Anomaly Detection in Embryo Development and Morphology Using Medical Computer Vision-Aided Swin Transformer with Boosted Dipper-Throated Optimization Algorithm
Abstract
1. Introduction
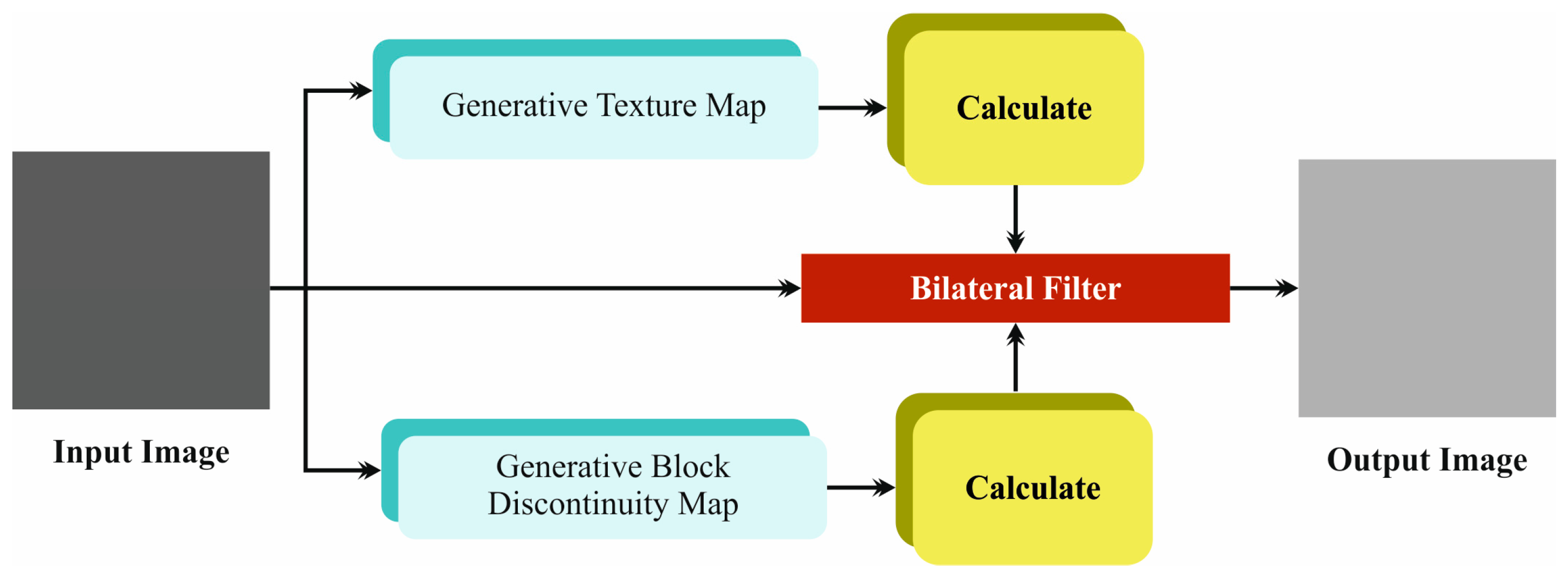
- The EDMCV-STBDTO technique utilizes the BF model to efficiently mitigate noise in embryo images, which improves overall image quality. This preprocessing step crucially enhances the reliability of subsequent feature extraction, allowing for more precise classification outcomes in the evaluation of embryo development.
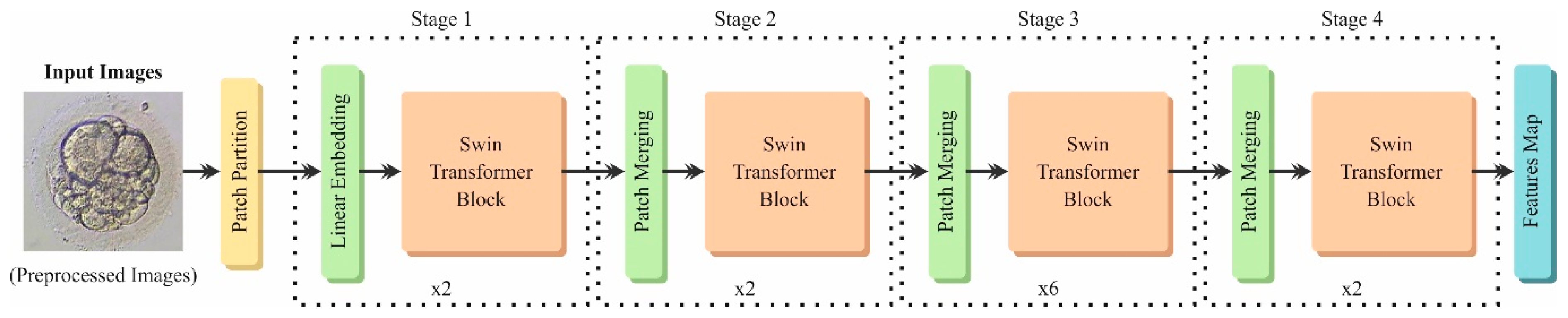
- The ST method is employed by the EDMCV-STBDTO technique to enable advanced feature representation, effectually capturing complex patterns within the embryo image data. This methodology improves the capability of the approach to discern subtle differences in embryo quality, ultimately resulting in an enhanced classification accuracy. The integration of this cutting-edge architecture emphasizes the significance of robust feature extraction in DL applications.
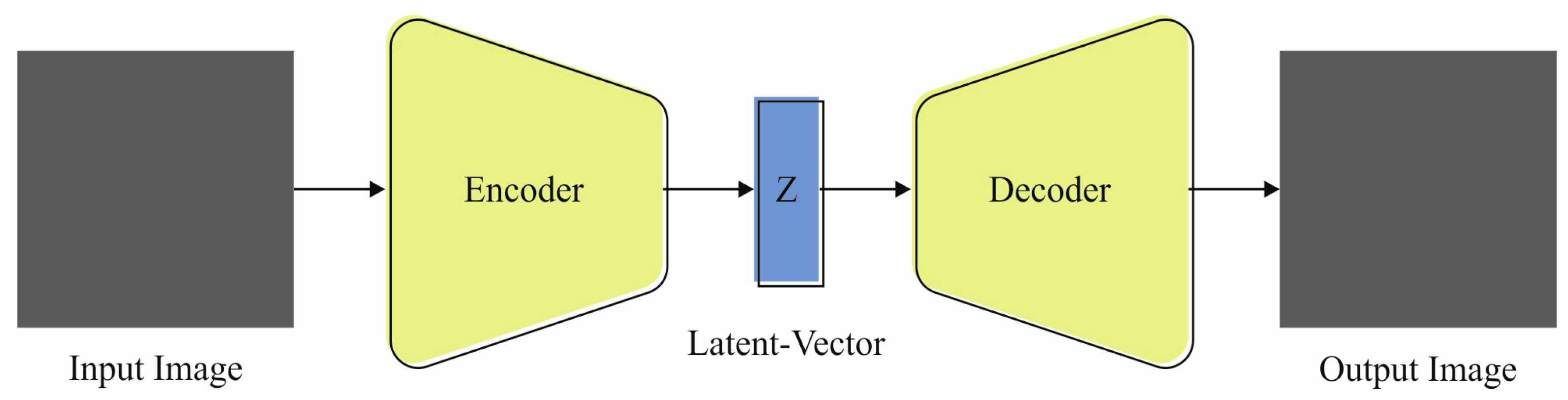
- The EDMCV-STBDTO model employs a VAE method to classify human embryo development, capitalizing on its capacity to learn intrinsic data dispersions. This methodology allows for efficient modeling of the underlying characteristics of embryo images, facilitating precise differentiation between quality classes. By incorporating the VAE, the approach improves the overall predictive performance of the classification task.
- The BDTO model is implemented by the EDMCV-STBDTO technique for the effectual selection of hyperparameters in the VAE method, which improves the performance and accuracy of the approach. This optimization model streamlines the tuning process, allowing for a more efficient exploration of the hyperparameter space. By enhancing the VAE’s configuration, the approach results in improved classification outcomes in embryo quality analysis.
- The incorporation of an ST with a VAE model for embryo classification depicts a novel methodology, integrating advanced DL techniques to substantially improve predictive capabilities in reproductive science. This integration allows for an enhanced feature extraction and representation, effectually addressing intrinsic data patterns in embryo images. By employing these advanced techniques, the model not only enhances classification accuracy but also contributes to a deeper understanding of embryo quality evaluation.
2. Literature Review
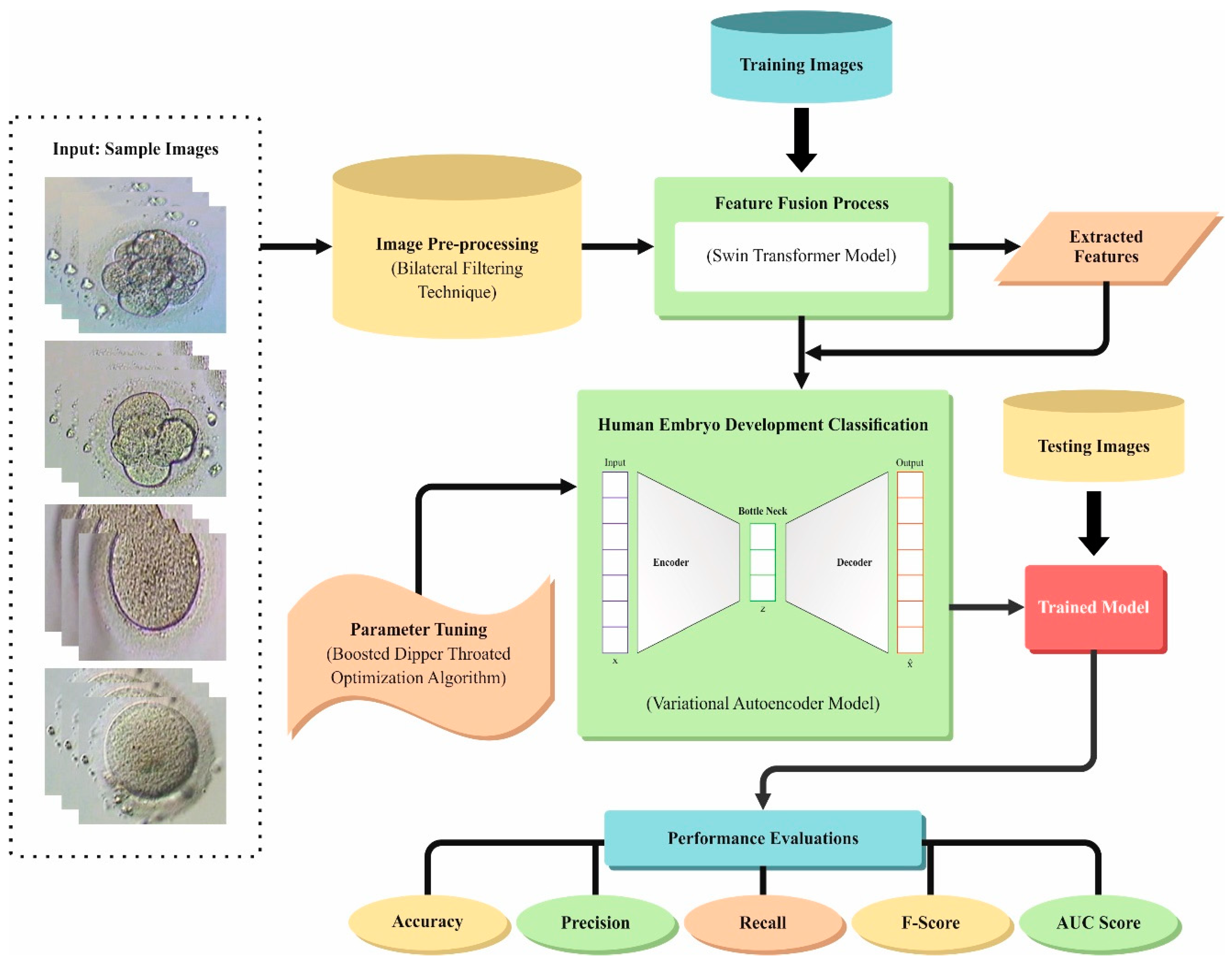
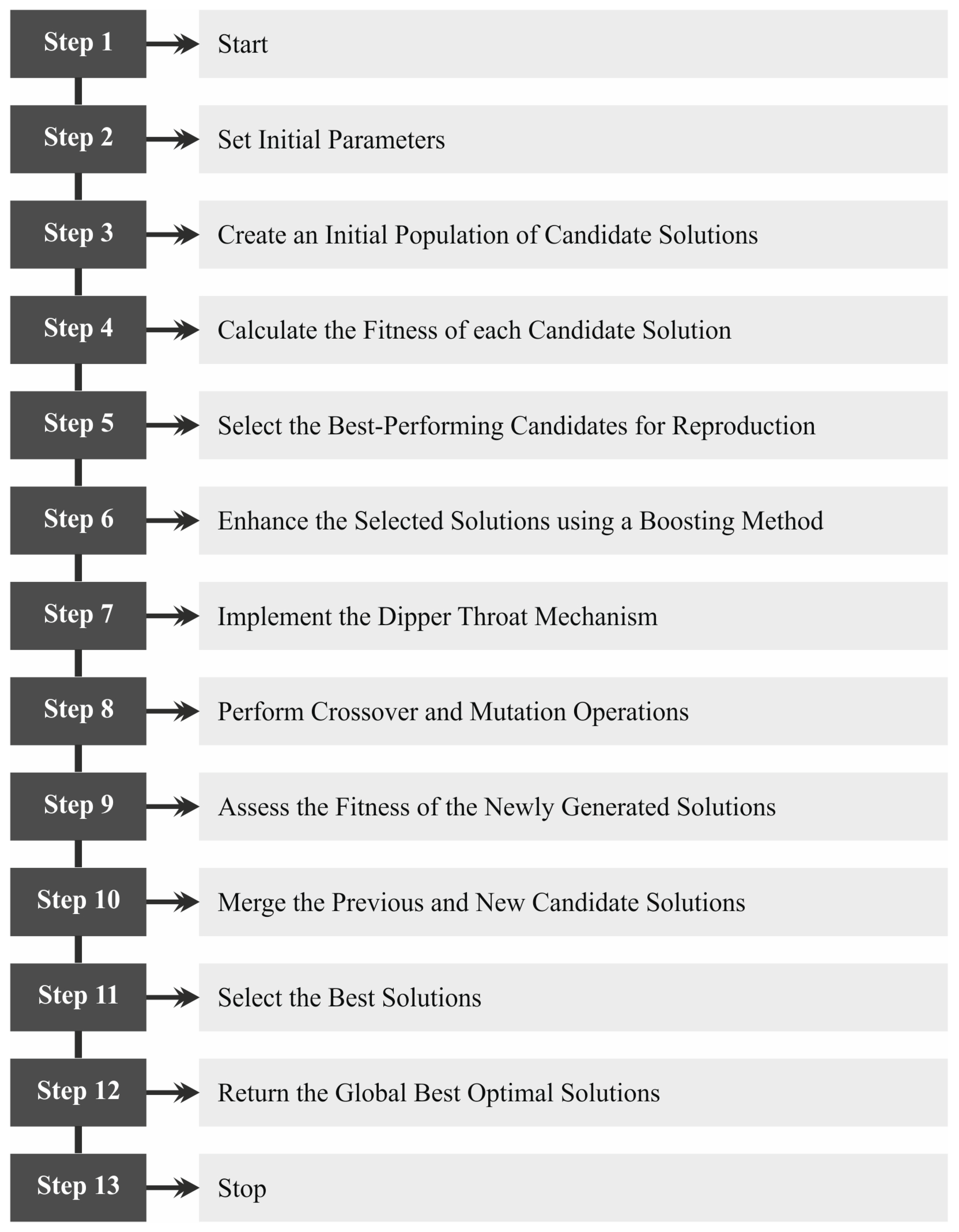
3. Proposed Method
3.1. Noise Reduction
3.2. Feature Extraction Using Swin Transformation
3.2.1. Phase 1: Early Transformation and Embedding
3.2.2. Phase 2: Hierarchical Representation
3.2.3. Phases 3 and 4: Additional Hierarchical Representation
- SW-MSA.
- A dual-layer function of multilayer perceptron (MLP) with Gaussian Error Linear Unit (GELU).
- Normalization layers (LNs) are used before every MSA and MLP element.
- Residual connections are used next to every module.
3.3. Classification Using VAE Model
3.4. BDTO-Based Parameter Tuning
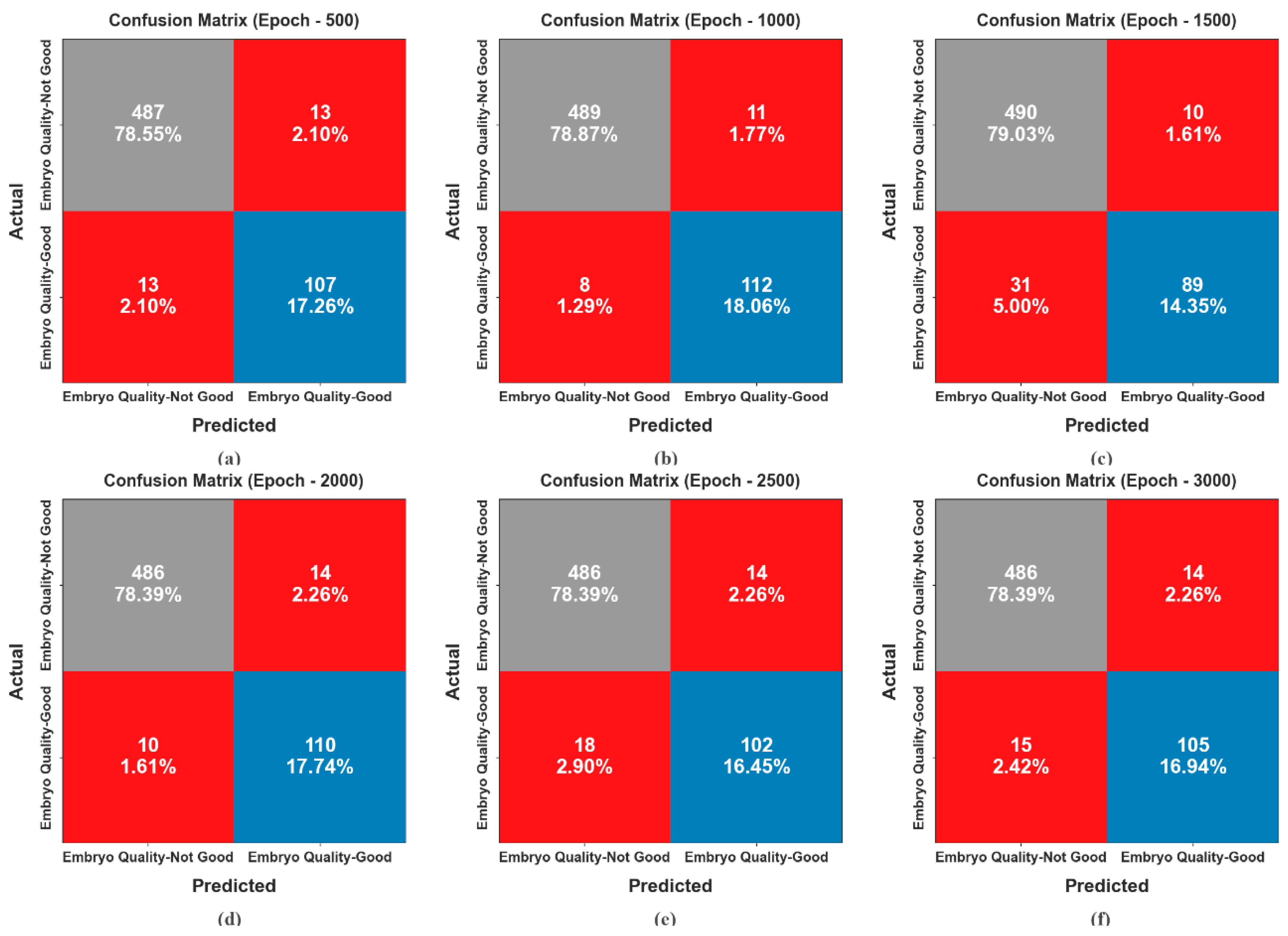
4. Experimental Validation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Mahmoudinia, M.; Sovizi, B.; Ebadi, S.M.R.; Zakerinasab, F.; Sadeghi, T.; Mahmoudinia, M. Live Birth after Cleavage-Stage versus Blastocyst-Stage Embryo Transfer in Assisted Reproductive Technology: A Randomised Controlled Study. Int. J. Fertil. Steril. 2024, 18 (Suppl. S1), 10–16. [Google Scholar] [PubMed]
- Kragh, M.F.; Rimestad, J.; Lassen, J.T.; Berntsen, J.; Karstoft, H. Predicting Embryo Viability Based on Self-Supervised Alignment of Time-Lapse Videos. IEEE Trans. Med. Imaging 2022, 41, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadis, I.; Zaninovic, N.; Badiola, A.C.; Bormann, C.L. Artificial intelligence in the embryology laboratory: A review. Reprod. Biomed. Online 2022, 44, 435–448. [Google Scholar] [CrossRef]
- VerMilyea, M.; Hall, J.M.M.; Diakiw, S.M.; Johnston, A.; Nguyen, T.; Perugini, D.; Miller, A.; Picou, A.; Murphy, A.P.; Perugini, M. Development of an artificial intelligence-based assessment model for prediction of embryo viability using static images captured by optical light microscopy during IVF. Hum. Reprod. 2020, 35, 770–784. [Google Scholar] [CrossRef]
- Liao, Q.; Zhang, Q.; Feng, X.; Huang, H.; Xu, H.; Tian, B.; Liu, J.; Yu, Q.; Guo, N.; Liu, Q.; et al. Development of deep learning algorithms for predicting blastocyst formation and quality by time-lapse monitoring. Commun. Biol. 2021, 4, 415. [Google Scholar] [CrossRef]
- Berntsen, J.; Rimestad, J.; Lassen, J.T.; Tran, D.; Kragh, M.F. Robust and generalizable embryo selection based on artificial intelligence and time-lapse image sequences. PLoS ONE 2022, 17, e0262661. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Badiola, A.; Flores-Saiffe Farias, A.; Mendizabal-Ruiz, G.; Garcia-Sanchez, R.; Drakeley, A.J.; Garcia-Sandoval, J.P. Predicting pregnancy test results after embryo transfer by image feature extraction and analysis using machine learning. Sci. Rep. 2020, 10, 4394. [Google Scholar] [CrossRef]
- Bormann, C.L.; Kanakasabapathy, M.K.; Thirumalaraju, P.; Gupta, R.; Pooniwala, R.; Kandula, H.; Hariton, E.; Souter, I.; Dimitriadis, I.; Ramirez, L.B.; et al. Performance of a deep learning based neural network in the selection of human blastocysts for implantation. eLife 2020, 9, e55301. [Google Scholar] [CrossRef]
- Gao, J.; Yuan, Y.; Li, J.; Tian, T.; Lian, Y.; Liu, P.; Li, R.; Qiao, J.; Long, X.; Wang, H. Sequential embryo transfer versus double cleavage-stage embryo or double blastocyst transfer in patients with recurrent implantation failure with frozen-thawed embryo transfer cycles: A cohort study. Front. Endocrinol. 2023, 14, 1238251. [Google Scholar] [CrossRef]
- Chavez-Badiola, A.; Flores-Saiffe-Farías, A.; Mendizabal-Ruiz, G.; Drakeley, A.J.; Cohen, J. Embryo Ranking Intelligent Classification Algorithm (ERICA): Artificial intelligence clinical assistant predicting embryo ploidy and implantation. Reprod. Biomed. Online 2020, 41, 585–593. [Google Scholar] [CrossRef]
- Liao, Z.; Yan, C.; Wang, J.; Zhang, N.; Yang, H.; Lin, C.; Zhang, H.; Wang, W.; Li, W. A clinical consensus-compliant deep learning approach to quantitatively evaluate human in vitro fertilization early embryonic development with optical microscope images. Artif. Intell. Med. 2024, 149, 102773. [Google Scholar] [CrossRef]
- Sharma, A.; Alawad, F.; Kakulavarapu, R.; Iliceto, M.; Riegler, M.A.; Stensen, M.H.; Hammer, H.L. Exploring Embryo Development at the Morula Stage-an AI-based Approach to Determine Whether to Use or Discard an Embryo. In Proceedings of the 2024 4th International Conference on Applied Artificial Intelligence (ICAPAI), Halden, Norway, 16 April 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 1–8. [Google Scholar]
- Yang, L.; Leynes, C.; Pawelka, A.; Lorenzo, I.; Chou, A.; Lee, B.; Heaney, J.D. Machine learning in time-lapse imaging to differentiate embryos from young vs old mice. Biol. Reprod. 2024, 110, 1115–1124. [Google Scholar] [CrossRef]
- Raymahapatra, P.; Khang, A.; Chaudhuri, A.K. A Novel Human Embryo Microscope Image Classification Technique Based on ConvNeXtLarge Model. In Medical Robotics and AI-Assisted Diagnostics for a High-Tech Healthcare Industry; IGI Global: Hershey, PA, USA, 2024; pp. 224–238. [Google Scholar]
- Einy, S.; Sen, E.; Saygin, H.; Hivehchi, H.; Dorostkar Navaei, Y. Local Binary Convolutional Neural Networks’ Long Short-Term Memory Model for Human Embryos’ Anomaly Detection. Sci. Program. 2023, 2023, 2426601. [Google Scholar] [CrossRef]
- Sharma, A.; Dorobantiu, A.; Ali, S.; Iliceto, M.; Stensen, M.H.; Delbarre, E.; Riegler, M.A.; Hammer, H.L. Deep learning methods to forecasting human embryo development in time-lapse videos. bioRxiv 2024. [Google Scholar] [CrossRef]
- Weatherbee, B.A.; Weberling, A.; Gantner, C.W.; Iwamoto-Stohl, L.K.; Barnikel, Z.; Barrie, A.; Campbell, A.; Cunningham, P.; Drezet, C.; Efstathiou, P.; et al. Distinct pathways drive anterior hypoblast specification in the implanting human embryo. Nat. Cell Biol. 2024, 26, 353–365. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Lam, S.M.; Chen, L.; Gao, Y.; Wang, W.; Xu, Y.; Tan, T.; Yu, H.; Zhang, M.; et al. Low-input lipidomics reveals lipid metabolism remodelling during early mammalian embryo development. Nat. Cell Biol. 2024, 26, 278–293. [Google Scholar] [CrossRef]
- Singh, A.V.; Varma, M.; Rai, M.; Pratap Singh, S.; Bansod, G.; Laux, P.; Luch, A. Advancing Predictive Risk Assessment of Chemicals via Integrating Machine Learning, Computational Modeling, and Chemical/Nano-Quantitative Structure-Activity Relationship Approaches. Adv. Intell. Syst. 2024, 6, 2300366. [Google Scholar] [CrossRef]
- Sarker, M.M.K.; Singh, V.K.; Alsharid, M.; Hernandez-Cruz, N.; Papageorghiou, A.T.; Noble, J.A. COMFormer: Classification of maternal-fetal and brain anatomy using a residual cross-covariance attention guided transformer in ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2023, 70, 1417–1427. [Google Scholar] [CrossRef]
- Yang, X.; Liu, L.; Yan, Z.; Yu, J.; Hu, X.; Yu, X.; Dong, C.; Chen, J.; Liu, H.; Yu, Z.; et al. Hierarchical online contrastive anomaly detection for fetal arrhythmia diagnosis in ultrasound. Med. Image Anal. 2024, 97, 103229. [Google Scholar] [CrossRef]
- Zhao, L.; Tan, G.; Pu, B.; Wu, Q.; Ren, H.; Li, K. TransFSM: Fetal anatomy segmentation and biometric measurement in ultrasound images using a hybrid transformer. IEEE J. Biomed. Health Inform. 2024, 28, 285–296. [Google Scholar] [CrossRef]
- Sindhu, K.G.; Annamalai, R. Enhanced Multi-Class Fetal Plane Detection with Limb Localization in Ultrasound Images. In Proceedings of the 2024 IEEE International Conference on Contemporary Computing and Communications (InC4), Bangalore, India, 15–16 March 2024; IEEE: Piscataway, NJ, USA, 2024; Volume 1, pp. 1–6. [Google Scholar]
- Tang, J.; Han, J.; Xie, B.; Xue, J.; Zhou, H.; Jiang, Y.; Hu, L.; Chen, C.; Zhang, K.; Zhu, F.; et al. The Two-Stage Ensemble Learning Model Based on Aggregated Facial Features in Screening for Fetal Genetic Diseases. Int. J. Environ. Res. Public Health 2023, 20, 2377. [Google Scholar] [CrossRef]
- Degala, S.K.B.; Tewari, R.P.; Kamra, P.; Kasiviswanathan, U.; Pandey, R. Segmentation and Estimation of Fetal Biometric Parameters using an Attention Gate Double U-Net with Guided Decoder Architecture. Comput. Biol. Med. 2024, 180, 109000. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, S.; Xia, W.; Fenster, A.; Gan, H.; Zhou, R. LDW-RS Loss: Label Density-Weighted Loss with Ranking Similarity Regularization for Imbalanced Deep Fetal Brain Age Regression. In International Conference on Neural Information Processing; Springer Nature: Singapore, 2023; pp. 125–137. [Google Scholar]
- Li, F.; Li, P.; Wu, X.; Zeng, P.; Lyu, G.; Fan, Y.; Liu, P.; Song, H.; Liu, Z. FHUSP-NET: A multi-task model for fetal heart ultrasound standard plane recognition and key anatomical structures detection. Comput. Biol. Med. 2024, 168, 107741. [Google Scholar] [CrossRef]
- Abirami, P.; Rajini, S.N.S. Detection of tuberculosis using optimized deep learning approach with enhanced selective median (esmf) filter. Afr. J. Biol. Sci. 2024, 6, 193–210. [Google Scholar]
- Khadidos, A.O. Advancements in remote sensing: Harnessing the power of artificial intelligence for scene image classification. AIMS Math. 2024, 9, 10235–10254. [Google Scholar] [CrossRef]
- Falola, Y.; Churilova, P.; Liu, R.; Huang, C.K.; Delgado, J.F.; Misra, S. Generating extremely low-dimensional representation of subsurface earth models using vector quantization and deep Autoencoder. Pet. Res. 2024. [Google Scholar] [CrossRef]
- Tang, X.; Sheykhahmad, F.R. Boosted dipper throated optimization algorithm-based Xception neural network for skin cancer diagnosis: An optimal approach. Heliyon 2024, 10, e26415. [Google Scholar] [CrossRef]
- Embryo Classification Based on Microscopic Images. Available online: https://www.kaggle.com/competitions/world-championship-2023-embryo-classification/data (accessed on 21 September 2024).
- Wu, C.; Yan, W.; Li, H.; Li, J.; Wang, H.; Chang, S.; Yu, T.; Jin, Y.; Ma, C.; Luo, Y.; et al. A classification system of day 3 human embryos using deep learning. Biomed. Signal Process. Control 2021, 70, 102943. [Google Scholar] [CrossRef]
- Thirumalaraju, P.; Kanakasabapathy, M.K.; Bormann, C.L.; Gupta, R.; Pooniwala, R.; Kandula, H.; Souter, I.; Dimitriadis, I.; Shafiee, H. Evaluation of deep convolutional neural networks in classifying human embryo images based on their morphological quality. Heliyon 2021, 7, e06298. [Google Scholar] [CrossRef]
- Liu, Z.; Huang, B.; Cui, Y.; Xu, Y.; Zhang, B.; Zhu, L.; Wang, Y.; Jin, L.; Wu, D. Multi-task deep learning with dynamic programming for embryo early development stage classification from time-lapse videos. IEEE Access 2019, 7, 122153–122163. [Google Scholar] [CrossRef]
- Aburass, S.; Dorgham, O.; Al Shaqsi, J. A hybrid machine learning model for classifying gene mutations in cancer using LSTM, BiLSTM, CNN, GRU, and GloVe. Syst. Soft Comput. 2024, 6, 200110. [Google Scholar] [CrossRef]
- Dai, Y.; Itai, T.; Pei, G.; Yan, F.; Chu, Y.; Jiang, X.; Weinberg, S.M.; Mukhopadhyay, N.; Marazita, M.L.; Simon, L.M.; et al. DeepFace: Deep learning-based framework to contextualize orofacial cleft-related variants during human embryonic craniofacial development. Hum. Genet. Genom. Adv. 2024, 5, 100312. [Google Scholar] [CrossRef] [PubMed]
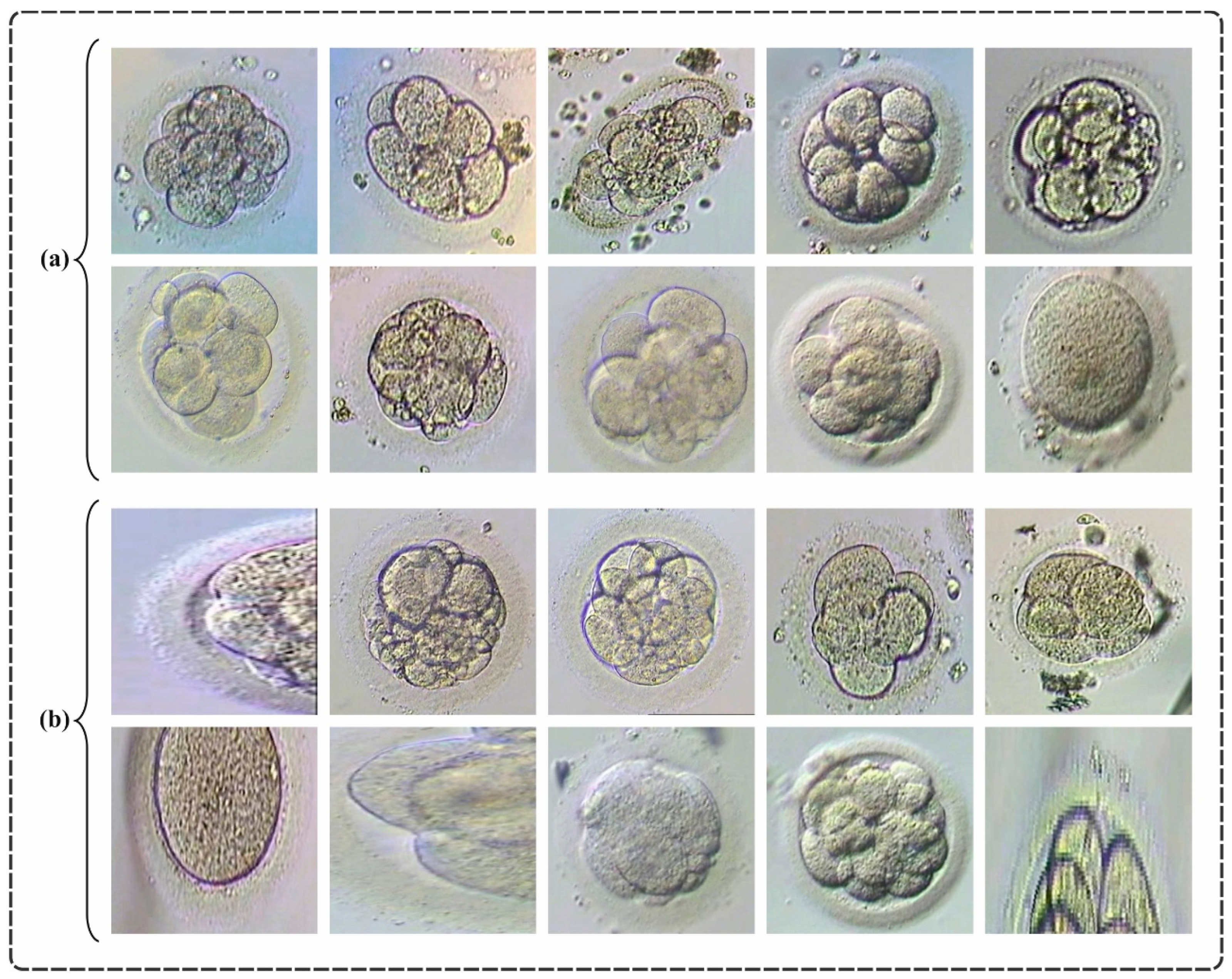

| Classes | No. of Images |
|---|---|
| Embryo Quality-Not Good | 500 |
| Embryo Quality-Good | 120 |
| Total Images | 620 |
| Class | |||||
|---|---|---|---|---|---|
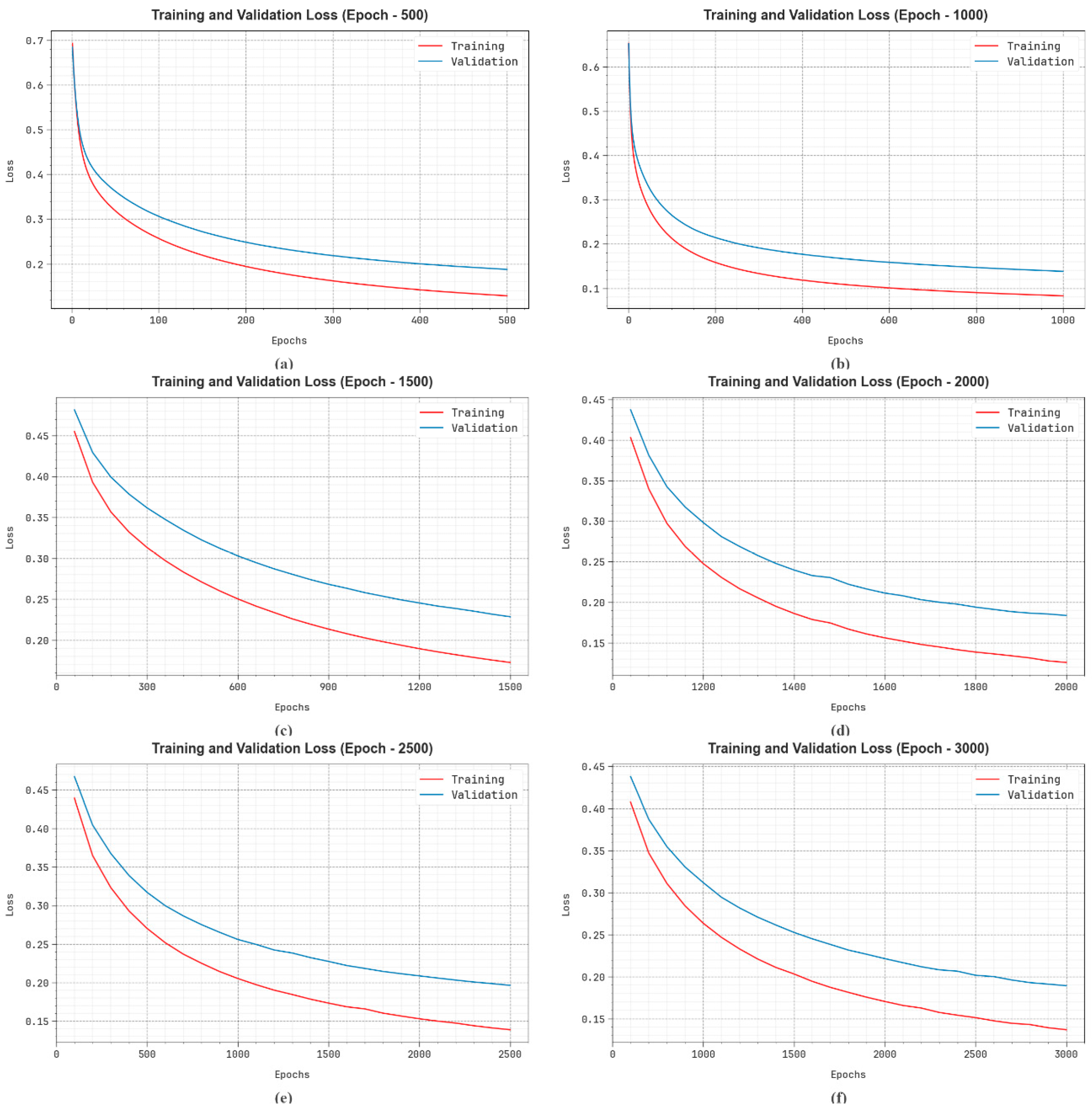
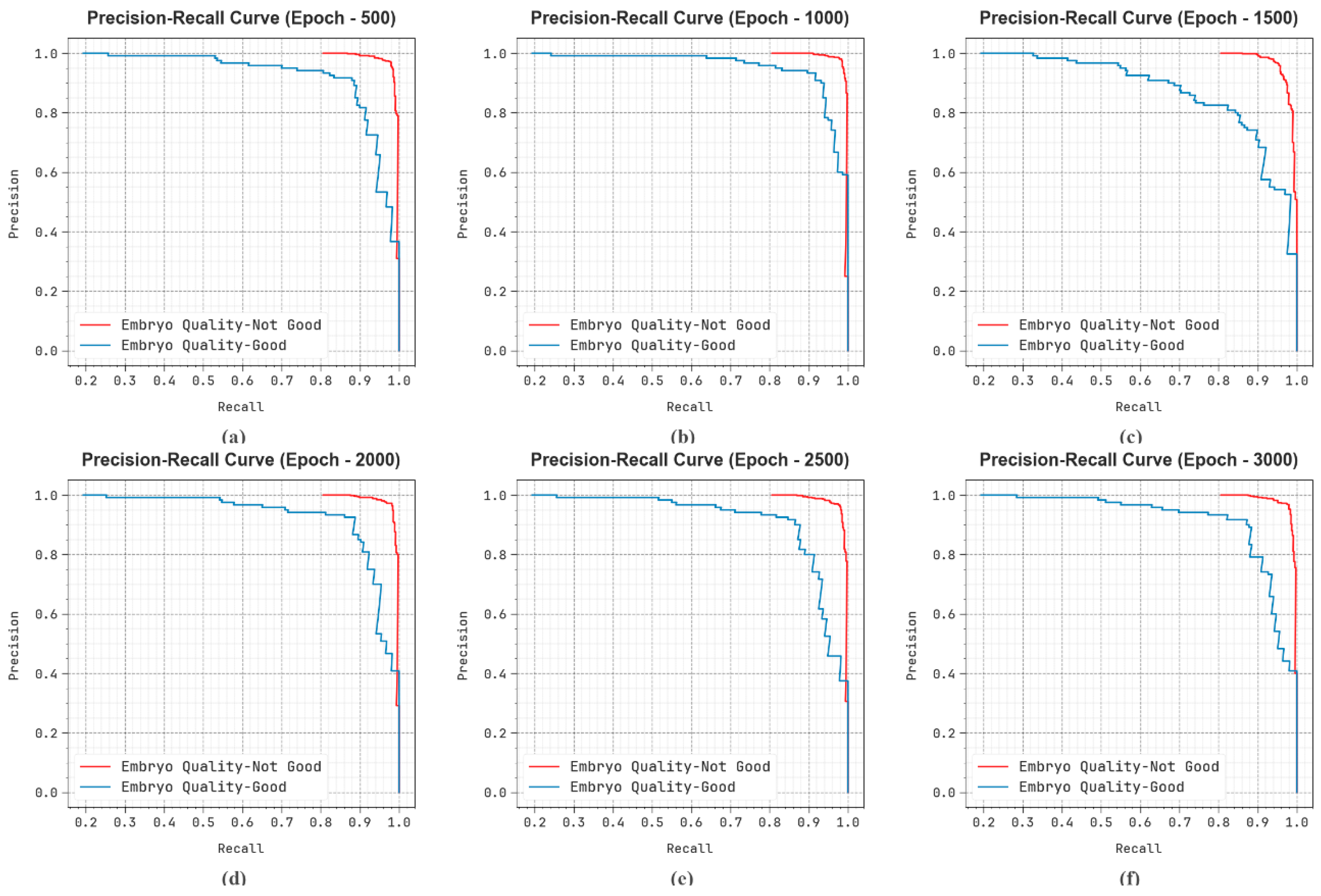
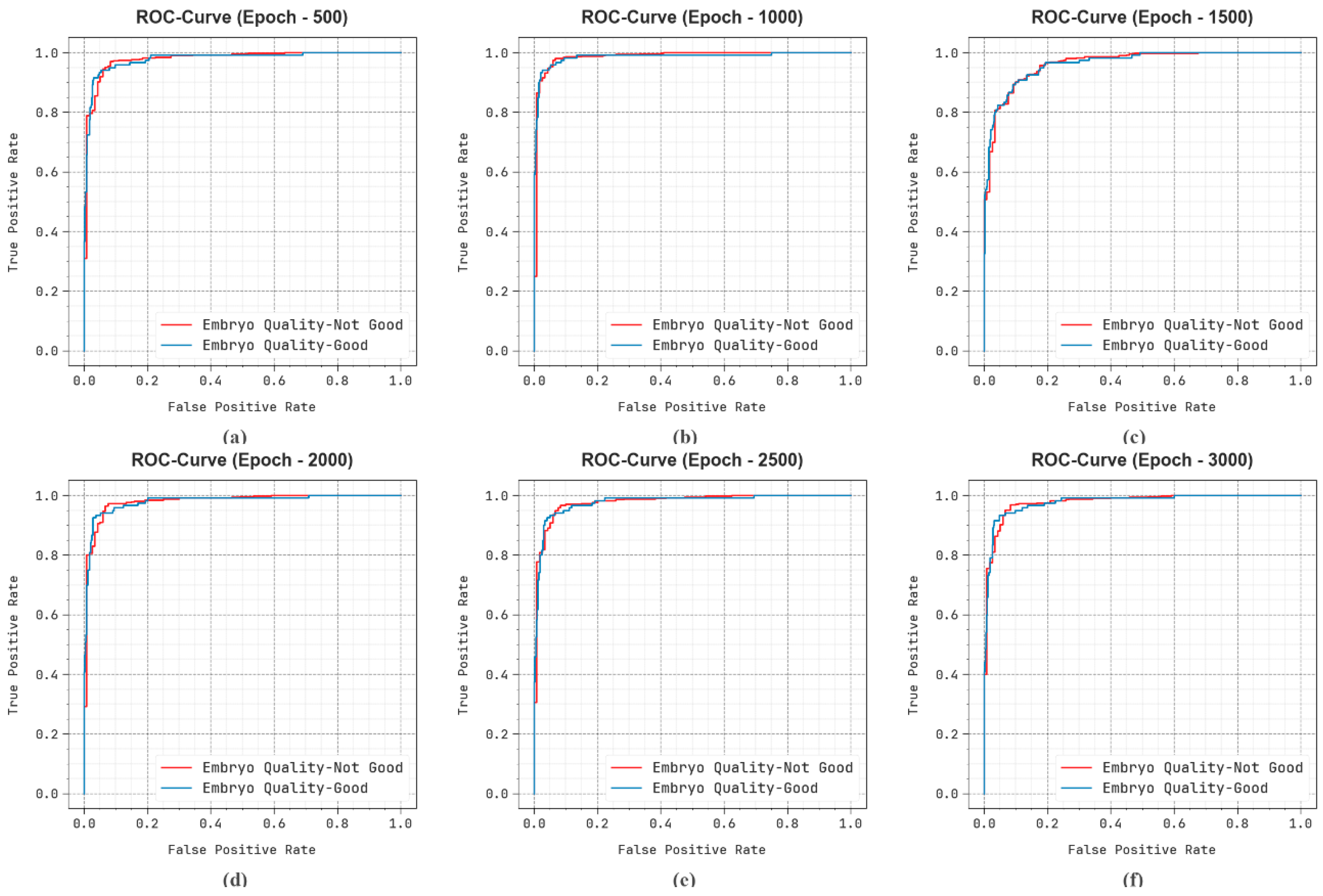
| Epoch-500 | |||||
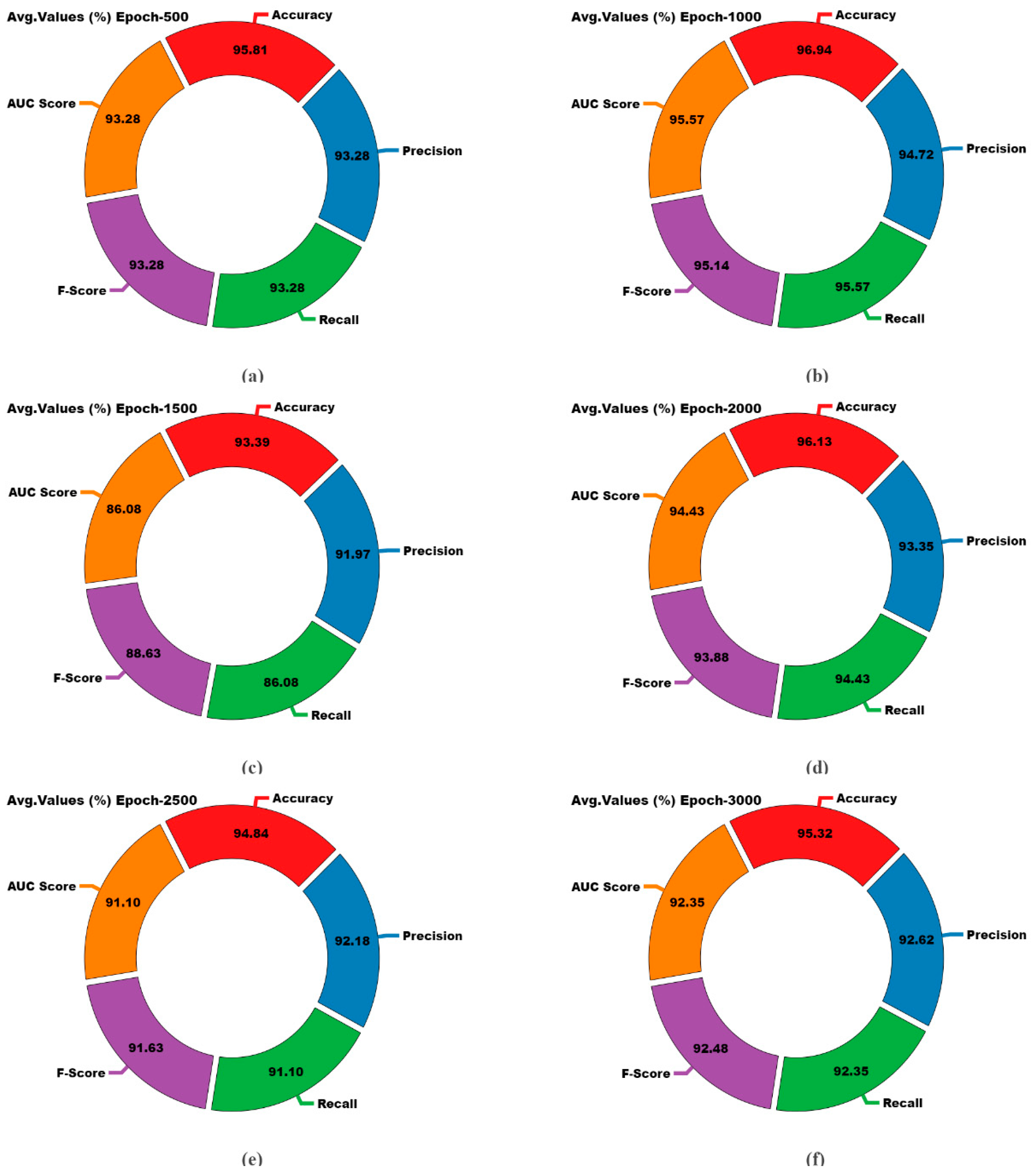
| Embryo Quality-Not Good | 95.81 | 97.40 | 97.40 | 97.40 | 93.28 |
| Embryo Quality-Good | 95.81 | 89.17 | 89.17 | 89.17 | 93.28 |
| Average | 95.81 | 93.28 | 93.28 | 93.28 | 93.28 |
| Epoch-1000 | |||||
| Embryo Quality-Not Good | 96.94 | 98.39 | 97.80 | 98.09 | 95.57 |
| Embryo Quality-Good | 96.94 | 91.06 | 93.33 | 92.18 | 95.57 |
| Average | 96.94 | 94.72 | 95.57 | 95.14 | 95.57 |
| Epoch-1500 | |||||
| Embryo Quality-Not Good | 93.39 | 94.05 | 98.00 | 95.98 | 86.08 |
| Embryo Quality-Good | 93.39 | 89.90 | 74.17 | 81.28 | 86.08 |
| Average | 93.39 | 91.97 | 86.08 | 88.63 | 86.08 |
| Epoch-2000 | |||||
| Embryo Quality-Not Good | 96.13 | 97.98 | 97.20 | 97.59 | 94.43 |
| Embryo Quality-Good | 96.13 | 88.71 | 91.67 | 90.16 | 94.43 |
| Average | 96.13 | 93.35 | 94.43 | 93.88 | 94.43 |
| Epoch-2500 | |||||
| Embryo Quality-Not Good | 94.84 | 96.43 | 97.20 | 96.81 | 91.10 |
| Embryo Quality-Good | 94.84 | 87.93 | 85.00 | 86.44 | 91.10 |
| Average | 94.84 | 92.18 | 91.10 | 91.63 | 91.10 |
| Epoch-3000 | |||||
| Embryo Quality-Not Good | 95.32 | 97.01 | 97.20 | 97.10 | 92.35 |
| Embryo Quality-Good | 95.32 | 88.24 | 87.50 | 87.87 | 92.35 |
| Average | 95.32 | 92.62 | 92.35 | 92.48 | 92.35 |
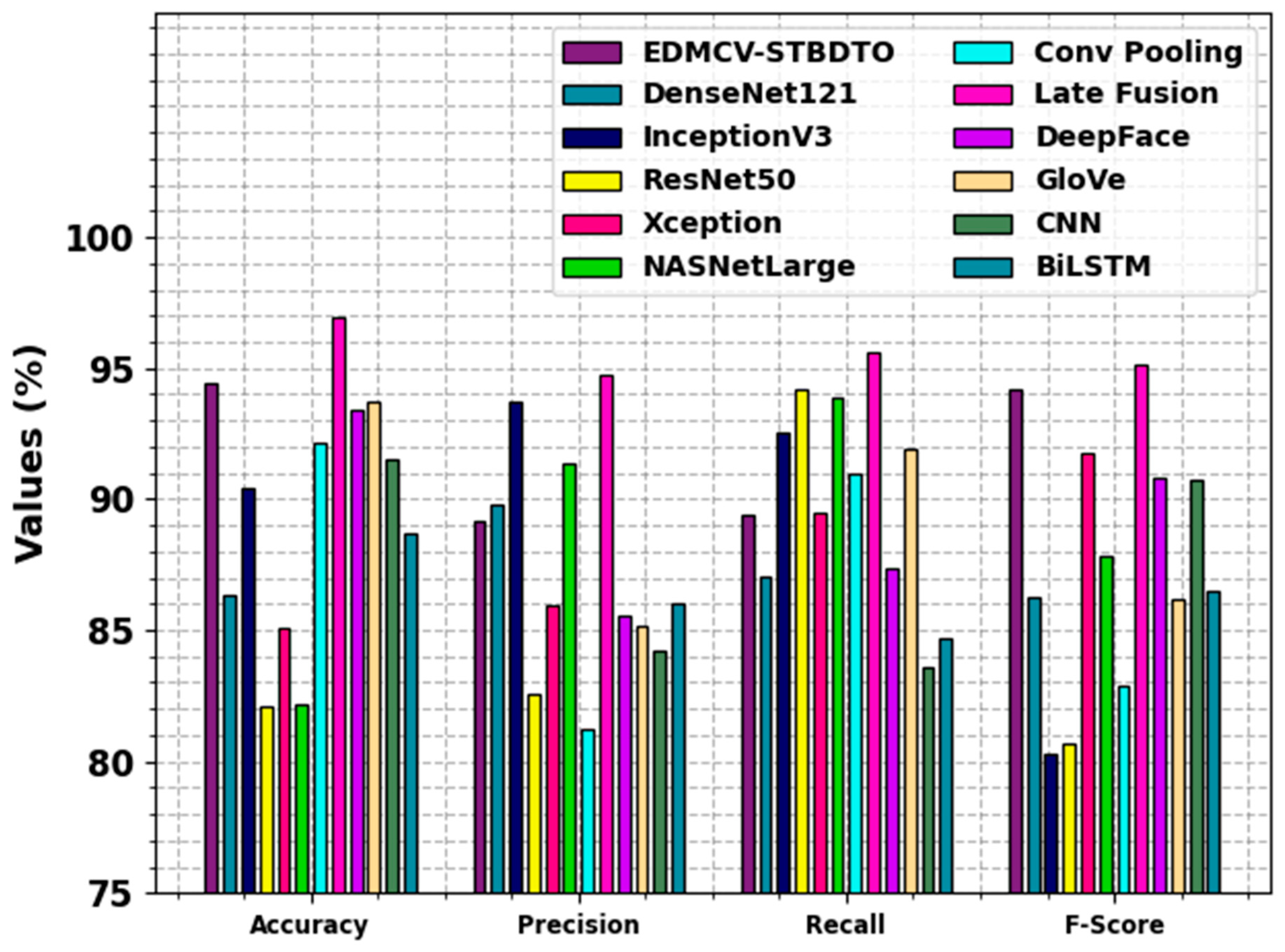
| Methodology | ||||
|---|---|---|---|---|
| EDMCV-STBDTO | 94.42 | 89.15 | 89.37 | 94.18 |
| DenseNet121 | 86.31 | 89.78 | 87.06 | 86.29 |
| InceptionV3 | 90.42 | 93.70 | 92.55 | 80.29 |
| ResNet50 | 82.11 | 82.53 | 94.18 | 80.67 |
| Xception | 85.09 | 85.91 | 89.48 | 91.77 |
| NASNetLarge | 82.14 | 91.33 | 93.89 | 87.83 |
| Conv Pooling | 92.15 | 81.23 | 91.00 | 82.89 |
| Late Fusion | 96.94 | 94.72 | 95.57 | 95.14 |
| DeepFace | 93.37 | 85.53 | 87.36 | 90.82 |
| GloVe | 93.75 | 85.15 | 91.94 | 86.19 |
| CNN | 91.51 | 84.25 | 83.57 | 90.73 |
| BiLSTM | 88.67 | 86.03 | 84.70 | 86.51 |
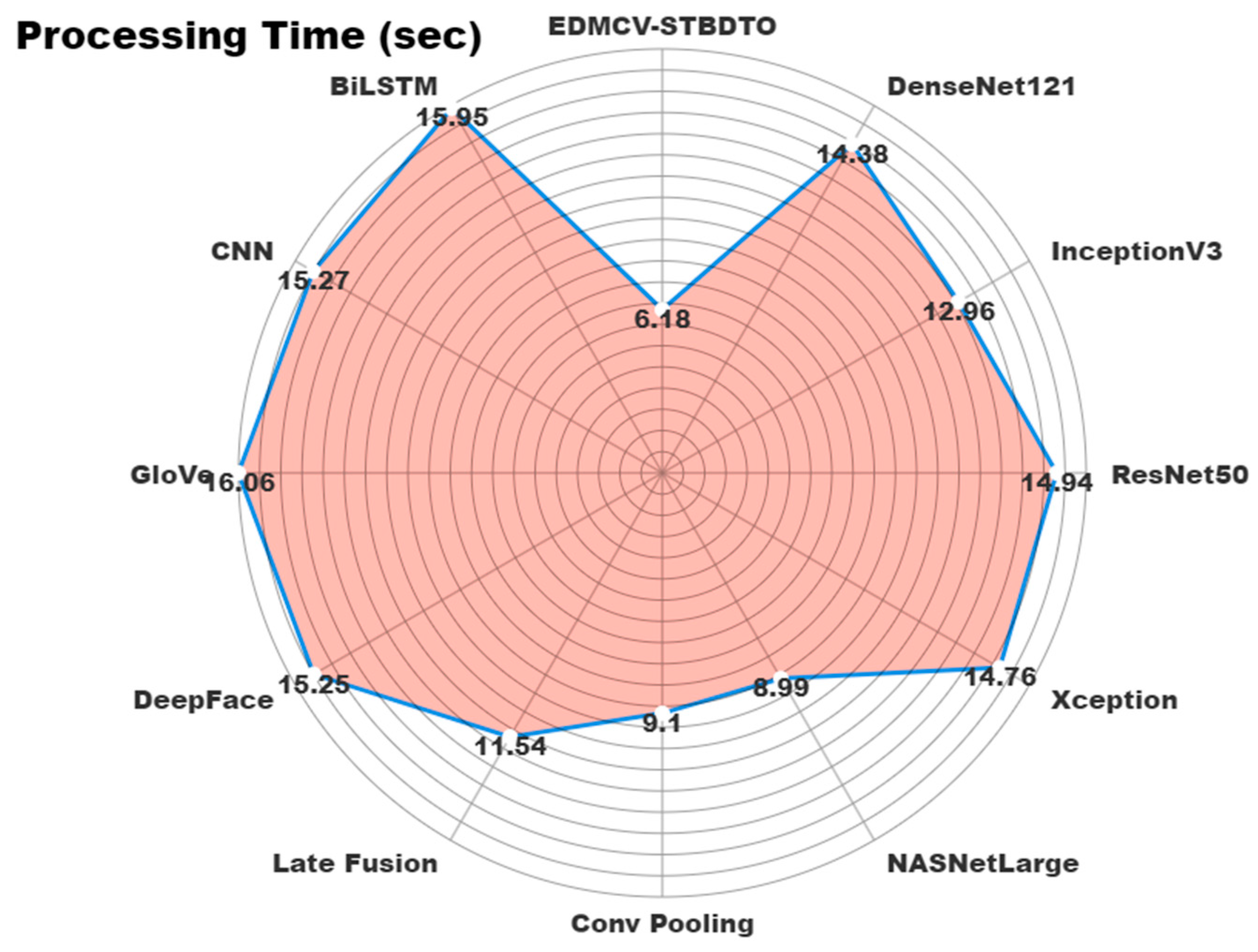
| Methodology | Processing Time (s) |
|---|---|
| EDMCV-STBDTO | 6.18 |
| DenseNet121 | 14.38 |
| InceptionV3 | 12.96 |
| ResNet50 | 14.94 |
| Xception | 14.76 |
| NASNetLarge | 8.99 |
| Conv Pooling | 9.10 |
| Late Fusion | 11.54 |
| DeepFace | 15.25 |
| GloVe | 16.06 |
| CNN | 15.27 |
| BiLSTM | 15.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazroa, A.A.; Maashi, M.; Said, Y.; Maray, M.; Alzahrani, A.A.; Alkharashi, A.; Al-Sharafi, A.M. Anomaly Detection in Embryo Development and Morphology Using Medical Computer Vision-Aided Swin Transformer with Boosted Dipper-Throated Optimization Algorithm. Bioengineering 2024, 11, 1044. https://doi.org/10.3390/bioengineering11101044
Mazroa AA, Maashi M, Said Y, Maray M, Alzahrani AA, Alkharashi A, Al-Sharafi AM. Anomaly Detection in Embryo Development and Morphology Using Medical Computer Vision-Aided Swin Transformer with Boosted Dipper-Throated Optimization Algorithm. Bioengineering. 2024; 11(10):1044. https://doi.org/10.3390/bioengineering11101044
Chicago/Turabian StyleMazroa, Alanoud Al, Mashael Maashi, Yahia Said, Mohammed Maray, Ahmad A. Alzahrani, Abdulwhab Alkharashi, and Ali M. Al-Sharafi. 2024. "Anomaly Detection in Embryo Development and Morphology Using Medical Computer Vision-Aided Swin Transformer with Boosted Dipper-Throated Optimization Algorithm" Bioengineering 11, no. 10: 1044. https://doi.org/10.3390/bioengineering11101044
APA StyleMazroa, A. A., Maashi, M., Said, Y., Maray, M., Alzahrani, A. A., Alkharashi, A., & Al-Sharafi, A. M. (2024). Anomaly Detection in Embryo Development and Morphology Using Medical Computer Vision-Aided Swin Transformer with Boosted Dipper-Throated Optimization Algorithm. Bioengineering, 11(10), 1044. https://doi.org/10.3390/bioengineering11101044







