Systematic Meta-Analysis of Computer-Aided Detection of Breast Cancer Using Hyperspectral Imaging
Abstract
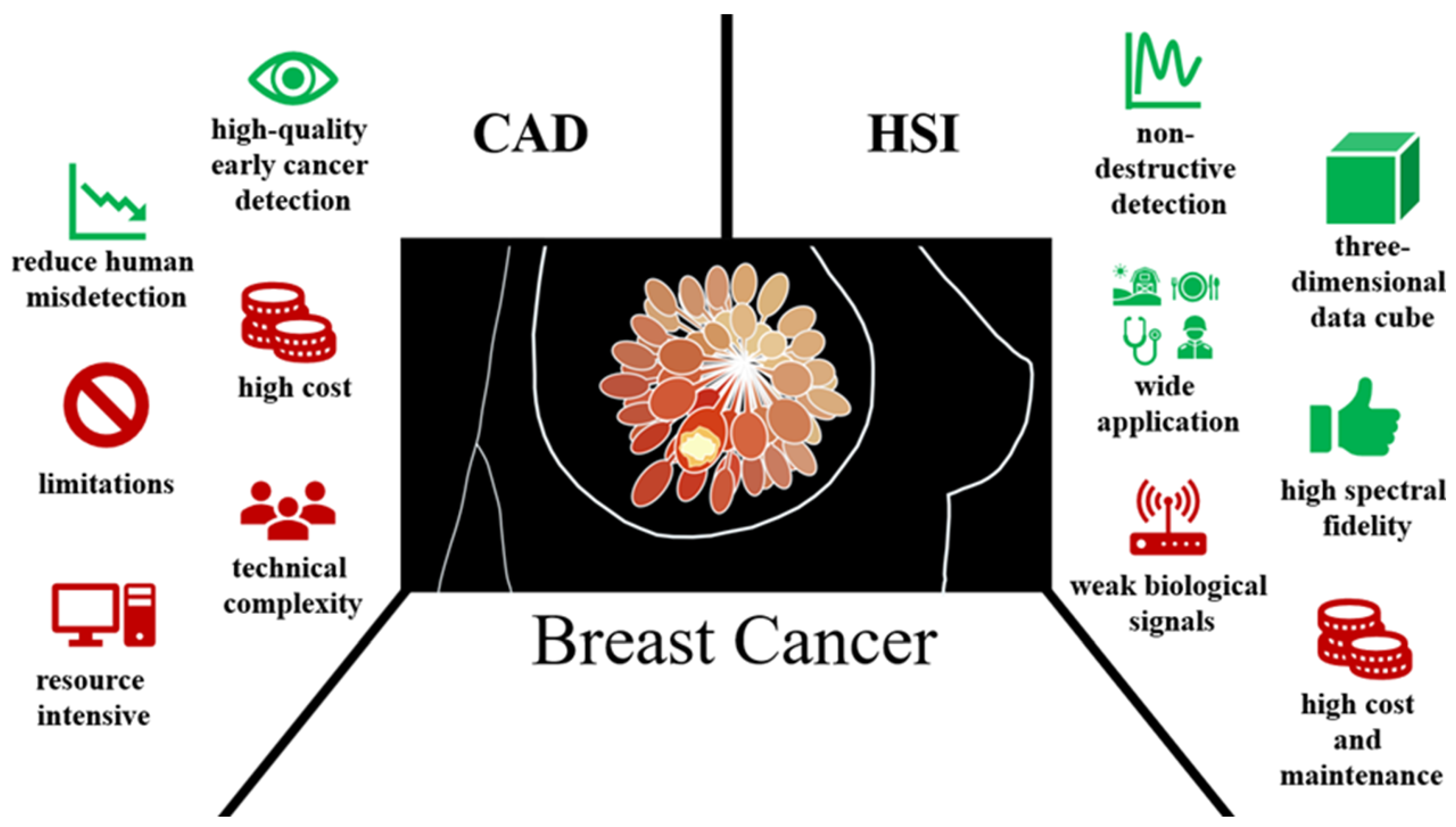
1. Introduction
2. QUADAS-2 Assessment
2.1. Study Selection Criteria
2.2. QUADAS-2 Results
3. Results
3.1. Studies under Clinical Feature Observation
3.2. Meta-Analysis of the Studies
3.3. Meta-Analysis of Subgroup
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tretli, S. Height and weight in relation to breast cancer morbidity and mortality. A prospective study of 570,000 women in Norway. Int. J. Cancer 1989, 44, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Hicks, C. Breast cancer type classification using machine learning. J. Pers. Med. 2021, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Chen, Z.; Tan, M.; Elingarami, S.; Liu, Y.; Li, T.; Deng, Y.; He, N.; Li, S.; Fu, J. A review on methods for diagnosis of breast cancer cells and tissues. Cell Prolif. 2020, 53, e12822. [Google Scholar] [CrossRef]
- Lee, H.-B.; Han, W. Unique features of young age breast cancer and its management. J. Breast Cancer 2014, 17, 301–307. [Google Scholar] [CrossRef]
- Zheng, B.; Yoon, S.W.; Lam, S.S. Breast cancer diagnosis based on feature extraction using a hybrid of K-means and support vector machine algorithms. Expert Syst. Appl. 2014, 41, 1476–1482. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast cancer treatment. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Colditz, G.A.; Rosner, B.A.; Speizer, F.E.; Nurses’ Health Study Research Group. Risk factors for breast cancer according to family history of breast cancer. JNCI J. Natl. Cancer Inst. 1996, 88, 365–371. [Google Scholar] [CrossRef]
- Dupont, W.D.; Page, D.L. Risk factors for breast cancer in women with proliferative breast disease. N. Engl. J. Med. 1985, 312, 146–151. [Google Scholar] [CrossRef]
- McTiernan, A. Behavioral risk factors in breast cancer: Can risk be modified? Oncol. 2003, 8, 326–334. [Google Scholar] [CrossRef]
- Yuan, J.-M.; Yu, M.C.; Ross, R.K.; Gao, Y.-T.; Henderson, B.E. Risk factors for breast cancer in Chinese women in Shanghai. Cancer Res. 1988, 48, 1949–1953. [Google Scholar] [PubMed]
- Kamińska, M.; Ciszewski, T.; Łopacka-Szatan, K.; Miotła, P.; Starosławska, E.J. Breast cancer risk factors. Menopause Rev./Przegląd Menopauzalny 2015, 14, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Momenimovahed, Z.; Salehiniya, H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer Targets Ther. 2019, 11, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Singletary, S.E. Rating the risk factors for breast cancer. Ann. Surg. 2003, 237, 474. [Google Scholar] [CrossRef]
- Helmrich, S.P.; Shapiro, S.; Rosenberg, L.; Kaufman, D.W.; Slone, D.; Bain, C.; Miettinen, O.S.; Stolley, P.D.; Rosenshein, N.B.; Knapp, R.C. Risk factors for breast cancer. Am. J. Epidemiol. 1983, 117, 35–45. [Google Scholar] [CrossRef]
- Martin, A.-M.; Weber, B.L. Genetic and hormonal risk factors in breast cancer. J. Natl. Cancer Inst. 2000, 92, 1126–1135. [Google Scholar] [CrossRef]
- Yari, Y.; Nguyen, T.V.; Nguyen, H.T.J.I.A. Deep learning applied for histological diagnosis of breast cancer. IEEE Access 2020, 8, 162432–162448. [Google Scholar] [CrossRef]
- Yassin, N.I.; Omran, S.; El Houby, E.M.; Allam, H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: A systematic review. Comput. Methods Programs Biomed. 2018, 156, 25–45. [Google Scholar] [CrossRef]
- Kasik, D.J.; Buxton, W.; Ferguson, D.R. Ten CAD challenges. IEEE Comput. Graph. Appl. 2005, 25, 81–92. [Google Scholar] [CrossRef]
- Kaushal, C.; Bhat, S.; Koundal, D.; Singla, A.J. Recent trends in computer assisted diagnosis (CAD) system for breast cancer diagnosis using histopathological images. IRBM 2019, 40, 211–227. [Google Scholar] [CrossRef]
- Lim, T.S.; Tay, K.G.; Huong, A.; Lim, X.Y. Breast cancer diagnosis system using hybrid support vector machine-artificial neural network. Int. J. Electr. Comput. Eng. 2021, 11, 3059. [Google Scholar] [CrossRef]
- Baltzer, P.A.; Kapetas, P.; Marino, M.A.; Clauser, P. New diagnostic tools for breast cancer. Memo-Mag. Eur. Med. Oncol. 2017, 10, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Ragab, D.A.; Sharkas, M.; Attallah, O. Breast cancer diagnosis using an efficient CAD system based on multiple classifiers. Diagnostics 2019, 9, 165. [Google Scholar] [CrossRef]
- Chan, H.P.; Hadjiiski, L.M.; Samala, R.K. Computer-aided diagnosis in the era of deep learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef]
- Fujita, H. AI-based computer-aided diagnosis (AI-CAD): The latest review to read first. Radiol. Phys. Technol. 2020, 13, 6–19. [Google Scholar] [CrossRef]
- Chan, H.-P.; Samala, R.K.; Hadjiiski, L.M. CAD and AI for breast cancer—Recent development and challenges. Br. J. Radiol. 2019, 93, 20190580. [Google Scholar] [CrossRef]
- Alam, N.; Islam, M.J. Pectoral muscle elimination on mammogram using K-means clustering approach. Int. J. Comput. Vis. Signal Process. 2014, 4, 11–21. [Google Scholar]
- Mokni, R.; Gargouri, N.; Damak, A.; Sellami, D.; Feki, W.; Mnif, Z. An automatic Computer-Aided Diagnosis system based on the Multimodal fusion of Breast Cancer (MF-CAD). Biomed. Signal Process. Control. 2021, 69, 102914. [Google Scholar] [CrossRef]
- Henriksen, E.L.; Carlsen, J.F.; Vejborg, I.M.; Nielsen, M.B.; Lauridsen, C.A. The efficacy of using computer-aided detection (CAD) for detection of breast cancer in mammography screening: A systematic review. Acta Radiol. 2019, 60, 13–18. [Google Scholar] [CrossRef]
- Salama, M.S.; Eltrass, A.S.; Elkamchouchi, H.M. An improved approach for computer-aided diagnosis of breast cancer in digital mammography. In Proceedings of the 2018 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Rome, Italy, 11–13 June 2018; pp. 1–5. [Google Scholar]
- Xu, L.; Shoaie, N.; Jahanpeyma, F.; Zhao, J.; Azimzadeh, M.; Al−Jamal, K.T. Optical, electrochemical and electrical (nano)biosensors for detection of exosomes: A comprehensive overview. Biosens. Bioelectron. 2020, 161, 112222. [Google Scholar] [CrossRef]
- Hsiao, Y.-P.; Mukundan, A.; Chen, W.-C.; Wu, M.-T.; Hsieh, S.-C.; Wang, H.-C. Design of a Lab-On-Chip for Cancer Cell Detection through Impedance and Photoelectrochemical Response Analysis. Biosensors 2022, 12, 405. [Google Scholar] [CrossRef] [PubMed]
- Zare, Y.; Rhee, K.Y. Effect of contact resistance on the electrical conductivity of polymer graphene nanocomposites to optimize the biosensors detecting breast cancer cells. Sci. Rep. 2022, 12, 5406. [Google Scholar] [CrossRef] [PubMed]
- Chupradit, S.; Jasim, S.A.; Bokov, D.; Mahmoud, M.Z.; Roomi, A.B.; Hachem, K.; Rudiansyah, M.; Suksatan, W.; Bidares, R. Recent advances in biosensor devices for HER-2 cancer biomarker detection. Anal. Methods 2022, 14, 1301–1310. [Google Scholar] [CrossRef]
- Salahandish, R.; Ghaffarinejad, A.; Naghib, S.M.; Majidzadeh-A, K.; Zargartalebi, H.; Sanati-Nezhad, A. Nano-biosensor for highly sensitive detection of HER2 positive breast cancer. Biosens. Bioelectron. 2018, 117, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Liu, X.; Xie, Y.; Chen, M.; Zheng, C.; Zhong, H.; Li, M. Integrated SERS-Vertical Flow Biosensor Enabling Multiplexed Quantitative Profiling of Serological Exosomal Proteins in Patients for Accurate Breast Cancer Subtyping. ACS Nano 2023, 17, 4077–4088. [Google Scholar] [CrossRef]
- Zheng, Z.; Wu, L.; Li, L.; Zong, S.; Wang, Z.; Cui, Y. Simultaneous and highly sensitive detection of multiple breast cancer biomarkers in real samples using a SERS microfluidic chip. Talanta 2018, 188, 507–515. [Google Scholar] [CrossRef]
- Tourassi, G.D.; Frederick, E.D.; Markey, M.K.; Floyd, C.E., Jr. Application of the mutual information criterion for feature selection in computer-aided diagnosis. Med. Phys. 2001, 28, 2394–2402. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef]
- Kenway, G.; Kennedy, G.; Martins, J.R. A CAD-free approach to high-fidelity aerostructural optimization. In Proceedings of the 13th AIAA/ISSMO Multidisciplinary Analysis Optimization Conference, Ft. Worth, TX, USA, 13–15 September 2010; p. 9231. [Google Scholar]
- Ekpo, E.U.; Alakhras, M.; Brennan, P. Errors in mammography cannot be solved through technology alone. Asian Pac. J. Cancer Prev. 2018, 19, 291. [Google Scholar]
- Singh, B.K.; Verma, K.; Panigrahi, L.; Thoke, A. Integrating radiologist feedback with computer aided diagnostic systems for breast cancer risk prediction in ultrasonic images: An experimental investigation in machine learning paradigm. Expert Syst. Appl. 2017, 90, 209–223. [Google Scholar] [CrossRef]
- Hakimian, F.; Ghourchian, H. Ultrasensitive electrochemical biosensor for detection of microRNA-155 as a breast cancer risk factor. Anal. Chim. Acta 2020, 1136, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, V.S.A.; Das, A.B.; Saxena, U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Shen, Q. Spectral–spatial classification of hyperspectral imagery with 3D convolutional neural network. Remote Sens. 2017, 9, 67. [Google Scholar] [CrossRef]
- Goetz, A.F.; Vane, G.; Solomon, J.E.; Rock, B.N. Imaging spectrometry for earth remote sensing. Science 1985, 228, 1147–1153. [Google Scholar] [CrossRef]
- ElMasry, G.; Sun, D.-W. Principles of hyperspectral imaging technology. In Hyperspectral Imaging for Food Quality Analysis and Control; Elsevier: Amsterdam, The Netherlands, 2010; pp. 3–43. [Google Scholar]
- Manolakis, D.; Shaw, G. Detection algorithms for hyperspectral imaging applications. IEEE Signal Process. Mag. 2002, 19, 29–43. [Google Scholar] [CrossRef]
- Serranti, S.; Bonifazi, G. Hyperspectral imaging and its applications. In Optical Sensing and Detection IV; SPIE: Bellingham, WA, USA, 2016; pp. 146–165. [Google Scholar]
- Rehman, A.u.; Qureshi, S.A. A review of the medical hyperspectral imaging systems and unmixing algorithms’ in biological tissues. Photodiagnosis Photodyn. Ther. 2021, 33, 102165. [Google Scholar] [CrossRef] [PubMed]
- Signoroni, A.; Savardi, M.; Baronio, A.; Benini, S. Deep learning meets hyperspectral image analysis: A multidisciplinary review. J. Imaging 2019, 5, 52. [Google Scholar] [CrossRef]
- Lv, W.; Wang, X. Overview of hyperspectral image classification. J. Sens. 2020, 2020, 4817234. [Google Scholar] [CrossRef]
- Fabelo, H.; Ortega, S.; Ravi, D.; Kiran, B.R.; Sosa, C.; Bulters, D.; Callicó, G.M.; Bulstrode, H.; Szolna, A.; Piñeiro, J.F. Spatio-spectral classification of hyperspectral images for brain cancer detection during surgical operations. PLoS ONE 2018, 13, e0193721. [Google Scholar] [CrossRef]
- Hazlyna, H.N.; Mashor, M.Y.; Mokhtar, N.R.; Salihah, A.A.; Hassan, R.; Raof, R.A.A.; Osman, M.K. Comparison of acute leukemia Image segmentation using HSI and RGB color space. In Proceedings of the 10th International Conference on Information Science, Signal Processing and their Applications (ISSPA 2010), Kuala Lumpur, Malaysia, 10–13 May 2010; pp. 749–752. [Google Scholar]
- Jau, U.L.; Teh, C.S.; Ng, G.W. A comparison of RGB and HSI color segmentation in real-time video images: A preliminary study on road sign detection. In Proceedings of the 2008 International Symposium on Information Technology, Kuala Lumpur, Malaysia, 26–28 August 2008; pp. 1–6. [Google Scholar]
- Taghizadeh, M.; Gowen, A.A.; O’Donnell, C.P. Comparison of hyperspectral imaging with conventional RGB imaging for quality evaluation of Agaricus bisporus mushrooms. Biosyst. Eng. 2011, 108, 191–194. [Google Scholar] [CrossRef]
- Li, K.; Dai, D.; Gool, L.V. Hyperspectral Image Super-Resolution with RGB Image Super-Resolution as an Auxiliary Task. In Proceedings of the 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2022; pp. 4039–4048. [Google Scholar]
- Yan, L.; Wang, X.; Zhao, M.; Kaloorazi, M.; Chen, J.; Rahardja, S. Reconstruction of Hyperspectral Data From RGB Images With Prior Category Information. IEEE Trans. Comput. Imaging 2020, 6, 1070–1081. [Google Scholar] [CrossRef]
- Tao, C.; Zhu, H.; Sun, P.; Wu, R.; Zheng, Z. Hyperspectral image recovery based on fusion of coded aperture snapshot spectral imaging and RGB images by guided filtering. Opt. Commun. 2020, 458, 124804. [Google Scholar] [CrossRef]
- Akhtar, N.; Mian, A. Hyperspectral Recovery from RGB Images using Gaussian Processes. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Manickavasagan, A. Machine learning techniques for analysis of hyperspectral images to determine quality of food products: A review. Curr. Res. Food Sci. 2021, 4, 28–44. [Google Scholar] [CrossRef]
- Paulus, S.; Mahlein, A.-K. Technical workflows for hyperspectral plant image assessment and processing on the greenhouse and laboratory scale. GigaScience 2020, 9, giaa090. [Google Scholar] [CrossRef] [PubMed]
- Ortega, S.; Guerra, R.; Diaz, M.; Fabelo, H.; López, S.; Callico, G.M.; Sarmiento, R. Hyperspectral push-broom microscope development and characterization. IEEE Access 2019, 7, 122473–122491. [Google Scholar] [CrossRef]
- Jia, B.; Wang, W.; Ni, X.; Lawrence, K.C.; Zhuang, H.; Yoon, S.-C.; Gao, Z. Essential processing methods of hyperspectral images of agricultural and food products. Chemom. Intell. Lab. Syst. 2020, 198, 103936. [Google Scholar] [CrossRef]
- Ma, J.; Sun, D.-W.; Pu, H.; Wei, Q.; Wang, X. Protein content evaluation of processed pork meats based on a novel single shot (snapshot) hyperspectral imaging sensor. J. Food Eng. 2019, 240, 207–213. [Google Scholar] [CrossRef]
- Stergar, J.; Hren, R.; Milanič, M. Design and validation of a custom-made hyperspectral microscope imaging system for biomedical applications. Sensors 2023, 23, 2374. [Google Scholar] [CrossRef]
- Pu, H.; Lin, L.; Sun, D.W. Principles of hyperspectral microscope imaging techniques and their applications in food quality and safety detection: A review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 853–866. [Google Scholar] [CrossRef]
- Constantinou, P.; Nicklee, T.; Hedley, D.W.; Damaskinos, S.; Wilson, B.C. A high-resolution MACROscope with differential phase contrast, transmitted light, confocal fluorescence, and hyperspectral capabilities for large-area tissue imaging. IEEE J. Sel. Top. Quantum Electron. 2005, 11, 766–777. [Google Scholar] [CrossRef]
- Xu, Z.; Jiang, Y.; He, S. Multi-mode microscopic hyperspectral imager for the sensing of biological samples. Appl. Sci. 2020, 10, 4876. [Google Scholar] [CrossRef]
- Leon, R.; Gelado, S.H.; Fabelo, H.; Ortega, S.; Quintana, L.; Szolna, A.; Piñeiro, J.F.; Balea-Fernandez, F.; Morera, J.; Clavo, B. Hyperspectral imaging for in-vivo/ex-vivo tissue analysis of human brain cancer. In Proceedings of the Medical Imaging 2022: Image-Guided Procedures, Robotic Interventions, and Modeling, San Diego, CA, USA, 20–23 February 2022; pp. 525–534. [Google Scholar]
- Halicek, M.; Fabelo, H.; Ortega, S.; Callico, G.M.; Fei, B. In-vivo and ex-vivo tissue analysis through hyperspectral imaging techniques: Revealing the invisible features of cancer. Cancers 2019, 11, 756. [Google Scholar] [CrossRef]
- Liu, N.; Gonzalez, J.M.; Ottestad, S.; Hernandez, J. Application of hyperspectral imaging for cocoa bean grading with machine learning approaches. In Proceedings of the Hyperspectral Imaging and Applications II, Birmingham, UK, 6–7 December 2022; pp. 38–43. [Google Scholar]
- Lu, B.; Dao, P.D.; Liu, J.; He, Y.; Shang, J. Recent advances of hyperspectral imaging technology and applications in agriculture. Remote Sens. 2020, 12, 2659. [Google Scholar] [CrossRef]
- Liu, S.; Marinelli, D.; Bruzzone, L.; Bovolo, F. A review of change detection in multitemporal hyperspectral images: Current techniques, applications, and challenges. IEEE Geosci. Remote Sens. Mag. 2019, 7, 140–158. [Google Scholar] [CrossRef]
- He, J.; Zhao, L.; Yang, H.; Zhang, M.; Li, W. HSI-BERT: Hyperspectral image classification using the bidirectional encoder representation from transformers. IEEE Trans. Geosci. Remote Sens. 2019, 58, 165–178. [Google Scholar] [CrossRef]
- Chen, C.-W.; Tseng, Y.-S.; Mukundan, A.; Wang, H.-C. Air Pollution: Sensitive Detection of PM2.5 and PM10 Concentration Using Hyperspectral Imaging. Appl. Sci. 2021, 11, 4543. [Google Scholar] [CrossRef]
- Mukundan, A.; Huang, C.-C.; Men, T.-C.; Lin, F.-C.; Wang, H.-C. Air Pollution Detection Using a Novel Snap-Shot Hyperspectral Imaging Technique. Sensors 2022, 22, 6231. [Google Scholar] [CrossRef]
- Wu, D.; Sun, D.-W. Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: A review—Part I: Fundamentals. Innov. Food Sci. Emerg. Technol. 2013, 19, 1–14. [Google Scholar] [CrossRef]
- Huang, H.; Liu, L.; Ngadi, M.O. Recent developments in hyperspectral imaging for assessment of food quality and safety. Sensors 2014, 14, 7248–7276. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.-J.; Mukundan, A.; Tsao, Y.-M.; Huang, C.-W.; Wang, H.-C. Identification of Early Esophageal Cancer by Semantic Segmentation. J. Pers. Med. 2022, 12, 1204. [Google Scholar] [CrossRef]
- Offerhaus, H.L.; Bohndiek, S.E.; Harvey, A.R. Hyperspectral imaging in biomedical applications. J. Opt. 2019, 21, 010202. [Google Scholar] [CrossRef]
- Tiwari, K.C.; Arora, M.K.; Singh, D. An assessment of independent component analysis for detection of military targets from hyperspectral images. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 730–740. [Google Scholar] [CrossRef]
- Shimoni, M.; Haelterman, R.; Perneel, C. Hypersectral imaging for military and security applications: Combining myriad processing and sensing techniques. IEEE Geosci. Remote Sens. Mag. 2019, 7, 101–117. [Google Scholar] [CrossRef]
- Mukundan, A.; Tsao, Y.-M.; Lin, F.-C.; Wang, H.-C. Portable and low-cost hologram verification module using a snapshot-based hyperspectral imaging algorithm. Sci. Rep. 2022, 12, 18475. [Google Scholar] [CrossRef]
- Liu, Y.; Lee, Y.H.; Zhang, Q.; Cui, Y.; Ling, X.Y. Plasmonic nanopillar arrays encoded with multiplex molecular information for anti-counterfeiting applications. J. Mater. Chem. C 2016, 4, 4312–4319. [Google Scholar] [CrossRef]
- Nieke, J.; Rast, M. Towards the copernicus hyperspectral imaging mission for the environment (CHIME). In Proceedings of the Igarss 2018-2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; pp. 157–159. [Google Scholar]
- Govender, M.; Chetty, K.; Bulcock, H. A review of hyperspectral remote sensing and its application in vegetation and water resource studies. Water Sa 2007, 33, 145–151. [Google Scholar] [CrossRef]
- Barducci, A.; Guzzi, D.; Marcoionni, P.; Pippi, I. Aerospace wetland monitoring by hyperspectral imaging sensors: A case study in the coastal zone of San Rossore Natural Park. J. Environ. Manag. 2009, 90, 2278–2286. [Google Scholar] [CrossRef]
- Dmitriev, E.V.; Kozoderov, V.; Dementyev, A.; Safonova, A. Combining classifiers in the problem of thematic processing of hyperspectral aerospace images. Optoelectron. Instrum. Data Process. 2018, 54, 213–221. [Google Scholar] [CrossRef]
- Ertürk, A.; Plaza, A. Informative change detection by unmixing for hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1252–1256. [Google Scholar] [CrossRef]
- Lu, Y.; Perez, D.; Dao, M.; Kwan, C.; Li, J. Deep learning with synthetic hyperspectral images for improved soil detection in multispectral imagery. In Proceedings of the 2018 9th IEEE annual ubiquitous computing, Electronics & Mobile Communication Conference (UEMCON), New York, NY, USA, 8–10 November 2018; pp. 666–672. [Google Scholar]
- Xue, Q.; Qi, M.; Li, Z.; Yang, B.; Li, W.; Wang, F.; Li, Q. Fluorescence hyperspectral imaging system for analysis and visualization of oil sample composition and thickness. Appl. Opt. 2021, 60, 8349–8359. [Google Scholar] [CrossRef]
- Nalepa, J. Recent Advances in Multi- and Hyperspectral Image Analysis. Sensors 2021, 21, 6002. [Google Scholar] [CrossRef]
- Gomez, R.B. Hyperspectral imaging: A useful technology for transportation analysis. Opt. Eng. 2002, 41, 2137–2143. [Google Scholar] [CrossRef]
- Manolakis, D. Taxonomy of detection algorithms for hyperspectral imaging applications. Opt. Eng. 2005, 44, 066403. [Google Scholar] [CrossRef]
- Grahn, H.; Geladi, P. Techniques and Applications of Hyperspectral Image Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Ortega, S.; Fabelo, H.; Camacho, R.; De la Luz Plaza, M.; Callicó, G.M.; Sarmiento, R. Detecting brain tumor in pathological slides using hyperspectral imaging. Biomed. Opt. Express 2018, 9, 818–831. [Google Scholar] [CrossRef]
- Martinez, B.; Leon, R.; Fabelo, H.; Ortega, S.; Piñeiro, J.F.; Szolna, A.; Hernandez, M.; Espino, C.; J. O’Shanahan, A.; Carrera, D. Most relevant spectral bands identification for brain cancer detection using hyperspectral imaging. Sensors 2019, 19, 5481. [Google Scholar] [CrossRef]
- Florimbi, G.; Fabelo, H.; Torti, E.; Ortega, S.; Marrero-Martin, M.; Callico, G.M.; Danese, G.; Leporati, F. Towards real-time computing of intraoperative hyperspectral imaging for brain cancer detection using multi-GPU platforms. IEEE Access 2020, 8, 8485–8501. [Google Scholar] [CrossRef]
- Torti, E.; Florimbi, G.; Castelli, F.; Ortega, S.; Fabelo, H.; Callicó, G.M.; Marrero-Martin, M.; Leporati, F. Parallel K-means clustering for brain cancer detection using hyperspectral images. Electronics 2018, 7, 283. [Google Scholar] [CrossRef]
- Maktabi, M.; Wichmann, Y.; Köhler, H.; Ahle, H.; Lorenz, D.; Bange, M.; Braun, S.; Gockel, I.; Chalopin, C.; Thieme, R. Tumor cell identification and classification in esophageal adenocarcinoma specimens by hyperspectral imaging. Sci. Rep. 2022, 12, 1–14. [Google Scholar] [CrossRef]
- Schröder, A.; Maktabi, M.; Thieme, R.; Jansen–Winkeln, B.; Gockel, I.; Chalopin, C. Evaluation of artificial neural networks for the detection of esophagus tumor cells in microscopic hyperspectral images. In Proceedings of the 2022 25th Euromicro Conference on Digital System Design (DSD), Maspalomas, Spain, 31 August–2 September 2022; pp. 827–834. [Google Scholar]
- Collins, T.; Maktabi, M.; Barberio, M.; Bencteux, V.; Jansen-Winkeln, B.; Chalopin, C.; Marescaux, J.; Hostettler, A.; Diana, M.; Gockel, I. Automatic recognition of colon and esophagogastric cancer with machine learning and hyperspectral imaging. Diagnostics 2021, 11, 1810. [Google Scholar] [CrossRef]
- Song, J.; Hu, M.; Wang, J.; Zhou, M.; Sun, L.; Qiu, S.; Li, Q.; Sun, Z.; Wang, Y. ALK positive lung cancer identification and targeted drugs evaluation using microscopic hyperspectral imaging technique. Infrared Phys. Technol. 2019, 96, 267–275. [Google Scholar] [CrossRef]
- Amreddy, N.; Muralidharan, R.; Babu, A.; Mehta, M.; Johnson, E.V.; Zhao, Y.D.; Munshi, A.; Ramesh, R. Tumor-targeted and pH-controlled delivery of doxorubicin using gold nanorods for lung cancer therapy. Int. J. Nanomed. 2015, 10, 6773. [Google Scholar]
- Cui, X.; Zhang, Z.; Li, Z.; Cheng, X.-Y.; Qi, Z.-M. Micro-hyperspectral imaging methodology for characterization of lung cancer cell. In Proceedings of the Ninth Symposium on Novel Photoelectronic Detection Technology and Applications, Hefei, China, 21–23 April 2023; pp. 1016–1021. [Google Scholar]
- Calin, M.A.; Parasca, S.V.; Savastru, D.; Manea, D. Hyperspectral imaging in the medical field: Present and future. Appl. Spectrosc. Rev. 2014, 49, 435–447. [Google Scholar] [CrossRef]
- Chen, H.; Li, Q.; Hu, Q.; Jiao, X.; Ren, W.; Wang, S.; Peng, G. Double spiral chip-embedded micro-trapezoid filters (SMT filters) for the sensitive isolation of CTCs of prostate cancer by spectral detection. Nanoscale Adv. 2022, 4, 5392–5403. [Google Scholar] [CrossRef]
- Zarei, N.; Bakhtiari, A.; Gallagher, P.; Keys, M.; MacAulay, C. Automated prostate glandular and nuclei detection using hyperspectral imaging. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 1028–1031. [Google Scholar]
- Liu, S.; Wang, Q.; Zhang, G.; Du, J.; Hu, B.; Zhang, Z. Using hyperspectral imaging automatic classification of gastric cancer grading with a shallow residual network. Anal. Methods 2020, 12, 3844–3853. [Google Scholar] [CrossRef]
- Kiyotoki, S.; Nishikawa, J.; Okamoto, T.; Hamabe, K.; Saito, M.; Goto, A.; Fujita, Y.; Hamamoto, Y.; Takeuchi, Y.; Satori, S.; et al. New method for detection of gastric cancer by hyperspectral imaging: A pilot study. J. Biomed. Opt. 2013, 18, 026010. [Google Scholar] [CrossRef]
- Goto, A.; Nishikawa, J.; Kiyotoki, S.; Nakamura, M.; Nishimura, J.; Okamoto, T.; Ogihara, H.; Fujita, Y.; Hamamoto, Y.; Sakaida, I. Use of hyperspectral imaging technology to develop a diagnostic support system for gastric cancer. J. Biomed. Opt. 2015, 20, 016017. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Hsiao, Y.-P.; Mukundan, A.; Tsao, Y.-M.; Chang, W.-Y.; Wang, H.-C. Classification of Skin Cancer Using Novel Hyperspectral Imaging Engineering via YOLOv5. J. Clin. Med. 2023, 12, 1134. [Google Scholar] [CrossRef]
- Leon, R.; Martinez-Vega, B.; Fabelo, H.; Ortega, S.; Melian, V.; Castaño, I.; Carretero, G.; Almeida, P.; Garcia, A.; Quevedo, E. Non-invasive skin cancer diagnosis using hyperspectral imaging for in-situ clinical support. J. Clin. Med. 2020, 9, 1662. [Google Scholar] [CrossRef]
- Zherdeva, L.A.; Bratchenko, I.A.; Myakinin, O.O.; Moryatov, A.A.; Kozlov, S.V.; Zakharov, V.P. In vivo hyperspectral imaging and differentiation of skin cancer. In Optics in Health Care and Biomedical Optics VII; SPIE: Bellingham, WA, USA, 2016; pp. 658–665. [Google Scholar]
- McCormack, D.R.; Walsh, A.J.; Sit, W.; Arteaga, C.L.; Chen, J.; Cook, R.S.; Skala, M.C. In vivo hyperspectral imaging of microvessel response to trastuzumab treatment in breast cancer xenografts. Biomed. Opt. Express 2014, 5, 2247–2261. [Google Scholar] [CrossRef]
- Hou, Y.; Ren, Z.; Liu, G.; Zeng, L.; Huang, Z. Design of a novel LD-induced hyper-spectral imager for breast cancer diagnosis based on VHT grating. In Proceedings of the 2011 Symposium on Photonics and Optoelectronics (SOPO), Wuhan, China, 16–18 May 2011; pp. 1–4. [Google Scholar]
- Lee, J.; Mulder, F.; Leeflang, M.; Wolff, R.; Whiting, P.; Bossuyt, P.M. QUAPAS: An Adaptation of the QUADAS-2 Tool to Assess Prognostic Accuracy Studies. Ann. Intern. Med. 2022, 175, 1010–1018. [Google Scholar] [CrossRef]
- Wade, R.; Corbett, M.; Eastwood, A. Quality assessment of comparative diagnostic accuracy studies: Our experience using a modified version of the QUADAS-2 tool. Res. Synth. Methods 2013, 4, 280–286. [Google Scholar] [CrossRef]
- Yang, B.; Mallett, S.; Takwoingi, Y.; Davenport, C.F.; Hyde, C.J.; Whiting, P.F.; Deeks, J.J.; Leeflang, M.M.; Group†, Q.-C. QUADAS-C: A tool for assessing risk of bias in comparative diagnostic accuracy studies. Ann. Intern. Med. 2021, 174, 1592–1599. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Aboughaleb, I.H.; Aref, M.H.; El-Sharkawy, Y.H. Hyperspectral imaging for diagnosis and detection of ex-vivo breast cancer. Photodiagnosis Photodyn. Ther. 2020, 31, 101922. [Google Scholar] [CrossRef]
- Kho, E.; Dashtbozorg, B.; De Boer, L.L.; Van de Vijver, K.K.; Sterenborg, H.J.; Ruers, T.J. Broadband hyperspectral imaging for breast tumor detection using spectral and spatial information. Biomed. Opt. Express 2019, 10, 4496–4515. [Google Scholar] [CrossRef]
- Jong, L.-J.S.; de Kruif, N.; Geldof, F.; Veluponnar, D.; Sanders, J.; Peeters, M.-J.T.V.; van Duijnhoven, F.; Sterenborg, H.J.; Dashtbozorg, B.; Ruers, T.J. Discriminating healthy from tumor tissue in breast lumpectomy specimens using deep learning-based hyperspectral imaging. Biomed. Opt. Express 2022, 13, 2581–2604. [Google Scholar] [CrossRef]
- Ortega Sarmiento, S.; Halicek, M.; Fabelo Gómez, H.A.; Guerra Hernández, R.C.; López, C.; Lejeune, M.; Godtliebsen, F.; Marrero Callicó, G.I.; Fei, B. Hyperspectral imaging and deep learning for the detection of breast cancer cells in digitized histological images. Proc. SPIE Int. Soc. Opt. Eng. 2020, 11320, 113200V. [Google Scholar]
- Khouj, Y.; Dawson, J.; Coad, J.; Vona-Davis, L. Hyperspectral Imaging and K-Means Classification for Histologic Evaluation of Ductal Carcinoma In Situ. Front. Oncol. 2018, 8, 17. [Google Scholar] [CrossRef]
- Aref, M.H.; El-Gohary, M.; Elrewainy, A.; Mahmoud, A.; Aboughaleb, I.H.; Hussein, A.A.; Abd El-Ghaffar, S.; Mahran, A.; El-Sharkawy, Y.H. Emerging technology for intraoperative margin assessment and post-operative tissue diagnosis for breast-conserving surgery. Photodiagnosis Photodyn. Ther. 2023, 42, 103507. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Tao, X.; Li, Q.; Sun, L.; Chen, J.; Zhou, M.; Hu, M.; Zhou, X. PCA-U-Net based breast cancer nest segmentation from microarray hyperspectral images. Fundam. Res. 2021, 1, 631–640. [Google Scholar] [CrossRef]
- Kho, E.; de Boer, L.L.; Van de Vijver, K.K.; van Duijnhoven, F.; Vrancken Peeters, M.-J.T.; Sterenborg, H.J.; Ruers, T.J. Hyperspectral Imaging for Resection Margin Assessment during Cancer SurgeryHyperspectral Imaging for Resection Margin Assessment. Clin. Cancer Res. 2019, 25, 3572–3580. [Google Scholar] [CrossRef]
- Deeks, J.J.; Macaskill, P.; Irwig, L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J. Clin. Epidemiol. 2005, 58, 882–893. [Google Scholar] [CrossRef]
- Sterne, J.A.; Sutton, A.J.; Ioannidis, J.P.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.; Rücker, G.; Harbord, R.M.; Schmid, C.H. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011, 343, d4002. [Google Scholar] [CrossRef]
- Andrade, C. Understanding the basics of meta-analysis and how to read a forest plot: As simple as it gets. J. Clin. Psychiatry 2020, 81, 21858. [Google Scholar] [CrossRef]
- Martin, M.E.; Wabuyele, M.B.; Chen, K.; Kasili, P.; Panjehpour, M.; Phan, M.; Overholt, B.; Cunningham, G.; Wilson, D.; DeNovo, R.C. Development of an advanced hyperspectral imaging (HSI) system with applications for cancer detection. Ann. Biomed. Eng. 2006, 34, 1061–1068. [Google Scholar] [CrossRef]
- Fischer, C.; Kakoulli, I. Multispectral and hyperspectral imaging technologies in conservation: Current research and potential applications. Stud. Conserv. 2006, 51, 3–16. [Google Scholar] [CrossRef]
- Sersa, G.; Simoncic, U.; Milanic, M. Imaging perfusion changes in oncological clinical applications by hyperspectral imaging: A literature review. Radiol. Oncol. 2022, 56, 420–429. [Google Scholar]
- Hren, R.; Stergar, J.; Simončič, U.; Serša, G.; Milanič, M. Assessing Perfusion Changes in Clinical Oncology Applications Using Hyperspectral Imaging. In Proceedings of the European Medical and Biological Engineering Conference, Portorož, Slovenia, 9–13 June 2024; pp. 122–129. [Google Scholar]



| Study | Risk of Bias | Applicability Concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard | |
| Aboughaleb et al./2020 [122] | + | + | + | + | + | + | + |
| Kho et al./2019 [123] | + | + | + | + | + | + | + |
| Jong et al./2022 [124] | + | ? | + | + | + | ? | + |
| Ortega et al./2020 [125] | + | ? | + | + | + | ? | + |
| Khouj et al./2018 [126] | + | + | + | + | + | + | + |
| Aref et al./2023 [127] | + | + | + | + | + | + | + |
| Wang et al./2021 [128] | + | + | ? | ? | + | + | + |
| Kho et al./2019 [129] | + | + | + | + | + | + | + |
| Study | Nationality | Index Number | Method | Numberof Patients | Sensitivity (%) | Specificity (%) | Accuracy (%) | Band |
|---|---|---|---|---|---|---|---|---|
| Aboughaleb et al./2020 [122] | Western | 1 | K-mean | 10 | 95.00 | 96.00 | NA | VIS |
| Kho et al./2019 [123] | Western | 2 | LDA + SNV | 42 | 76.00 | 92.00 | NA | VIS + NIR |
| 3 | U-NET + SNV | 42 | 80.00 | 93.00 | ||||
| Jong et al./2022 [124] | Western | 4 | BCCE + 1D-CNN | 29 | 67.00 | 97.00 | 92.00 | VIS + NIR |
| 5 | BCCE + DC-CNN | 62.00 | 95.00 | 89.00 | ||||
| 6 | BCCE + 3D-CNN | 0.00 | 36.00 | 80.00 | ||||
| 7 | PDE + 1D-CNN | 87.00 | 90.00 | 90.00 | ||||
| 8 | PDE + DC-CNN | 78.00 | 94.00 | 91.00 | ||||
| 9 | PDE + 3D-CNN | 72.00 | 84.00 | 82.00 | ||||
| Ortega et al./2020 [125] | Western | 10 | 2D-CNN | 112 | 92.00 | 87.00 | 88.00 | VIS + NIR |
| Khouj et al./2018 [126] | Western | 11 | K-means | 10 | 85.45 | 94.64 | 80.27 | VIS |
| Aref et al./2023 [127] | Western | 12 | K-means | 30 | 98.95 | 98.44 | NA | VIS + NIR |
| Wang et al./2021 [128] | Asian | 13 | PCA + U-Net | 30 | 84.12 | 84.12 | 87.14 | UV + VIS + NIR |
| Kho et al./2019 [129] | Western | 14 | SVM | 8 | 94.00 | 94.00 | 93.00 | NIR |
| 15 | 9 | 96.00 | 96.00 | 84.00 |
| Subgroup | Number of Studies | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|
| Average meta-analysis | 8 | 77.83 | 88.75 | 86.95 |
| Nationality | ||||
| Western | 7 | 77.39 | 89.08 | 86.93 |
| Asian | 1 | 84.12 | 84.12 | 87.14 |
| Methods | ||||
| K-mean | 3 | 93.13 | 96.36 | 80.27 |
| CNN | 7 | 65.43 | 83.29 | 87.43 |
| SNV | 2 | 78.00 | 92.50 | NA |
| PCA | 1 | 84.12 | 84.12 | 87.14 |
| SVM | 2 | 95.00 | 95.00 | 88.50 |
| Wavelength range bands | ||||
| VIS + NIR | 10 | 71.30 | 86.64 | 87.43 |
| VIS | 2 | 90.23 | 95.32 | 80.27 |
| NIR | 2 | 95.00 | 95.00 | 88.50 |
| UV + VIS + NIR | 1 | 84.12 | 84.12 | 87.14 |
| Published Years | ||||
| Before 2019 | 1 | 85.45 | 94.64 | 80.27 |
| 2019–2021 | 5 | 88.16 | 91.73 | 88.04 |
| 2022–2023 | 2 | 66.42 | 84.92 | 87.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leung, J.-H.; Karmakar, R.; Mukundan, A.; Thongsit, P.; Chen, M.-M.; Chang, W.-Y.; Wang, H.-C. Systematic Meta-Analysis of Computer-Aided Detection of Breast Cancer Using Hyperspectral Imaging. Bioengineering 2024, 11, 1060. https://doi.org/10.3390/bioengineering11111060
Leung J-H, Karmakar R, Mukundan A, Thongsit P, Chen M-M, Chang W-Y, Wang H-C. Systematic Meta-Analysis of Computer-Aided Detection of Breast Cancer Using Hyperspectral Imaging. Bioengineering. 2024; 11(11):1060. https://doi.org/10.3390/bioengineering11111060
Chicago/Turabian StyleLeung, Joseph-Hang, Riya Karmakar, Arvind Mukundan, Pacharasak Thongsit, Meei-Maan Chen, Wen-Yen Chang, and Hsiang-Chen Wang. 2024. "Systematic Meta-Analysis of Computer-Aided Detection of Breast Cancer Using Hyperspectral Imaging" Bioengineering 11, no. 11: 1060. https://doi.org/10.3390/bioengineering11111060
APA StyleLeung, J.-H., Karmakar, R., Mukundan, A., Thongsit, P., Chen, M.-M., Chang, W.-Y., & Wang, H.-C. (2024). Systematic Meta-Analysis of Computer-Aided Detection of Breast Cancer Using Hyperspectral Imaging. Bioengineering, 11(11), 1060. https://doi.org/10.3390/bioengineering11111060











