task requirements, improving the flexibility and prediction performance of the model.
- Improving prediction performance: Through experimental validation on the miRNA-disease association prediction task, our DGNMDA method demonstrates significant performance improvements on multiple benchmark datasets. Our results demonstrate the efficacy and advantages of our method, offering novel insights and resources for future studies in this domain.
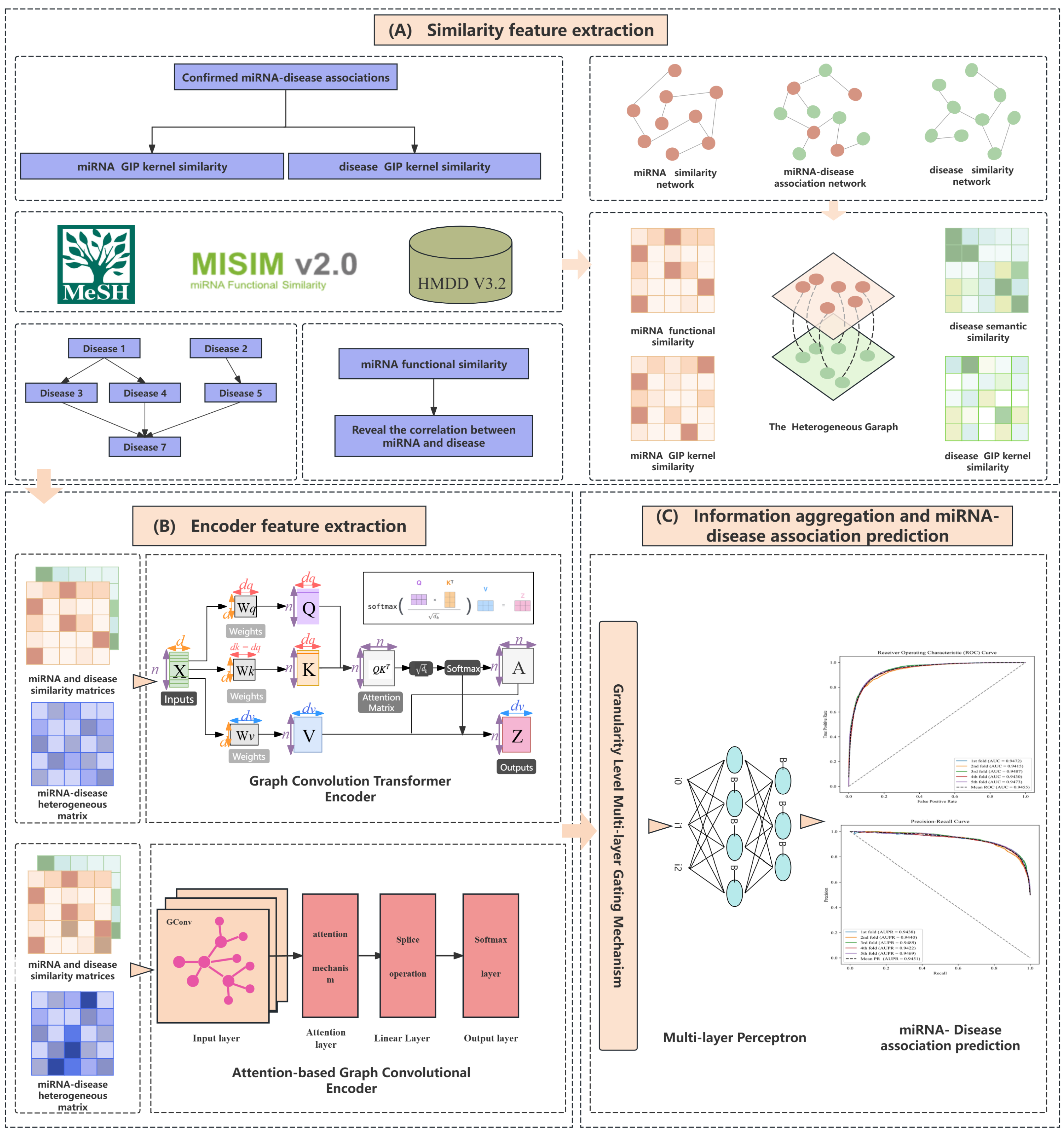
2. Materials and Methods
2.1. Experimental Data
2.2. Building miRNA-Disease Resemblance Graphs
2.3. Graph Convolutional Attention Network (GCAN) Encoder
2.4. Graph Convolutional Transformer Encoder
2.5. Fine-Grained Multi-Layer Feature Interaction Gating
3. Results and Discussion
3.1. Comparative Analysis with State-Of-The-Art Methods
- NIMCGCN [29]: employs GCNs to derive features from similarity graphs and integrates a neural inductive matrix completion model to generate a complete miRNA-disease association matrix.
- AGAEMD [30]: considers the attention distribution between nodes in the heterogeneous network and dynamically refines the miRNA functional resemblance profile.
- HGANMDA [23]: leverages attention mechanisms at both node and semantic levels to capture the significance of adjacent nodes and meta-paths, reconstructing the associations between miRNAs and diseases.
- MMGCN [22]: combines GCNs and multi-channel attention mechanisms to extract feature information, adaptively capturing the importance of different features.
- AMHMDA [31]: harnesses GCNs to derive multi-faceted node features from various similarity networks, forming a hypergraph, which is then fused via attention to enable miRNA-disease association inference.
3.2. Ablation Study
3.3. Comparison of Single-Source and Multi-Source Features
3.4. Case Study
3.5. Parameter Analysis
3.5.1. Impact of Feature-Embedding Dimension
3.5.2. Experiments on the Number of Multi-Layer Gating Layers
3.5.3. Impact of Graph Convolutional Layers and Attention Heads
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Mendell, J.T.; Olson, E.N. MicroRNAs in stress signaling and human disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef]
- Zhou, Q.; Cui, F.; Lei, C.; Ma, S.; Huang, J.; Wang, X.; Qian, H.; Zhang, D.; Yang, Y. ATG7-mediated autophagy involves in miR-138-5p regulated self-renewal and invasion of lung cancer stem-like cells derived from A549 cells. Anti-Cancer Drugs 2021, 32, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, M.X.; Yan, G.Y. RWRMDA: Predicting novel human microRNA–disease associations. Mol. BioSyst. 2012, 8, 2792–2798. [Google Scholar] [CrossRef]
- Xie, X.; Wang, Y.; Sheng, N.; Zhang, S.; Cao, Y.; Fu, Y. Predicting miRNA-disease associations based on multi-view information fusion. Front. Genet. 2022, 13, 979815. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Liu, Z.P.; Gao, R. Predicting multiple types of MicroRNA-disease associations based on tensor factorization and label propagation. Comput. Biol. Med. 2022, 146, 105558. [Google Scholar] [CrossRef]
- Wang, W.; Chen, H. Predicting miRNA-disease associations based on graph attention networks and dual Laplacian regularized least squares. Brief. Bioinform. 2022, 23, bbac292. [Google Scholar] [CrossRef]
- Chen, X.; Sun, L.G.; Zhao, Y. NCMCMDA: miRNA–disease association prediction through neighborhood constraint matrix completion. Brief. Bioinform. 2021, 22, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wang, S.; Wu, Q.; Azim, R.; Li, W. Predicting potential miRNA-disease associations by combining gradient boosting decision tree with logistic regression. Comput. Biol. Chem. 2020, 85, 107200. [Google Scholar] [CrossRef] [PubMed]
- Xuan, P.; Dong, Y.; Guo, Y.; Zhang, T.; Liu, Y. Dual convolutional neural network based method for predicting disease-related miRNAs. Int. J. Mol. Sci. 2018, 19, 3732. [Google Scholar] [CrossRef] [PubMed]
- Scarselli, F.; Gori, M.; Tsoi, A.C.; Hagenbuchner, M.; Monfardini, G. The graph neural network model. IEEE Trans. Neural Netw. 2008, 20, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Li, J.; Zheng, C.; Liu, J.; Zhang, Q. HGTMDA: A Hypergraph Learning Approach with Improved GCN-Transformer for miRNA–Disease Association Prediction. Bioengineering 2024, 11, 680. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Z.; Wang, X.; Meng, Z.; Cui, W. Mhtan-dti: Metapath-based hierarchical transformer and attention network for drug–target interaction prediction. Brief. Bioinform. 2023, 24, bbad079. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Z.; Wang, K.; Gao, X.; Wang, G. End-to-end interpretable disease–gene association prediction. Brief. Bioinform. 2023, 24, bbad118. [Google Scholar] [CrossRef]
- Gu, P.; Wu, T.; Zou, M.; Pan, Y.; Guo, J.; Xiahou, J.; Peng, X.; Li, H.; Ma, J.; Zhang, L. Multi-head self-attention model for classification of temporal lobe epilepsy subtypes. Front. Physiol. 2020, 11, 604764. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Casanova, P.; Lio, A.R.P.; Bengio, Y. Graph Attention networks. arXiv 2017, arXiv:1710.10903. [Google Scholar]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Philip, S.Y. A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4–24. [Google Scholar] [CrossRef]
- Tang, X.; Luo, J.; Shen, C.; Lai, Z. Multi-view multichannel attention graph convolutional network for miRNA–disease association prediction. Brief. Bioinform. 2021, 22, bbab174. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhong, T.; Huang, D.; You, Z.H.; Nie, R. Hierarchical graph attention network for miRNA-disease association prediction. Mol. Ther. 2022, 30, 1775–1786. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Ji, B.; Zhang, M.; Liu, F.; Xie, X.; Peng, S. MHGTMDA: Molecular heterogeneous graph transformer based on biological entity graph for miRNA-disease associations prediction. Mol. Ther.-Nucleic Acids 2024, 23, 102139. [Google Scholar] [CrossRef]
- Torres, L.H.; Ribeiro, B.; Arrais, J.P. Few-shot learning with transformers via graph embeddings for molecular property prediction. Expert Syst. Appl. 2023, 225, 120005. [Google Scholar] [CrossRef]
- Zheng, M.; Gao, P.; Zhang, R.; Li, K.; Wang, X.; Li, H.; Dong, H. End-to-end object detection with adaptive clustering transformer. arXiv 2020, arXiv:2011.09315. [Google Scholar]
- Zhang, C.; Zhao, Y.; Wang, J. Transformer-based dynamic fusion clustering network. Knowl.-Based Syst. 2022, 258, 109984. [Google Scholar] [CrossRef]
- Guo, M.; Zhang, Y.; Liu, T. Gaussian transformer: A lightweight approach for natural language inference. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 6489–6496. [Google Scholar]
- Li, J.; Zhang, S.; Liu, T.; Ning, C.; Zhang, Z.; Zhou, W. Neural inductive matrix completion with graph convolutional networks for miRNA-disease association prediction. Bioinformatics 2020, 36, 2538–2546. [Google Scholar] [CrossRef]
- Zhang, H.; Fang, J.; Sun, Y.; Xie, G.; Lin, Z.; Gu, G. Predicting miRNA-disease associations via node-level attention graph auto-encoder. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 20, 1308–1318. [Google Scholar] [CrossRef]
- Ning, Q.; Zhao, Y.; Gao, J.; Chen, C.; Li, X.; Li, T.; Yin, M. AMHMDA: Attention aware multi-view similarity networks and hypergraph learning for miRNA–disease associations identification. Brief. Bioinform. 2023, 24, bbad094. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Costinean, S.; Zanesi, N.; Pekarsky, Y.; Tili, E.; Volinia, S.; Heerema, N.; Croce, C.M. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7024–7029. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M.; Calin, G.A. miRNAs, cancer, and stem cell division. Cell 2005, 122, 6–7. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted deletion reveals essential and overlapping functions of the miR-17 92 family of miRNA clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef]
- Tilly, H.; Da Silva, M.G.; Vitolo, U.; Jack, A.; Meignan, M.; Lopez-Guillermo, A.; Walewski, J.; André, M.; Johnson, P.; Pfreundschuh, M.; et al. Diffuse large B-cell lymphoma (DLBCL): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, v116–v125. [Google Scholar] [CrossRef]
- Smith, A.; Crouch, S.; Lax, S.; Li, J.; Painter, D.; Howell, D.; Patmore, R.; Jack, A.; Roman, E. Lymphoma incidence, survival and prevalence 2004–2014: Sub-type analyses from the UK’s Haematological Malignancy Research Network. Br. J. Cancer 2015, 112, 1575–1584. [Google Scholar] [CrossRef]
- Shankland, K.R.; Armitage, J.O.; Hancock, B.W. Non-hodgkin lymphoma. Lancet 2012, 380, 848–857. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Murray, T.; Xu, J.; Smigal, C.; Thun, M.J. Cancer statistics, 2006. CA-ATLANTA 2006, 56, 106. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Blenkiron, C.; Goldstein, L.D.; Thorne, N.P.; Spiteri, I.; Chin, S.F.; Dunning, M.J.; Barbosa-Morais, N.L.; Teschendorff, A.E.; Green, A.R.; Ellis, I.O.; et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007, 8, 1–16. [Google Scholar] [CrossRef]
- Xu, F.; Wang, Y.; Ling, Y.; Zhou, C.; Wang, H.; Teschendorff, A.E.; Zhao, Y.; Zhao, H.; He, Y.; Zhang, G.; et al. dbDEMC 3.0: Functional exploration of differentially expressed miRNAs in cancers of human and model organisms. Genom. Proteom. Bioinform. 2022, 20, 446–454. [Google Scholar] [CrossRef]






| Methods | ACC | F1-Score | Recall | Precision | AUC | AUPRC |
|---|---|---|---|---|---|---|
| NIMCGCN | 0.8131 | 0.8148 | 0.8220 | 0.8076 | 0.8945 | 0.8926 |
| AGAEMD | 0.8502 | 0.8507 | 0.8544 | 0.8481 | 0.9270 | 0.9286 |
| HGANMDA | 0.8489 | 0.8481 | 0.8433 | 0.8529 | 0.9265 | 0.9253 |
| MAGCN | 0.8483 | 0.8473 | 0.8425 | 0.8533 | 0.9245 | 0.9268 |
| AMHMDA | 0.8648 | 0.8623 | 0.8539 | 0.8755 | 0.9411 | 0.9403 |
| DGNMDA | 0.8773 | 0.8800 | 0.8768 | 0.8896 | 0.9455 | 0.9451 |
| Methods | DGN-A | DGN-B | DGN-C | DGN-D | DGNMDA |
|---|---|---|---|---|---|
| AUC | 0.9367 | 0.9398 | 0.9392 | 0.9382 | 0.9455 |
| AUPR | 0.9358 | 0.9374 | 0.9383 | 0.9367 | 0.9451 |
| Metrics | MS+DS | MS+DG | MG+DS | MG+DG | ALL |
|---|---|---|---|---|---|
| AUC | 0.9406 | 0.9411 | 0.9403 | 0.9421 | 0.9455 |
| AUPR | 0.9399 | 0.9408 | 0.9400 | 0.9418 | 0.9451 |
| Cancer: Lymphoma | |||||
|---|---|---|---|---|---|
| Rank | miRNA | Evidence | Rank | miRNA | Evidence |
| 1 | hsa-mir-21 | dbDEMC | 11 | hsa-mir-150 | dbDEMC |
| 2 | hsa-mir-34a | dbDEMC | 12 | hsa-mir-29b | dbDEMC |
| 3 | hsa-mir-17 | dbDEMC | 13 | hsa-mir-222 | dbDEMC |
| 4 | hsa-mir-92a | dbDEMC | 14 | hsa-mir-181a | dbDEMC |
| 5 | hsa-mir-145 | dbDEMC | 15 | hsa-mir-29c | dbDEMC |
| 6 | hsa-mir-19a | dbDEMC | 16 | hsa-mir-132 | dbDEMC |
| 7 | hsa-mir-126 | dbDEMC | 17 | hsa-let-7g | dbDEMC |
| 8 | hsa-mir-146a | dbDEMC | 18 | hsa-mir-200a | dbDEMC |
| 9 | hsa-let-7b | dbDEMC | 19 | hsa-mir-26a | dbDEMC |
| 10 | hsa-mir-221 | dbDEMC | 20 | hsa-mir-181b | dbDEMC |
| Cancer: Lung cancer | |||||
| Rank | miRNA | Evidence | Rank | miRNA | Evidence |
| 1 | hsa-mir-21 | dbDEMC | 11 | hsa-mir-145 | dbDEMC |
| 2 | hsa-mir-155 | dbDEMC | 12 | hsa-mir-125b | dbDEMC |
| 3 | hsa-mir-17 | dbDEMC | 13 | hsa-mir-16 | dbDEMC |
| 4 | hsa-mir-34a | dbDEMC | 14 | hsa-mir-29a | dbDEMC |
| 5 | hsa-mir-146a | dbDEMC | 15 | hsa-mir-31 | dbDEMC |
| 6 | hsa-mir-15a | dbDEMC | 16 | hsa-mir-122 | dbDEMC |
| 7 | hsa-mir-223 | dbDEMC | 17 | hsa-mir-150 | dbDEMC |
| 8 | hsa-mir-200b | dbDEMC | 18 | hsa-mir-29c | dbDEMC |
| 9 | hsa-let-7d | dbDEMC | 19 | hsa-mir-92a | dbDEMC |
| 10 | hsa-mir-106a | dbDEMC | 20 | hsa-mir-124 | dbDEMC |
| Cancer: Breast cancer | |||||
| Rank | miRNA | Evidence | Rank | miRNA | Evidence |
| 1 | hsa-mir-21 | dbDEMC | 11 | hsa-mir-126 | dbDEMC |
| 2 | hsa-mir-155 | dbDEMC | 12 | hsa-let-7e | dbDEMC |
| 3 | hsa-mir-17 | dbDEMC | 13 | hsa-let-7f | dbDEMC |
| 4 | hsa-mir-29a | dbDEMC | 14 | hsa-mir-31 | dbDEMC |
| 5 | hsa-mir-205 | dbDEMC | 15 | hsa-mir-210 | dbDEMC |
| 6 | hsa-mir-145 | dbDEMC | 16 | hsa-mir-34c | dbDEMC |
| 7 | hsa-mir-200c | dbDEMC | 17 | hsa-mir-206 | dbDEMC |
| 8 | hsa-mir-429 | dbDEMC | 18 | hsa-mir-27a | dbDEMC |
| 9 | hsa-mir-18a | dbDEMC | 19 | hsa-mir-125b | dbDEMC |
| 10 | hsa-mir-19b | dbDEMC | 20 | hsa-mir-199a | dbDEMC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, D.; Zhang, Q.; Zheng, C.; Li, J.; Yin, Z. DGNMDA: Dual Heterogeneous Graph Neural Network Encoder for miRNA-Disease Association Prediction. Bioengineering 2024, 11, 1132. https://doi.org/10.3390/bioengineering11111132
Lu D, Zhang Q, Zheng C, Li J, Yin Z. DGNMDA: Dual Heterogeneous Graph Neural Network Encoder for miRNA-Disease Association Prediction. Bioengineering. 2024; 11(11):1132. https://doi.org/10.3390/bioengineering11111132
Chicago/Turabian StyleLu, Daying, Qi Zhang, Chunhou Zheng, Jian Li, and Zhe Yin. 2024. "DGNMDA: Dual Heterogeneous Graph Neural Network Encoder for miRNA-Disease Association Prediction" Bioengineering 11, no. 11: 1132. https://doi.org/10.3390/bioengineering11111132
APA StyleLu, D., Zhang, Q., Zheng, C., Li, J., & Yin, Z. (2024). DGNMDA: Dual Heterogeneous Graph Neural Network Encoder for miRNA-Disease Association Prediction. Bioengineering, 11(11), 1132. https://doi.org/10.3390/bioengineering11111132





