Advances in Medical Image Segmentation: A Comprehensive Review of Traditional, Deep Learning and Hybrid Approaches
Abstract
1. Introduction
2. Traditional Segmentation Techniques
2.1. Thresholding Techniques
- I(x,y) is the pixel value of the grayscale image at position (x,y);
- B(x,y) is the pixel value of the binary image at position (x,y);
- T is the chosen threshold.
2.1.1. Global Thresholding Techniques
2.1.2. Local Thresholding Techniques
2.2. Edge-Based Segmentation
2.3. Region-Based Segmentation
2.3.1. Seeded Region Growing
- Seed Point Selection: Initiate by identifying one or more seed points within the image. These points are chosen based on specific attributes such as brightness, color, or texture, which serve as a foundation for region growth;
- Criteria Determination: Define the parameters that will guide the region’s growth. This could involve setting thresholds for color differences, brightness similarities, or texture consistencies between adjacent pixels;
- Regional Expansion: Evaluate the pixels surrounding each seed point. Based on the established criteria, decide whether to integrate these pixels into the current region. This inclusion leads to a gradual expansion of the region;
- Iterative Process: Continue this procedure iteratively, adding new pixels to regions as appropriate. The process concludes when no further pixels can be assimilated into any region.
2.3.2. Split and Merge
- 5.
- Let represent the whole image with different objects. Segmentation can be thought of as dividing into subregions , , …, as a process;
- 6.
- Split any region into four almost equal regions where ;
- 7.
- If , then consider any two or more neighboring subregions , , …, , and then merge the regions into a single region;
- 8.
- Repeat steps 2–3 until no further splitting and merging is possible.
2.4. Clustering-Based Segmentation
2.4.1. Hierarchical Clustering
2.4.2. Partitional Clustering
2.5. Graphic-Based Segmentation
2.5.1. Minimal Spanning Tree (MST) Based Methods
2.5.2. Graph Cuts
2.5.3. Markov Random Fields (MRF)
2.5.4. Shortest Path Methods
3. Deep Learning in Medical Image Segmentation
3.1. Overview of Deep Learning Techniques
3.1.1. Convolutional Neural Networks (CNNs)
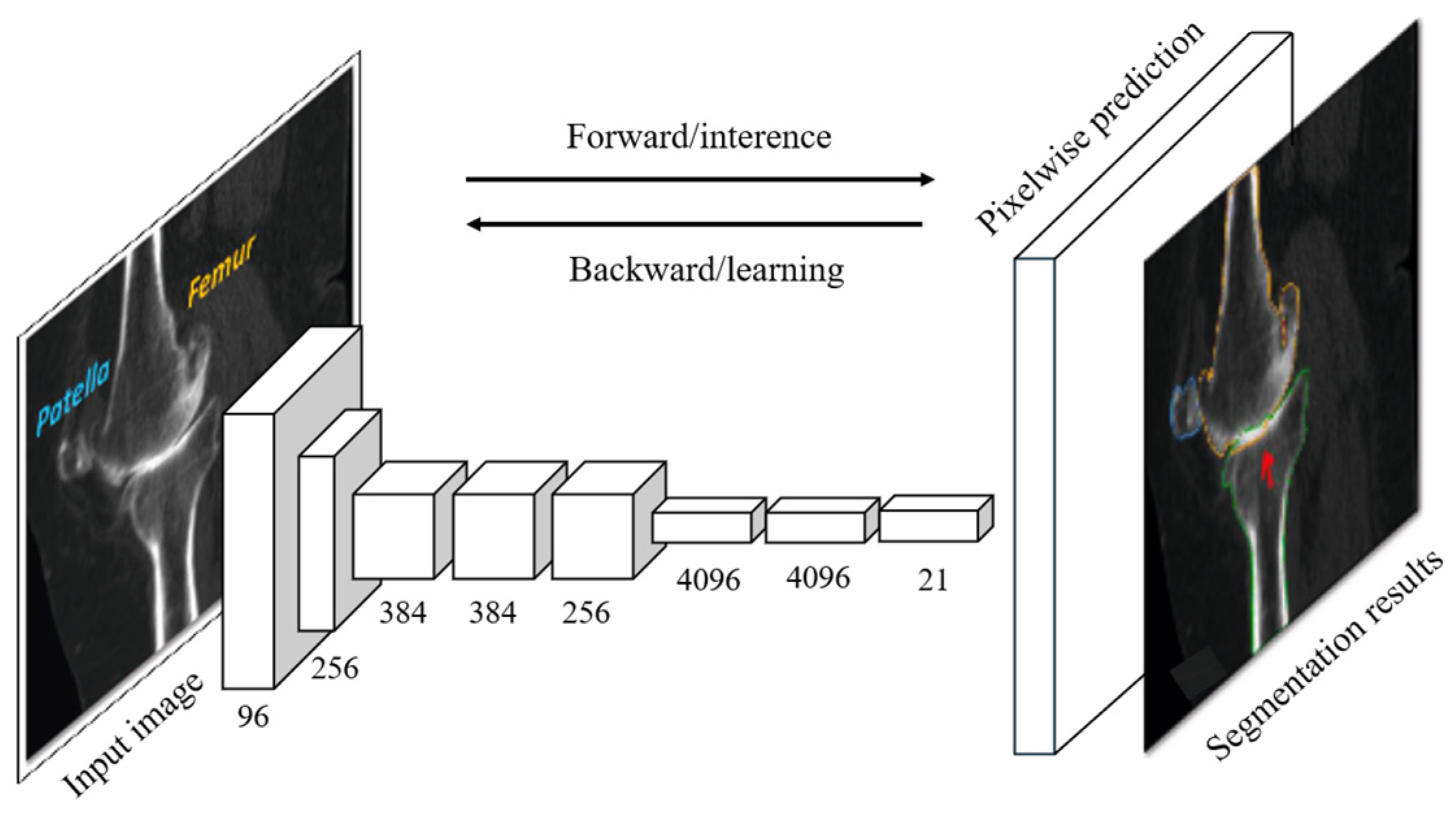
3.1.2. Fully Convolutional Networks (FCNs)
3.1.3. U-Net Architecture
3.1.4. Recurrent Neural Networks (RNNs)
3.1.5. Generative Adversarial Networks (GANs)
3.1.6. Autoencoders (AEs)
3.2. Challenges and Limitations of Deep Learning
3.3. Applications of Deep Learning in Medical Image Segmentation
3.3.1. Brain Tumour
3.3.2. Breast
3.3.3. Liver Tumor
3.3.4. Lung
3.3.5. Prostate
3.3.6. Retinal Vessel
3.3.7. Skin Lesion
4. Integration of Deep Learning with Traditional Techniques
4.1. Combining Thresholding Techniques with Deep Learning
4.2. Combining Edge-Based Methods with Deep Learning
4.3. Combining Region-Based Methods with Deep Learning
4.4. Combining Clustering-Based Methods with Deep Learning
4.5. Combining Graph-Based Methods with Deep Learning
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Panayides, A.S.; Amini, A.; Filipovic, N.D.; Sharma, A.; Tsaftaris, S.A.; Young, A.; Foran, D.; Do, N.; Golemati, S.; Kurc, T.; et al. AI in Medical Imaging Informatics: Current Challenges and Future Directions. IEEE J. Biomed. Health Inform. 2020, 24, 1837–1857. [Google Scholar] [CrossRef] [PubMed]
- Abdou, M.A. Literature Review: Efficient Deep Neural Networks Techniques for Medical Image Analysis. Neural Comput. Appl. 2022, 34, 5791–5812. [Google Scholar] [CrossRef]
- Alirr, O.I.; Rahni, A.A.A. Survey on Liver Tumour Resection Planning System: Steps, Techniques, and Parameters. J. Digit. Imaging 2020, 33, 304–323. [Google Scholar] [CrossRef] [PubMed]
- Nyo, M.T.; Mebarek-Oudina, F.; Hlaing, S.S.; Khan, N.A. Otsu’s Thresholding Technique for MRI Image Brain Tumor Segmentation. Multimed. Tools Appl. 2022, 81, 43837–43849. [Google Scholar] [CrossRef]
- Abdel-Gawad, A.H.; Said, L.A.; Radwan, A.G. Optimized Edge Detection Technique for Brain Tumor Detection in MR Images. IEEE Access 2020, 8, 136243–136259. [Google Scholar] [CrossRef]
- Khalid, N.E.A.; Ibrahim, S.; Manaf, M.; Ngah, U.K. Seed-Based Region Growing Study for Brain Abnormalities Segmentation. In Proceedings of the 2010 International Symposium on Information Technology, Kuala Lumpur, Malaysia, 15–17 June 2010; Volume 2, pp. 856–860. [Google Scholar]
- Mittal, H.; Pandey, A.C.; Saraswat, M.; Kumar, S.; Pal, R.; Modwel, G. A Comprehensive Survey of Image Segmentation: Clustering Methods, Performance Parameters, and Benchmark Datasets. Multimed. Tools Appl. 2022, 81, 35001–35026. [Google Scholar] [CrossRef]
- Bağci, U.; Yao, J.; Caban, J.; Turkbey, E.; Aras, O.; Mollura, D.J. A Graph-Theoretic Approach for Segmentation of PET Images. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 8479–8482. [Google Scholar]
- Mohd Sagheer, S.V.; George, S.N. A Review on Medical Image Denoising Algorithms. Biomed. Signal Process. Control 2020, 61, 102036. [Google Scholar] [CrossRef]
- Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3D Deep Learning on Medical Images: A Review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef]
- Wang, J.; Wei, L.; Wang, L.; Zhou, Q.; Zhu, L.; Qin, J. Boundary-Aware Transformers for Skin Lesion Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021, Strasbourg, France, 27 September–1 October 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 206–216. [Google Scholar]
- Khan, M.Z.; Gajendran, M.K.; Lee, Y.; Khan, M.A. Deep Neural Architectures for Medical Image Semantic Segmentation: Review. IEEE Access 2021, 9, 83002–83024. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent Advances in Convolutional Neural Networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Altini, N.; Brunetti, A.; Puro, E.; Taccogna, M.G.; Saponaro, C.; Zito, F.A.; De Summa, S.; Bevilacqua, V. NDG-CAM: Nuclei Detection in Histopathology Images with Semantic Segmentation Networks and Grad-CAM. Bioengineering 2022, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2015, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Özcan, F.; Uçan, O.N.; Karaçam, S.; Tunçman, D. Fully Automatic Liver and Tumor Segmentation from CT Image Using an AIM-Unet. Bioengineering 2023, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning Phrase Representations Using RNN Encoder–Decoder for Statistical Machine Translation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; Moschitti, A., Pang, B., Daelemans, W., Eds.; Association for Computational Linguistics: Doha, Qatar, 2014; pp. 1724–1734. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; Curran Associates, Inc.: Red Hook, NY, USA, 2014; Volume 27. [Google Scholar]
- Kingma, D.P.; Welling, M. An Introduction to Variational Autoencoders. Found. Trends® Mach. Learn. 2019, 12, 307–392. [Google Scholar] [CrossRef]
- Li, F.; Wang, C.; Liu, X.; Peng, Y.; Jin, S. A Composite Model of Wound Segmentation Based on Traditional Methods and Deep Neural Networks. Comput. Intell. Neurosci. 2018, 2018, 4149103. [Google Scholar] [CrossRef]
- Senthilkumaran, N.; Vaithegi, S. Image Segmentation By Using Thresholding Techniques For Medical Images. Comput. Sci. Eng. Int. J. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Goh, T.Y.; Basah, S.N.; Yazid, H.; Aziz Safar, M.J.; Ahmad Saad, F.S. Performance Analysis of Image Thresholding: Otsu Technique. Measurement 2018, 114, 298–307. [Google Scholar] [CrossRef]
- Perez, A.; Gonzalez, R.C. An Iterative Thresholding Algorithm for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 1987, PAMI-9, 742–751. [Google Scholar] [CrossRef]
- Sujji, G.E.; Lakshmi, Y.V.S.; Jiji, G.W. MRI Brain Image Segmentation Based on Thresholding. Int. J. Adv. Comput. Res. 2013, 3, 97. [Google Scholar]
- Kittler, J.; Illingworth, J. Minimum Error Thresholding. Pattern Recognit. 1986, 19, 41–47. [Google Scholar] [CrossRef]
- Yin, P.-Y. Maximum Entropy-Based Optimal Threshold Selection Using Deterministic Reinforcement Learning with Controlled Randomization. Signal Process. 2002, 82, 993–1006. [Google Scholar] [CrossRef]
- Jyothi, S.; Bhargavi, K. A Survey on Threshold Based Segmentation Technique in Image Processing. Int. J. Innov. Res. Dev. 2014, 3, 234–239. [Google Scholar]
- Lie, W.N. Automatic Target Segmentation by Locally Adaptive Image Thresholding. IEEE Trans. Image Process. 1995, 4, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Saxena, L.P. Niblack’s Binarization Method and Its Modifications to Real-Time Applications: A Review. Artif. Intell. Rev. 2019, 51, 673–705. [Google Scholar] [CrossRef]
- Sauvola, J.; Pietikäinen, M. Adaptive Document Image Binarization. Pattern Recognit. 2000, 33, 225–236. [Google Scholar] [CrossRef]
- Bernsen, J. Dynamic Thresholding of Grey-Level Images. In Multi-Pass Approach to Adaptive Thresholding Based Image Segmentation, Proceedings of the 8th International IEEE Conference CADSM, Oxford, UK, 27 July–1 August 1986; IEEE: Piscataway, NJ, USA, 1986; pp. 1251–1255. [Google Scholar]
- Kaur, D.; Kaur, Y. Various Image Segmentation Techniques: A Review. Int. J. Comput. Sci. Mob. Comput. 2014, 3, 809–814. [Google Scholar]
- Saini, S.; Arora, K. A Study Analysis on the Different Image Segmentation Techniques. Int. J. Inf. Comput. Technol. 2014, 4, 1445–1452. [Google Scholar]
- Abo-Zahhad, M.; Gharieb, R.R.; Ahmed, S.M.; Donkol, A.A.E.-B. Edge Detection with a Preprocessing Approach. J. Signal Inf. Process. 2014, 5, 123. [Google Scholar] [CrossRef]
- Lawrence, G. Roberts Machine Perception of Three-Dimensional Solids. Doctoral Dissertation, Massachusetts Institute of Technology, Cambridge, MA, USA, 1963. [Google Scholar]
- Lipkin, B.S. Picture Processing and Psychopictorics; Elsevier: Amsterdam, The Netherlands, 1970; ISBN 978-0-323-14685-2. [Google Scholar]
- Kanopoulos, N.; Vasanthavada, N.; Baker, R.L. Design of an Image Edge Detection Filter Using the Sobel Operator. IEEE J. Solid-State Circuits 1988, 23, 358–367. [Google Scholar] [CrossRef]
- Canny, J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, PAMI-8, 679–698. [Google Scholar] [CrossRef]
- Ding, L.; Goshtasby, A. On the Canny Edge Detector. Pattern Recognit. 2001, 34, 721–725. [Google Scholar] [CrossRef]
- Berzins, V. Accuracy of Laplacian Edge Detectors. Comput. Vis. Graph. Image Process. 1984, 27, 195–210. [Google Scholar] [CrossRef]
- Gunn, S.R. On the Discrete Representation of the Laplacian of Gaussian. Pattern Recognit. 1999, 32, 1463–1472. [Google Scholar] [CrossRef]
- Veelaert, P.; Teelen, K. Adaptive and Optimal Difference Operators in Image Processing. Pattern Recognit. 2009, 42, 2317–2326. [Google Scholar] [CrossRef]
- Maini, R. Study and Comparison of Various Image Edge Detection Techniques. Int. J. Image Process. IJIP 2009, 3, 1–11. [Google Scholar]
- Sharifi, M.; Fathy, M.; Mahmoudi, M.T. A Classified and Comparative Study of Edge Detection Algorithms. In Proceedings of the International Conference on Information Technology: Coding and Computing, Las Vegas, NV, USA, 8–10 April 2002; pp. 117–120. [Google Scholar]
- Acharjya, P.P.; Das, R.; Ghoshal, D. Study and Comparison of Different Edge Detectors for Image Segmentation. Glob. J. Comput. Sci. Technol. 2012, 12, 28–32. [Google Scholar]
- Heath, M.; Sarkar, S.; Sanocki, T.; Bowyer, K. Comparison of Edge Detectors: A Methodology and Initial Study. Comput. Vis. Image Underst. 1998, 69, 38–54. [Google Scholar] [CrossRef]
- Rashmi; Kumar, M.; Saxena, R. Algorithm and Technique on Various Edge Detection: A Survey. Signal Image Process. Int. J. 2013, 4, 65–75. [Google Scholar] [CrossRef]
- Mary Synthuja Jain Preetha, M.; Padma Suresh, L.; John Bosco, M. Image Segmentation Using Seeded Region Growing. In Proceedings of the 2012 International Conference on Computing, Electronics and Electrical Technologies (ICCEET), Nagercoil, India, 21–22 March 2012; pp. 576–583. [Google Scholar]
- Kaganami, H.G.; Beiji, Z. Region-Based Segmentation versus Edge Detection. In Proceedings of the 2009 Fifth International Conference on Intelligent Information Hiding and Multimedia Signal Processing, Kyoto, Japan, 12–14 September 2009; pp. 1217–1221. [Google Scholar]
- Adams, R.; Bischof, L. Seeded Region Growing. IEEE Trans. Pattern Anal. Mach. Intell. 1994, 16, 641–647. [Google Scholar] [CrossRef]
- Fan, J.; Zeng, G.; Body, M.; Hacid, M.-S. Seeded Region Growing: An Extensive and Comparative Study. Pattern Recognit. Lett. 2005, 26, 1139–1156. [Google Scholar] [CrossRef]
- Ikonomatakis, N.; Plataniotis, K.N.; Zervakis, M.; Venetsanopoulos, A.N. Region Growing and Region Merging Image Segmentation. In Proceedings of the 13th International Conference on Digital Signal Processing, Santorini, Greece, 2–4 July 1997; Volume 1, pp. 299–302. [Google Scholar]
- Horowitz, S.L.; Pavlidis, T. Picture Segmentation by a Tree Traversal Algorithm. J. ACM JACM 1976, 23, 368–388. [Google Scholar] [CrossRef]
- Faruquzzaman, A.B.M.; Paiker, N.R.; Arafat, J.; Karim, Z.; Ameer Ali, M. Object Segmentation Based on Split and Merge Algorithm. In Proceedings of the TENCON 2008—2008 IEEE Region 10 Conference, Hyderabad, India, 19–21 November 2008; pp. 1–4. [Google Scholar]
- Lu, Y.; Jiang, T.; Zang, Y. A Split–Merge-Based Region-Growing Method for fMRI Activation Detection. Hum. Brain Mapp. 2004, 22, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Prasad, M.; Gupta, A.; Bharill, N.; Patel, O.P.; Tiwari, A.; Er, M.J.; Ding, W.; Lin, C.-T. A Review of Clustering Techniques and Developments. Neurocomputing 2017, 267, 664–681. [Google Scholar] [CrossRef]
- Fraley, C.; Raftery, A.E. How Many Clusters? Which Clustering Method? Answers Via Model-Based Cluster Analysis. Comput. J. 1998, 41, 578–588. [Google Scholar] [CrossRef]
- Reddy, C.K.; Vinzamuri, B. A Survey of Partitional and Hierarchical Clustering Algorithms. In Data Clustering; Chapman and Hall/CRC: Boca Raton, FL, USA, 2014; ISBN 978-1-315-37351-5. [Google Scholar]
- Murtagh, F. A Survey of Recent Advances in Hierarchical Clustering Algorithms. Comput. J. 1983, 26, 354–359. [Google Scholar] [CrossRef]
- Ezugwu, A.E.; Ikotun, A.M.; Oyelade, O.O.; Abualigah, L.; Agushaka, J.O.; Eke, C.I.; Akinyelu, A.A. A Comprehensive Survey of Clustering Algorithms: State-of-the-Art Machine Learning Applications, Taxonomy, Challenges, and Future Research Prospects. Eng. Appl. Artif. Intell. 2022, 110, 104743. [Google Scholar] [CrossRef]
- Dunn, J.C. A Fuzzy Relative of the ISODATA Process and Its Use in Detecting Compact Well-Separated Clusters. J. Cybern. 1973, 3, 32–57. [Google Scholar] [CrossRef]
- Bezdek, J.C.; Ehrlich, R.; Full, W. FCM: The Fuzzy c-Means Clustering Algorithm. Comput. Geosci. 1984, 10, 191–203. [Google Scholar] [CrossRef]
- Ikotun, A.M.; Ezugwu, A.E.; Abualigah, L.; Abuhaija, B.; Heming, J. K-Means Clustering Algorithms: A Comprehensive Review, Variants Analysis, and Advances in the Era of Big Data. Inf. Sci. 2023, 622, 178–210. [Google Scholar] [CrossRef]
- Park, H.-S.; Jun, C.-H. A Simple and Fast Algorithm for K-Medoids Clustering. Expert Syst. Appl. 2009, 36, 3336–3341. [Google Scholar] [CrossRef]
- Razavi Zadegan, S.M.; Mirzaie, M.; Sadoughi, F. Ranked K-Medoids: A Fast and Accurate Rank-Based Partitioning Algorithm for Clustering Large Datasets. Knowl.-Based Syst. 2013, 39, 133–143. [Google Scholar] [CrossRef]
- Sisodia, D.; Singh, L.; Sisodia, S. Clustering Techniques: A Brief Survey of Different Clustering Algorithms. Int. J. Latest Trends Eng. Technol. 2012, 1, 82–87. [Google Scholar]
- Felzenszwalb, P.F.; Huttenlocher, D.P. Efficient Graph-Based Image Segmentation. Int. J. Comput. Vis. 2004, 59, 167–181. [Google Scholar] [CrossRef]
- Peng, B.; Zhang, L.; Zhang, D. A Survey of Graph Theoretical Approaches to Image Segmentation. Pattern Recognit. 2013, 46, 1020–1038. [Google Scholar] [CrossRef]
- Camilus, K.S.; Govindan, V.K. A Review on Graph Based Segmentation. Int. J. Image Graph. Signal Process. 2012, 4, 1–13. [Google Scholar] [CrossRef]
- Kruskal, J.B. On the Shortest Spanning Subtree of a Graph and the Traveling Salesman Problem. Proc. Am. Math. Soc. 1956, 7, 48–50. [Google Scholar] [CrossRef]
- Prim, R.C. Shortest Connection Networks and Some Generalizations. Bell Syst. Tech. J. 1957, 36, 1389–1401. [Google Scholar] [CrossRef]
- Jensen, P.M.; Jeppesen, N.; Dahl, A.B.; Dahl, V.A. Review of Serial and Parallel Min-Cut/Max-Flow Algorithms for Computer Vision. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 2310–2329. [Google Scholar] [CrossRef]
- Lv, X.; Wang, X.; Wang, Q.; Yu, J. 4D Light Field Segmentation from Light Field Super-Pixel Hypergraph Representation. IEEE Trans. Vis. Comput. Graph. 2021, 27, 3597–3610. [Google Scholar] [CrossRef]
- Wu, Z.; Leahy, R. Tissue Classification In MR Images Using Hierarchical Segmentation. In Proceedings of the 1990 IEEE Nuclear Science Symposium Conference Record, New York, NY, USA, 22–27 October 1990; pp. 1410–1414. [Google Scholar]
- Cox, I.J.; Rao, S.B.; Zhong, Y. “Ratio Regions”: A Technique for Image Segmentation. In Proceedings of the 13th International Conference on Pattern Recognition, Washington, DC, USA, 25–29 August 1996; Volume 2, pp. 557–564. [Google Scholar]
- Shi, J.; Malik, J. Normalized Cuts and Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 888–905. [Google Scholar] [CrossRef]
- Jermyn, I.H.; Ishikawa, H. Globally Optimal Regions and Boundaries. In Proceedings of the Seventh IEEE International Conference on Computer Vision, Corfu, Greece, 20–27 September 1999; IEEE: Kerkyra, Greece, 1999; Volume 2, pp. 904–910. [Google Scholar]
- Wang, S.; Siskind, J.M. Image Segmentation with Minimum Mean Cut. In Proceedings of the Proceedings Eighth IEEE International Conference on Computer Vision. ICCV 2001, Vancouver, BC, Canada, 7–14 July 2001; Volume 1, pp. 517–524. [Google Scholar]
- Wang, S.; Siskind, J.M. Image Segmentation with Ratio Cut. IEEE Trans. Pattern Anal. Mach. Intell. 2003, 25, 675–690. [Google Scholar] [CrossRef]
- Cigla, C.; Alatan, A.A. Region-Based Image Segmentation via Graph Cuts. In Proceedings of the 2008 15th IEEE International Conference on Image Processing, San Diego, CA, USA, 12–15 October 2008; pp. 2272–2275. [Google Scholar]
- Kohli, P.; Torr, P.H.S. Dynamic Graph Cuts for Efficient Inference in Markov Random Fields. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Geman, D.; Geman, S. Bayesian Image Analysis. In Disordered Systems and Biological Organization, Proceedings of the NATO Advanced Research Workshop on Disordered Systems and Biological Organization, Les Houches, France, 25 February–8 March 1985; Bienenstock, E., Soulié, F.F., Weisbuch, G., Eds.; Springer: Berlin/Heidelberg, Germany, 1986; pp. 301–319. [Google Scholar]
- Kato, Z.; Zerubia, J. Markov Random Fields in Image Segmentation. Found. Trends® Signal Process. 2012, 5, 1–155. [Google Scholar] [CrossRef]
- Geman, S.; Graffigne, C. Markov Random Field Image Models and Their Applications to Computer Vision. In Proceedings of the International Congress of Mathematicians, Berkeley, CA, USA, 3–11 August 1986; Volume 1, p. 2. [Google Scholar]
- Dijkstra, E.W. A Note on Two Problems in Connexion with Graphs. Numer. Math. 1959, 1, 269–271. [Google Scholar] [CrossRef]
- Bellman, R. On a Routing Problem. Q. Appl. Math. 1958, 16, 87–90. [Google Scholar] [CrossRef]
- Warshall, S. A Theorem on Boolean Matrices. J. ACM 1962, 9, 11–12. [Google Scholar] [CrossRef]
- Floyd, R.W. Algorithm 97: Shortest Path. Commun. ACM 1962, 5, 345. [Google Scholar] [CrossRef]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef]
- Fukushima, K. Neocognitron: A Self-Organizing Neural Network Model for a Mechanism of Pattern Recognition Unaffected by Shift in Position. Biol. Cybern. 1980, 36, 193–202. [Google Scholar] [CrossRef]
- Hubel, D.H.; Wiesel, T.N. Receptive Fields and Functional Architecture of Monkey Striate Cortex. J. Physiol. 1968, 195, 215–243. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 3523–3542. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. In Proceedings of the Computer Vision—ECCV 2014, Zurich, Switzerland, 6–14 September 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 818–833. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2015, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ciresan, D.; Giusti, A.; Gambardella, L.; Schmidhuber, J. Deep Neural Networks Segment Neuronal Membranes in Electron Microscopy Images. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25. [Google Scholar]
- Kaymak, R.; Kaymak, C.; Ucar, A. Skin Lesion Segmentation Using Fully Convolutional Networks: A Comparative Experimental Study. Expert Syst. Appl. 2020, 161, 113742. [Google Scholar] [CrossRef]
- Sun, J.; Peng, Y.; Guo, Y.; Li, D. Segmentation of the Multimodal Brain Tumor Image Used the Multi-Pathway Architecture Method Based on 3D FCN. Neurocomputing 2021, 423, 34–45. [Google Scholar] [CrossRef]
- Park, K.-B.; Choi, S.H.; Lee, J.Y. M-GAN: Retinal Blood Vessel Segmentation by Balancing Losses Through Stacked Deep Fully Convolutional Networks. IEEE Access 2020, 8, 146308–146322. [Google Scholar] [CrossRef]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A Review of Deep-Learning-Based Medical Image Segmentation Methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016, Athens, Greece, 17–21 October 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Zhang, Z.; Liu, Q.; Wang, Y. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning Representations by Back-Propagating Errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Sherstinsky, A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) Network. Phys. Nonlinear Phenom. 2020, 404, 132306. [Google Scholar] [CrossRef]
- Jabbar, A.; Li, X.; Omar, B. A Survey on Generative Adversarial Networks: Variants, Applications, and Training. ACM Comput. Surv. 2021, 54, 157. [Google Scholar] [CrossRef]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.Y.K.; Wang, Z.; Paul Smolley, S. Least Squares Generative Adversarial Networks. In Proceedings of the IEEE International Conference on Computer Vision 2017, Venice, Italy, 22–29 October 2017; pp. 2794–2802. [Google Scholar]
- Paszke, A.; Chaurasia, A.; Kim, S.; Culurciello, E. ENet: A Deep Neural Network Architecture for Real-Time Semantic Segmentation. arXiv 2016, arXiv:1606.02147. [Google Scholar]
- Bank, D.; Koenigstein, N.; Giryes, R. Autoencoders. In Machine Learning for Data Science Handbook: Data Mining and Knowledge Discovery Handbook; Rokach, L., Maimon, O., Shmueli, E., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 353–374. ISBN 978-3-031-24628-9. [Google Scholar]
- Karimpouli, S.; Tahmasebi, P. Segmentation of Digital Rock Images Using Deep Convolutional Autoencoder Networks. Comput. Geosci. 2019, 126, 142–150. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Jeyaseelan, L.; Li, Q.; Chiang, J.N.; Wu, Z.; Ding, X. Embracing Imperfect Datasets: A Review of Deep Learning Solutions for Medical Image Segmentation. Med. Image Anal. 2020, 63, 101693. [Google Scholar] [CrossRef]
- Laurer, M.; van Atteveldt, W.; Casas, A.; Welbers, K. Less Annotating, More Classifying: Addressing the Data Scarcity Issue of Supervised Machine Learning with Deep Transfer Learning and BERT-NLI. Polit. Anal. 2024, 32, 84–100. [Google Scholar] [CrossRef]
- Morid, M.A.; Borjali, A.; Del Fiol, G. A Scoping Review of Transfer Learning Research on Medical Image Analysis Using ImageNet. Comput. Biol. Med. 2021, 128, 104115. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Taylor, L.; Nitschke, G. Improving Deep Learning with Generic Data Augmentation. In Proceedings of the 2018 IEEE Symposium Series on Computational Intelligence (SSCI), Bangalore, India, 18–21 November 2018; pp. 1542–1547. [Google Scholar]
- Shin, H.-C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical Image Synthesis for Data Augmentation and Anonymization Using Generative Adversarial Networks. In Simulation and Synthesis in Medical Imaging, Proceedings of the Third International Workshop, SASHIMI 2018, Granada, Spain, 16 September 2018; Gooya, A., Goksel, O., Oguz, I., Burgos, N., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–11. [Google Scholar]
- Bhatt, C.; Kumar, I.; Vijayakumar, V.; Singh, K.U.; Kumar, A. The State of the Art of Deep Learning Models in Medical Science and Their Challenges. Multimed. Syst. 2021, 27, 599–613. [Google Scholar] [CrossRef]
- Han, S.; Mao, H.; Dally, W.J. Deep Compression: Compressing Deep Neural Networks with Pruning, Trained Quantization and Huffman Coding. arXiv 2016, arXiv:1510.00149. [Google Scholar]
- Li, H.; Wang, Z.; Yue, X.; Wang, W.; Tomiyama, H.; Meng, L. An Architecture-Level Analysis on Deep Learning Models for Low-Impact Computations. Artif. Intell. Rev. 2023, 56, 1971–2010. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2018, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Ben-Nun, T.; Hoefler, T. Demystifying Parallel and Distributed Deep Learning: An in-Depth Concurrency Analysis. ACM Comput. Surv. 2019, 52, 65. [Google Scholar] [CrossRef]
- Navarro, C.A.; Hitschfeld-Kahler, N.; Mateu, L. A Survey on Parallel Computing and Its Applications in Data-Parallel Problems Using GPU Architectures. Commun. Comput. Phys. 2014, 15, 285–329. [Google Scholar] [CrossRef]
- You, Y.; Zhang, Z.; Hsieh, C.-J.; Demmel, J.; Keutzer, K. Fast Deep Neural Network Training on Distributed Systems and Cloud TPUs. IEEE Trans. Parallel Distrib. Syst. 2019, 30, 2449–2462. [Google Scholar] [CrossRef]
- Vinuesa, R.; Sirmacek, B. Interpretable Deep-Learning Models to Help Achieve the Sustainable Development Goals. Nat. Mach. Intell. 2021, 3, 926. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. arXiv 2017. [Google Scholar] [CrossRef]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.-R.; Samek, W. On Pixel-Wise Explanations for Non-Linear Classifier Decisions by Layer-Wise Relevance Propagation. PLoS ONE 2015, 10, e0130140. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 1135–1144. [Google Scholar]
- Custode, L.L.; Iacca, G. Evolutionary Learning of Interpretable Decision Trees. IEEE Access 2023, 11, 6169–6184. [Google Scholar] [CrossRef]
- Liu, G.; Schulte, O.; Zhu, W.; Li, Q. Toward Interpretable Deep Reinforcement Learning with Linear Model U-Trees. In Proceedings of the Machine Learning and Knowledge Discovery in Databases, Dublin, Ireland, 10–14 September 2018; Berlingerio, M., Bonchi, F., Gärtner, T., Hurley, N., Ifrim, G., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 414–429. [Google Scholar]
- Hestness, J.; Narang, S.; Ardalani, N.; Diamos, G.; Jun, H.; Kianinejad, H.; Patwary, M.M.A.; Yang, Y.; Zhou, Y. Deep Learning Scaling Is Predictable, Empirically. arXiv 2017, arXiv:1712.00409. [Google Scholar]
- Rehman, M.U.; Cho, S.; Kim, J.H.; Chong, K.T. BU-Net: Brain Tumor Segmentation Using Modified U-Net Architecture. Electronics 2020, 9, 2203. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, C.; Zhou, T.; Feng, L.; Liu, L.; Zeng, Q.; Wang, G. A Deep Residual Attention-Based U-Net with a Biplane Joint Method for Liver Segmentation from CT Scans. Comput. Biol. Med. 2023, 152, 106421. [Google Scholar] [CrossRef] [PubMed]
- Daimary, D.; Bora, M.B.; Amitab, K.; Kandar, D. Brain Tumor Segmentation from MRI Images Using Hybrid Convolutional Neural Networks. Procedia Comput. Sci. 2020, 167, 2419–2428. [Google Scholar] [CrossRef]
- Lee, B.; Yamanakkanavar, N.; Choi, J.Y. Automatic Segmentation of Brain MRI Using a Novel Patch-Wise U-Net Deep Architecture. PLoS ONE 2020, 15, e0236493. [Google Scholar] [CrossRef]
- Tan, L.; Ma, W.; Xia, J.; Sarker, S. Multimodal Magnetic Resonance Image Brain Tumor Segmentation Based on ACU-Net Network. IEEE Access 2021, 9, 14608–14618. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, Z.; Dong, J.; Hou, Y.; Liu, B. Attention Gate ResU-Net for Automatic MRI Brain Tumor Segmentation. IEEE Access 2020, 8, 58533–58545. [Google Scholar] [CrossRef]
- Aboussaleh, I.; Riffi, J.; Mahraz, A.M.; Tairi, H. Inception-UDet: An Improved U-Net Architecture for Brain Tumor Segmentation. Ann. Data Sci. 2024, 11, 831–853. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, G.; Huang, H.; Yang, W.; Xu, X.; Liu, Y.; Lai, X. ME-Net: Multi-Encoder Net Framework for Brain Tumor Segmentation. Int. J. Imaging Syst. Technol. 2021, 31, 1834–1848. [Google Scholar] [CrossRef]
- Zhou, Z.; He, Z.; Jia, Y. AFPNet: A 3D Fully Convolutional Neural Network with Atrous-Convolution Feature Pyramid for Brain Tumor Segmentation via MRI Images. Neurocomputing 2020, 402, 235–244. [Google Scholar] [CrossRef]
- Byra, M.; Jarosik, P.; Szubert, A.; Galperin, M.; Ojeda-Fournier, H.; Olson, L.; O’Boyle, M.; Comstock, C.; Andre, M. Breast Mass Segmentation in Ultrasound with Selective Kernel U-Net Convolutional Neural Network. Biomed. Signal Process. Control 2020, 61, 102027. [Google Scholar] [CrossRef]
- Piantadosi, G.; Sansone, M.; Fusco, R.; Sansone, C. Multi-Planar 3D Breast Segmentation in MRI via Deep Convolutional Neural Networks. Artif. Intell. Med. 2020, 103, 101781. [Google Scholar] [CrossRef] [PubMed]
- Baccouche, A.; Garcia-Zapirain, B.; Castillo Olea, C.; Elmaghraby, A.S. Connected-UNets: A Deep Learning Architecture for Breast Mass Segmentation. npj Breast Cancer 2021, 7, 151. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Duan, X.; Wang, C.; Guo, H. Segmentation and Recognition of Breast Ultrasound Images Based on an Expanded U-Net. PLoS ONE 2021, 16, e0253202. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, L.; Xia, T.; Ni, H.; Li, J. Multi-Scale Fusion U-Net for the Segmentation of Breast Lesions. IEEE Access 2021, 9, 137125–137139. [Google Scholar] [CrossRef]
- Robin, M.; John, J.; Ravikumar, A. Breast Tumor Segmentation Using U-NET. In Proceedings of the 2021 5th International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 8–10 April 2021; pp. 1164–1167. [Google Scholar]
- Xue, C.; Zhu, L.; Fu, H.; Hu, X.; Li, X.; Zhang, H.; Heng, P.-A. Global Guidance Network for Breast Lesion Segmentation in Ultrasound Images. Med. Image Anal. 2021, 70, 101989. [Google Scholar] [CrossRef]
- Khaled, R.; Vidal, J.; Vilanova, J.C.; Martí, R. A U-Net Ensemble for Breast Lesion Segmentation in DCE MRI. Comput. Biol. Med. 2022, 140, 105093. [Google Scholar] [CrossRef]
- Ning, Z.; Zhong, S.; Feng, Q.; Chen, W.; Zhang, Y. SMU-Net: Saliency-Guided Morphology-Aware U-Net for Breast Lesion Segmentation in Ultrasound Image. IEEE Trans. Med. Imaging 2022, 41, 476–490. [Google Scholar] [CrossRef]
- Zhai, D.; Hu, B.; Gong, X.; Zou, H.; Luo, J. ASS-GAN: Asymmetric Semi-Supervised GAN for Breast Ultrasound Image Segmentation. Neurocomputing 2022, 493, 204–216. [Google Scholar] [CrossRef]
- Almotairi, S.; Kareem, G.; Aouf, M.; Almutairi, B.; Salem, M.A.-M. Liver Tumor Segmentation in CT Scans Using Modified SegNet. Sensors 2020, 20, 1516. [Google Scholar] [CrossRef]
- Budak, Ü.; Guo, Y.; Tanyildizi, E.; Şengür, A. Cascaded Deep Convolutional Encoder-Decoder Neural Networks for Efficient Liver Tumor Segmentation. Med. Hypotheses 2020, 134, 109431. [Google Scholar] [CrossRef]
- Sabir, M.W.; Khan, Z.; Saad, N.M.; Khan, D.M.; Al-Khasawneh, M.A.; Perveen, K.; Qayyum, A.; Azhar Ali, S.S. Segmentation of Liver Tumor in CT Scan Using ResU-Net. Appl. Sci. 2022, 12, 8650. [Google Scholar] [CrossRef]
- Tran, S.-T.; Cheng, C.-H.; Liu, D.-G. A Multiple Layer U-Net, Un-Net, for Liver and Liver Tumor Segmentation in CT. IEEE Access 2021, 9, 3752–3764. [Google Scholar] [CrossRef]
- Roy, S.S.; Roy, S.; Mukherjee, P.; Roy, A.H. An Automated Liver Tumour Segmentation and Classification Model by Deep Learning Based Approaches. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2023, 11, 638–650. [Google Scholar]
- Selvaraj, A.; Nithiyaraj, E. CEDRNN: A Convolutional Encoder-Decoder Residual Neural Network for Liver Tumour Segmentation. Neural Process. Lett. 2023, 55, 1605–1624. [Google Scholar] [CrossRef]
- Wang, J.; Peng, Y.; Jing, S.; Han, L.; Li, T.; Luo, J. A Deep-Learning Approach for Segmentation of Liver Tumors in Magnetic Resonance Imaging Using UNet++. BMC Cancer 2023, 23, 1060. [Google Scholar] [CrossRef]
- Gaál, G.; Maga, B.; Lukács, A. Attention U-Net Based Adversarial Architectures for Chest X-Ray Lung Segmentation. arXiv 2020, arXiv:2003.10304. [Google Scholar]
- Hu, Q.; de F. Souza, L.F.; Holanda, G.B.; Alves, S.S.A.; dos S. Silva, F.H.; Han, T.; Rebouças Filho, P.P. An Effective Approach for CT Lung Segmentation Using Mask Region-Based Convolutional Neural Networks. Artif. Intell. Med. 2020, 103, 101792. [Google Scholar] [CrossRef]
- Khanna, A.; Londhe, N.D.; Gupta, S.; Semwal, A. A Deep Residual U-Net Convolutional Neural Network for Automated Lung Segmentation in Computed Tomography Images. Biocybern. Biomed. Eng. 2020, 40, 1314–1327. [Google Scholar] [CrossRef]
- Munawar, F.; Azmat, S.; Iqbal, T.; Grönlund, C.; Ali, H. Segmentation of Lungs in Chest X-Ray Image Using Generative Adversarial Networks. IEEE Access 2020, 8, 153535–153545. [Google Scholar] [CrossRef]
- Xiao, Z.; Liu, B.; Geng, L.; Zhang, F.; Liu, Y. Segmentation of Lung Nodules Using Improved 3D-UNet Neural Network. Symmetry 2020, 12, 1787. [Google Scholar] [CrossRef]
- Chen, K.; Xuan, Y.; Lin, A.; Guo, S. Lung Computed Tomography Image Segmentation Based on U-Net Network Fused with Dilated Convolution. Comput. Methods Programs Biomed. 2021, 207, 106170. [Google Scholar] [CrossRef] [PubMed]
- Jalali, Y.; Fateh, M.; Rezvani, M.; Abolghasemi, V.; Anisi, M.H. ResBCDU-Net: A Deep Learning Framework for Lung CT Image Segmentation. Sensors 2021, 21, 268. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Jing, L.; Huo, Y.; Li, L.; Akin, O.; Tian, Y. LGAN: Lung Segmentation in CT Scans Using Generative Adversarial Network. Comput. Med. Imaging Graph. 2021, 87, 101817. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Luo, J.; Yang, Y.; Wang, W.; Deng, J.; Yu, L. Automatic Lung Segmentation in Chest X-Ray Images Using Improved U-Net. Sci. Rep. 2022, 12, 8649. [Google Scholar] [CrossRef] [PubMed]
- Astono, I.P.; Welsh, J.S.; Chalup, S.; Greer, P. Optimisation of 2D U-Net Model Components for Automatic Prostate Segmentation on MRI. Appl. Sci. 2020, 10, 2601. [Google Scholar] [CrossRef]
- Hambarde, P.; Talbar, S.; Mahajan, A.; Chavan, S.; Thakur, M.; Sable, N. Prostate Lesion Segmentation in MR Images Using Radiomics Based Deeply Supervised U-Net. Biocybern. Biomed. Eng. 2020, 40, 1421–1435. [Google Scholar] [CrossRef]
- Ushinsky, A.; Bardis, M.; Glavis-Bloom, J.; Uchio, E.; Chantaduly, C.; Nguyentat, M.; Chow, D.; Chang, P.D.; Houshyar, R.; Ushinsky, A.; et al. A 3D-2D Hybrid U-Net Convolutional Neural Network Approach to Prostate Organ Segmentation of Multiparametric MRI. Am. J. Roentgenol. 2020, 216, 111–116. [Google Scholar] [CrossRef]
- Chen, J.; Wan, Z.; Zhang, J.; Li, W.; Chen, Y.; Li, Y.; Duan, Y. Medical Image Segmentation and Reconstruction of Prostate Tumor Based on 3D AlexNet. Comput. Methods Programs Biomed. 2021, 200, 105878. [Google Scholar] [CrossRef]
- He, K.; Lian, C.; Adeli, E.; Huo, J.; Gao, Y.; Zhang, B.; Zhang, J.; Shen, D. MetricUNet: Synergistic Image- and Voxel-Level Learning for Precise Prostate Segmentation via Online Sampling. Med. Image Anal. 2021, 71, 102039. [Google Scholar] [CrossRef]
- He, K.; Lian, C.; Zhang, B.; Zhang, X.; Cao, X.; Nie, D.; Gao, Y.; Zhang, J.; Shen, D. HF-UNet: Learning Hierarchically Inter-Task Relevance in Multi-Task U-Net for Accurate Prostate Segmentation in CT Images. IEEE Trans. Med. Imaging 2021, 40, 2118–2128. [Google Scholar] [CrossRef]
- Jin, Y.; Yang, G.; Fang, Y.; Li, R.; Xu, X.; Liu, Y.; Lai, X. 3D PBV-Net: An Automated Prostate MRI Data Segmentation Method. Comput. Biol. Med. 2021, 128, 104160. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, G.; Wu, X.; Ding, X.; Cao, X.; Wang, L.; Zhang, J.; Wang, P. Automatic Segmentation of Prostate Magnetic Resonance Imaging Using Generative Adversarial Networks. Clin. Imaging 2021, 70, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.M.; Alhussein, M.; Aurangzeb, K.; Arsalan, M.; Naqvi, S.S.; Nawaz, S.J. Residual Connection-Based Encoder Decoder Network (RCED-Net) for Retinal Vessel Segmentation. IEEE Access 2020, 8, 131257–131272. [Google Scholar] [CrossRef]
- Chala, M.; Nsiri, B.; El yousfi Alaoui, M.H.; Soulaymani, A.; Mokhtari, A.; Benaji, B. An Automatic Retinal Vessel Segmentation Approach Based on Convolutional Neural Networks. Expert Syst. Appl. 2021, 184, 115459. [Google Scholar] [CrossRef]
- Guo, C.; Szemenyei, M.; Yi, Y.; Wang, W.; Chen, B.; Fan, C. SA-UNet: Spatial Attention U-Net for Retinal Vessel Segmentation. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 1236–1242. [Google Scholar]
- Li, X.; Jiang, Y.; Li, M.; Yin, S. Lightweight Attention Convolutional Neural Network for Retinal Vessel Image Segmentation. IEEE Trans. Ind. Inform. 2021, 17, 1958–1967. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Xu, X. Pyramid U-Net for Retinal Vessel Segmentation. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1125–1129. [Google Scholar]
- Dong, F.; Wu, D.; Guo, C.; Zhang, S.; Yang, B.; Gong, X. CRAUNet: A Cascaded Residual Attention U-Net for Retinal Vessel Segmentation. Comput. Biol. Med. 2022, 147, 105651. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, J.; Yang, L.; Bian, G.; Yu, H. ResDO-UNet: A Deep Residual Network for Accurate Retinal Vessel Segmentation from Fundus Images. Biomed. Signal Process. Control 2023, 79, 104087. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, J.; Yang, L.; Yu, H.; Bian, G. Wave-Net: A Lightweight Deep Network for Retinal Vessel Segmentation from Fundus Images. Comput. Biol. Med. 2023, 152, 106341. [Google Scholar] [CrossRef]
- Lei, B.; Xia, Z.; Jiang, F.; Jiang, X.; Ge, Z.; Xu, Y.; Qin, J.; Chen, S.; Wang, T.; Wang, S. Skin Lesion Segmentation via Generative Adversarial Networks with Dual Discriminators. Med. Image Anal. 2020, 64, 101716. [Google Scholar] [CrossRef]
- Öztürk, Ş.; Özkaya, U. Skin Lesion Segmentation with Improved Convolutional Neural Network. J. Digit. Imaging 2020, 33, 958–970. [Google Scholar] [CrossRef]
- Xie, F.; Yang, J.; Liu, J.; Jiang, Z.; Zheng, Y.; Wang, Y. Skin Lesion Segmentation Using High-Resolution Convolutional Neural Network. Comput. Methods Programs Biomed. 2020, 186, 105241. [Google Scholar] [CrossRef] [PubMed]
- Zafar, K.; Gilani, S.O.; Waris, A.; Ahmed, A.; Jamil, M.; Khan, M.N.; Sohail Kashif, A. Skin Lesion Segmentation from Dermoscopic Images Using Convolutional Neural Network. Sensors 2020, 20, 1601. [Google Scholar] [CrossRef] [PubMed]
- Thurnhofer-Hemsi, K.; Domínguez, E. A Convolutional Neural Network Framework for Accurate Skin Cancer Detection. Neural Process. Lett. 2021, 53, 3073–3093. [Google Scholar] [CrossRef]
- Alahmadi, M.D. Multiscale Attention U-Net for Skin Lesion Segmentation. IEEE Access 2022, 10, 59145–59154. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Koundal, D.; Nayak, S.R.; Barsocchi, P.; Bhoi, A.K. Modified U-NET Architecture for Segmentation of Skin Lesion. Sensors 2022, 22, 867. [Google Scholar] [CrossRef]
- Dai, D.; Dong, C.; Xu, S.; Yan, Q.; Li, Z.; Zhang, C.; Luo, N. Ms RED: A Novel Multi-Scale Residual Encoding and Decoding Network for Skin Lesion Segmentation. Med. Image Anal. 2022, 75, 102293. [Google Scholar] [CrossRef]
- Guan, X.; Yang, G.; Ye, J.; Yang, W.; Xu, X.; Jiang, W.; Lai, X. 3D AGSE-VNet: An Automatic Brain Tumor MRI Data Segmentation Framework. BMC Med. Imaging 2022, 22, 6. [Google Scholar] [CrossRef]
- Piantadosi, G.; Sansone, M.; Sansone, C. Breast Segmentation in MRI via U-Net Deep Convolutional Neural Networks. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3917–3922. [Google Scholar]
- Khaled, R.; Vidal, J.; Martí, R. Deep Learning Based Segmentation of Breast Lesions in DCE-MRI. In Proceedings of the Pattern Recognition. ICPR International Workshops and Challenges, Virtual Event, 10–15 January 2021; Del Bimbo, A., Cucchiara, R., Sclaroff, S., Farinella, G.M., Mei, T., Bertini, M., Escalante, H.J., Vezzani, R., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 417–430. [Google Scholar]
- Aghamohammadi, A.; Ranjbarzadeh, R.; Naiemi, F.; Mogharrebi, M.; Dorosti, S.; Bendechache, M. TPCNN: Two-Path Convolutional Neural Network for Tumor and Liver Segmentation in CT Images Using a Novel Encoding Approach. Expert Syst. Appl. 2021, 183, 115406. [Google Scholar] [CrossRef]
- Sirco, A.; Almisreb, A.; Tahir, N.M.; Bakri, J. Liver Tumour Segmentation Based on ResNet Technique. In Proceedings of the 2022 IEEE 12th International Conference on Control System, Computing and Engineering (ICCSCE), Penang, Malaysia, 21–22 October 2022; pp. 203–208. [Google Scholar]
- Sun, S.; Bauer, C.; Beichel, R. Automated 3-D Segmentation of Lungs with Lung Cancer in CT Data Using a Novel Robust Active Shape Model Approach. IEEE Trans. Med. Imaging 2012, 31, 449–460. [Google Scholar] [CrossRef]
- Hossain, M.R.I.; Ahmed, I.; Kabir, M.H. Automatic Lung Tumor Detection Based on GLCM Features. In Proceedings of the Computer Vision—ACCV 2014 Workshops, Singapore, 1–2 November 2014; Jawahar, C.V., Shan, S., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 109–121. [Google Scholar]
- Bradley, D.; Roth, G. Adaptive Thresholding Using the Integral Image. J. Graph. Tools 2007, 12, 13–21. [Google Scholar] [CrossRef]
- Haas, B.; Coradi, T.; Scholz, M.; Kunz, P.; Huber, M.; Oppitz, U.; André, L.; Lengkeek, V.; Huyskens, D.; van Esch, A.; et al. Automatic Segmentation of Thoracic and Pelvic CT Images for Radiotherapy Planning Using Implicit Anatomic Knowledge and Organ-Specific Segmentation Strategies. Phys. Med. Biol. 2008, 53, 1751. [Google Scholar] [CrossRef] [PubMed]
- Campadelli, P.; Casiraghi, E.; Pratissoli, S. A Segmentation Framework for Abdominal Organs from CT Scans. Artif. Intell. Med. 2010, 50, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Sujatha, P.; Sudha, K.K. Performance Analysis of Different Edge Detection Techniques for Image Segmentation. Indian J. Sci. Technol. 2015, 8, 1–6. [Google Scholar] [CrossRef]
- Khadidos, A. Medical Image Segmentation Using Edge-Based Active Contours. Ph.D. Thesis, University of Warwick, Coventry, UK, 2016. [Google Scholar]
- Banerjee, S.; Mitra, S.; Uma Shankar, B. Single Seed Delineation of Brain Tumor Using Multi-Thresholding. Inf. Sci. 2016, 330, 88–103. [Google Scholar] [CrossRef]
- Mostafa, A.; Elfattah, M.A.; Fouad, A.; Hassanien, A.E.; Hefny, H. Enhanced Region Growing Segmentation for CT Liver Images. In Proceedings of the The 1st International Conference on Advanced Intelligent System and Informatics (AISI2015), Beni Suef, Egypt, 28–30 November 2015; Gaber, T., Hassanien, A.E., El-Bendary, N., Dey, N., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 115–127. [Google Scholar]
- Liu, G.; Zhang, Y.; Wang, A. Incorporating Adaptive Local Information into Fuzzy Clustering for Image Segmentation. IEEE Trans. Image Process. 2015, 24, 3990–4000. [Google Scholar] [CrossRef]
- Devi, K.Y.; Sasikala, M. Labeling and Clustering-Based Level Set Method for Automated Segmentation of Lung Tumor Stages in CT Images. J. Ambient Intell. Humaniz. Comput. 2021, 12, 2299–2309. [Google Scholar] [CrossRef]
- Li, K.; Wu, X.; Chen, D.Z.; Sonka, M. Optimal Surface Segmentation in Volumetric Images-a Graph-Theoretic Approach. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 119–134. [Google Scholar] [CrossRef]
- Mayala, S.; Herdlevær, I.; Haugsøen, J.B.; Anandan, S.; Blaser, N.; Gavasso, S.; Brun, M. GUBS: Graph-Based Unsupervised Brain Segmentation in MRI Images. J. Imaging 2022, 8, 262. [Google Scholar] [CrossRef]
- Yan, Z.; Zhang, H.; Piramuthu, R.; Jagadeesh, V.; DeCoste, D.; Di, W.; Yu, Y. HD-CNN: Hierarchical Deep Convolutional Neural Networks for Large Scale Visual Recognition. In Proceedings of the IEEE International Conference on Computer Vision 2015, Santiago, Chile, 7–13 December 2015; pp. 2740–2748. [Google Scholar]
- Olimov, B.; Kim, J.; Paul, A. DCBT-Net: Training Deep Convolutional Neural Networks with Extremely Noisy Labels. IEEE Access 2020, 8, 220482–220495. [Google Scholar] [CrossRef]
- Christ, P.F.; Ettlinger, F.; Grün, F.; Elshaera, M.E.A.; Lipkova, J.; Schlecht, S.; Ahmaddy, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; et al. Automatic Liver and Tumor Segmentation of CT and MRI Volumes Using Cascaded Fully Convolutional Neural Networks. arXiv 2017, arXiv:1702.05970. [Google Scholar]
- Zhang, Y.; Chi, M. Mask-R-FCN: A Deep Fusion Network for Semantic Segmentation. IEEE Access 2020, 8, 155753–155765. [Google Scholar] [CrossRef]
- Dai, J.; Li, Y.; He, K.; Sun, J. R-FCN: Object Detection via Region-Based Fully Convolutional Networks. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; Curran Associates, Inc.: Red Hook, NY, USA, 2016; Volume 29. [Google Scholar]
- Feng, Z.; Yang, J.; Yao, L.; Qiao, Y.; Yu, Q.; Xu, X. Deep Retinal Image Segmentation: A FCN-Based Architecture with Short and Long Skip Connections for Retinal Image Segmentation. In Neural Information Processing, Proceedings of the 24th International Conference, ICONIP 2017, Guangzhou, China, 14–18 November 2017; Liu, D., Xie, S., Li, Y., Zhao, D., El-Alfy, E.-S.M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 713–722. [Google Scholar]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Pendse, M.; Thangarasa, V.; Chiley, V.; Holmdahl, R.; Hestness, J.; DeCoste, D. Memory Efficient 3D U-Net with Reversible Mobile Inverted Bottlenecks for Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 6th International Workshop, BrainLes 2020, Lima, Peru, 4 October 2020; Crimi, A., Bakas, S., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 388–397. [Google Scholar]
- Aldoj, N.; Biavati, F.; Michallek, F.; Stober, S.; Dewey, M. Automatic Prostate and Prostate Zones Segmentation of Magnetic Resonance Images Using DenseNet-like U-Net. Sci. Rep. 2020, 10, 14315. [Google Scholar] [CrossRef] [PubMed]
- Andreini, P.; Ciano, G.; Bonechi, S.; Graziani, C.; Lachi, V.; Mecocci, A.; Sodi, A.; Scarselli, F.; Bianchini, M. A Two-Stage GAN for High-Resolution Retinal Image Generation and Segmentation. Electronics 2022, 11, 60. [Google Scholar] [CrossRef]
- Wang, C.; Xu, C.; Yao, X.; Tao, D. Evolutionary Generative Adversarial Networks. IEEE Trans. Evol. Comput. 2019, 23, 921–934. [Google Scholar] [CrossRef]
- Zuo, Z.; Shuai, B.; Wang, G.; Liu, X.; Wang, X.; Wang, B.; Chen, Y. Convolutional Recurrent Neural Networks: Learning Spatial Dependencies for Image Representation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 2015, Boston, MA, USA, 7–12 June 2015; pp. 18–26. [Google Scholar]
- Pascanu, R.; Mikolov, T.; Bengio, Y. On the Difficulty of Training Recurrent Neural Networks. Available online: https://arxiv.org/abs/1211.5063 (accessed on 7 October 2024).
- Wang, Y.; Zhang, W. A Dense RNN for Sequential Four-Chamber View Left Ventricle Wall Segmentation and Cardiac State Estimation. Front. Bioeng. Biotechnol. 2021, 9, 696227. [Google Scholar] [CrossRef]
- Zheng, R.; Wang, Q.; Lv, S.; Li, C.; Wang, C.; Chen, W.; Wang, H. Automatic Liver Tumor Segmentation on Dynamic Contrast Enhanced MRI Using 4D Information: Deep Learning Model Based on 3D Convolution and Convolutional LSTM. IEEE Trans. Med. Imaging 2022, 41, 2965–2976. [Google Scholar] [CrossRef]
- Wickramasinghe, C.S.; Marino, D.L.; Manic, M. ResNet Autoencoders for Unsupervised Feature Learning from High-Dimensional Data: Deep Models Resistant to Performance Degradation. IEEE Access 2021, 9, 40511–40520. [Google Scholar] [CrossRef]
- Bercea, C.I.; Rueckert, D.; Schnabel, J.A. What Do AEs Learn? Challenging Common Assumptions in Unsupervised Anomaly Detection. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2023, Vancouver, BC, Canada, 8–12 October 2023; Greenspan, H., Madabhushi, A., Mousavi, P., Salcudean, S., Duncan, J., Syeda-Mahmood, T., Taylor, R., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2023; pp. 304–314. [Google Scholar]
- Chen, H.-J.; Ruan, S.-J.; Huang, S.-W.; Peng, Y.-T. Lung X-Ray Segmentation Using Deep Convolutional Neural Networks on Contrast-Enhanced Binarized Images. Mathematics 2020, 8, 545. [Google Scholar] [CrossRef]
- Venkateswarlu Isunuri, B.; Kakarla, J. Fast Brain Tumour Segmentation Using Optimized U-Net and Adaptive Thresholding. Autom. Časopis Za Autom. Mjer. Elektron. Račun. Komun. 2020, 61, 352–360. [Google Scholar] [CrossRef]
- Liu, W.; Yang, H.; Tian, T.; Cao, Z.; Pan, X.; Xu, W.; Jin, Y.; Gao, F. Full-Resolution Network and Dual-Threshold Iteration for Retinal Vessel and Coronary Angiograph Segmentation. IEEE J. Biomed. Health Inform. 2022, 26, 4623–4634. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.M.; Reddy, K.S.; Jayaram, M.; Venkata Maha Lakshmi, N.; Aluvalu, R.; Mahesh, T.R.; Kumar, V.V.; Stalin Alex, D. An Efficient Multilevel Thresholding Scheme for Heart Image Segmentation Using a Hybrid Generalized Adversarial Network. J. Sens. 2022, 2022, 4093658. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Cui, W.; Lei, B.; Kuang, X.; Zhang, T. Dual Encoder-Based Dynamic-Channel Graph Convolutional Network with Edge Enhancement for Retinal Vessel Segmentation. IEEE Trans. Med. Imaging 2022, 41, 1975–1989. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Yin, S.; Wang, K.; Teng, L.; Li, H. Multichannel Convolutional Neural Network-Based Fuzzy Active Contour Model for Medical Image Segmentation. Evol. Syst. 2022, 13, 535–549. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, X.; Zhang, X.; Lu, Y.; Huang, S.; Yang, D. EANet: Iterative Edge Attention Network for Medical Image Segmentation. Pattern Recognit. 2022, 127, 108636. [Google Scholar] [CrossRef]
- Gab Allah, A.M.; Sarhan, A.M.; Elshennawy, N.M. Edge U-Net: Brain Tumor Segmentation Using MRI Based on Deep U-Net Model with Boundary Information. Expert Syst. Appl. 2023, 213, 118833. [Google Scholar] [CrossRef]
- Liu, H.; Wang, H.; Wu, Y.; Xing, L. Superpixel Region Merging Based on Deep Network for Medical Image Segmentation. ACM Trans. Intell. Syst. Technol. 2020, 11, 39. [Google Scholar] [CrossRef]
- Ren, H.; Zhou, L.; Liu, G.; Peng, X.; Shi, W.; Xu, H.; Shan, F.; Liu, L. An Unsupervised Semi-Automated Pulmonary Nodule Segmentation Method Based on Enhanced Region Growing. Quant. Imaging Med. Surg. 2020, 10, 233–242. [Google Scholar] [CrossRef]
- Khan, H.A.; Gong, X.; Bi, F.; Ali, R. Novel Light Convolutional Neural Network for COVID Detection with Watershed Based Region Growing Segmentation. J. Imaging 2023, 9, 42. [Google Scholar] [CrossRef]
- Nithya, A.; Appathurai, A.; Venkatadri, N.; Ramji, D.R.; Anna Palagan, C. Kidney Disease Detection and Segmentation Using Artificial Neural Network and Multi-Kernel k-Means Clustering for Ultrasound Images. Measurement 2020, 149, 106952. [Google Scholar] [CrossRef]
- Khan, A.R.; Khan, S.; Harouni, M.; Abbasi, R.; Iqbal, S.; Mehmood, Z. Brain Tumor Segmentation Using K-Means Clustering and Deep Learning with Synthetic Data Augmentation for Classification. Microsc. Res. Tech. 2021, 84, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Mehmood, Z.; Nazir, T.; Naqvi, R.A.; Rehman, A.; Iqbal, M.; Saba, T. Skin Cancer Detection from Dermoscopic Images Using Deep Learning and Fuzzy K-Means Clustering. Microsc. Res. Tech. 2022, 85, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Fooladi, S.; Farsi, H.; Mohamadzadeh, S. Segmenting the Lesion Area of Brain Tumor Using Convolutional Neural Networks and Fuzzy K-Means Clustering. Int. J. Eng. 2023, 36, 1556–1568. [Google Scholar] [CrossRef]
- Li, L.; Wu, F.; Yang, G.; Xu, L.; Wong, T.; Mohiaddin, R.; Firmin, D.; Keegan, J.; Zhuang, X. Atrial Scar Quantification via Multi-Scale CNN in the Graph-Cuts Framework. Med. Image Anal. 2020, 60, 101595. [Google Scholar] [CrossRef]
- Mishra, Z.; Ganegoda, A.; Selicha, J.; Wang, Z.; Sadda, S.R.; Hu, Z. Automated Retinal Layer Segmentation Using Graph-Based Algorithm Incorporating Deep-Learning-Derived Information. Sci. Rep. 2020, 10, 9541. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, T.; Gay, H.; Zhang, W.; Sun, B. ARPM-Net: A Novel CNN-Based Adversarial Method with Markov Random Field Enhancement for Prostate and Organs at Risk Segmentation in Pelvic CT Images. Med. Phys. 2021, 48, 227–237. [Google Scholar] [CrossRef]








| Operator | Advantages | Disadvantages |
|---|---|---|
| Roberts Cross | The computation is simple and effective in noise minimization scenarios. | Sensitive to noise and inaccurate positioning of precise edges. |
| Prewitt | Particularly sensitive to edge orientation | Susceptible to noise and less crisp edges. |
| Sobel | Similar to Prewitt but provides better noise suppression while also providing moderate edge smoothing. | Edges may be too blurred and unsuitable for accurate edge detection tasks. |
| Canny | Canny boasts a high accuracy in edge detection, efficiently suppressing noise, which contributes to its ability to precisely position edges. | Canny is computationally complex, and its performance is highly dependent on parameter selection. |
| Laplacian | Accurately identifies the center of edges and is sensitive to fine detail. | Highly sensitive to noise and does not provide information about edge orientation. |
| Laplacian of Gaussian | Preliminary Gaussian smoothing reduces the effect of noise and provides good edge localization. | Higher computational complexity and may miss some fine edges. |
| Method | Cost Function | Optimization Method | Bias | Reference |
|---|---|---|---|---|
| Minimal cut | Gomory–Hu’s K-way maxflow algorithm | Short boundary | Wu and Leahy [75] | |
| Ratio regions | Minimize the ratio between the cost of the bounding contour and the benefit of the enclosed region | Smooth boundary | Cox et al. [76] | |
| Ncut | Measure both the total dissimilarity between the different groups as well as the total similarity within the groups | Similar weight partition | Shi and Malik [77] | |
| Minimum mean weight | Globally Optimal Regions and Boundaries | Jermyn and Ishikawa [78] | ||
| Mean cut | Global bipartition | None | Wang and Siskind [79] | |
| Ratio cut | Global bipartition | None | Wang and Siskind [80] | |
| Region-based Ncut | Consider the number of links on a Ncut basis | Similar weight partition | Cigla and Alatan [81] |
| Anatomical Region | Deep Learning Method | Dataset | DSC | IOU | Recall | Accuracy | SE | SP |
|---|---|---|---|---|---|---|---|---|
| Brain tumour | CNN models [140] | BraTS dataset | / | 0.633 | / | 0.927 | / | / |
| Patch-wise U-Net [141] | OASIS dataset | 0.93 | 0.877 | / | / | / | / | |
| BU-Net [138] | BraTS 2017 and 2018 datasets | TC: 0.901 WT: 0.837 ET: 0.788 | / | / | / | / | / | |
| ACU-Net [142] | BraTS 2015, 2018, and 2019 datasets | WT: 0.9273 ET: 0.8429 TC: 0.9580 | / | 0.9415 0.8119 0.7856 | 0.9652 0.9054 0.9248 | / | / | |
| AGResU-Net [143] | BraTS 2017–2019 datasets | TC: 0.787 WT: 0.876 ET: 0.745 | / | / | / | / | / | |
| Inception U-Det [144] | BraTS 2020, 2018, and 2017 datasets | 0.868 | 0.777 | / | 0.988 | / | / | |
| ME-Net [145] | BraTS 2020 dataset | TC: 0.74 WT: 0.88 ET: 0.70 | / | / | / | 0.742 0.905 0.724 | 0.999 0.999 0.999 | |
| AFP-Net [146] | BraTS 2013 dataset | TC: 0.73 WT: 0.85 ET: 0.67 | / | / | / | / | / | |
| Breast | SK U-Net CNNs [147] | UDIAT, OASBUD, and BUSI datasets | 0.701 | / | / | 0.965 | / | / |
| CNNs [148] | DSprivate42 and DSpublic88 datasets | 0.9578 | / | / | 0.9897 | 0.9549 | 0.8549 | |
| Connected U-Nets [149] | CBIS-DDSM, INbreast datasets | 0.9109 | 0.839 | / | / | / | / | |
| Expanded U-Net [150] | Breast ultrasound image | 0.905 | 0.827 | / | / | / | / | |
| MF U-Net [151] | BUSIS datasets | 0.9535 | 0.9112 | 0.9421 | / | / | / | |
| U-Net [152] | Kaggle dataset | / | / | / | 0.942 | / | / | |
| GG-Net [153] | DAF’s dataset | 0.954 | 0.912 | 0.957 | 0.951 | / | / | |
| U-Net [154] | TCGA-BRCA dataset | 0.680 | / | / | / | / | / | |
| SMU-Net [155] | BUS datasets | 0.8078 | 0.7012 | / | 0.7474 | 0.8933 | / | |
| ASSGAN [156] | DBUI, OASBUI, SPDBUI and SDBUI | 0.8319 | 0.7123 | / | 0.9589 | / | / | |
| Liver tumour | Modified SegNet [157] | 3D-IRACDb dataset | / | 0.937 | / | 0.988 | / | / |
| CEDCNN [158] | 3DIRCADb dataset | 0.9522 | / | / | / | / | / | |
| ResU-Net [159] | 3D-IRCADb01 dataset | 0.97 | / | / | / | / | 0.9618 | |
| Dilated convolutions [160] | 3DIRCADb dataset | 0.9638 | / | / | / | / | / | |
| DRAUNet [139] | LiTS, 3DIRCADb, and Sliver07 | 0.972 | / | / | / | / | / | |
| Mask-RCNN [161] | CT images | / | / | / | 0.878 | / | / | |
| CEDRNN [162] | 3DIRCADb dataset | 0.952 | 0.91 | / | 0.957 | / | / | |
| UNet++ network [163] | MRI images | 0.915 | / | / | / | / | / | |
| Lung | Attention U-Net [164] | JSRT dataset | 0.975 | / | / | / | / | / |
| Mask R-CNN [165] | CT scans | 0.97 | / | / | 0.9768 | 0.8772 | 0.8670 | |
| Residual U-Net [166] | LUNA16 VESSEL12, and HUG-ILD | 0.9898 | 0.9798 | 0.9898 | 0.9898 | / | / | |
| GAN-based model [167] | CXR datasets | 0.9740 | 0.943 | / | / | / | / | |
| 3D-Res2Unet [168] | LUNA16 public dataset | 0.953 | / | 0.991 | / | / | / | |
| DC-U-Net model [169] | CT images | 0.9743 | 0.9627 | 0.9699 | 0.9731 | / | ||
| ResBCDU-Net [170] | LIDC-IDRI database | 0.9731 | / | 0.9701 | 0.9758 | / | / | |
| LGAN [171] | LIDC-IDRI dataset | 0.985 | 0.978 | / | / | / | / | |
| EfficientNet-B4 [172] | Benchmark lung segmentation | 0.978 | 0.957 | / | 0.989 | 0.9795 | 0.989 | |
| Prostate | 2D U-Net model [173] | Public dataset PROMISE12 | 0.89 | / | / | / | / | / |
| Deeply supervised U-Net [174] | Promise12 public dataset | 0.9067 | / | / | / | / | / | |
| 3D–2D Hybrid UNet [175] | Prostate mpMRI | 0.898 | / | / | / | / | / | |
| 3D AlexNet [176] | MRI images | 0.9768 | / | / | / | 0.896 | 0.902 | |
| MetricUNet [177] | PROMISE 2012 dataset | 0.9 | / | / | / | 0.89 | / | |
| HF-Unet [178] | CT images | 0.88 | / | / | / | 0.88 | / | |
| 3D PBV-Net [179] | PROMISE 12 and TPHOH | 0.9613 | / | / | 0.9797 | / | / | |
| SegDGAN [180] | PROMISE12 public dataset | 0.8869 | / | / | / | / | / | |
| Retinal Vessel | RCED-Net [181] | DRIVE, CHASE_DB1, and STARE | / | / | / | 0.9333 | 0.8419 | 0.9801 |
| M-GAN [103] | DRIVE, STARE, HRF, and CHASE-DB1datasets | / | 0.7198 | 0.8324 | 0.9761 | 0.8324 | 0.9938 | |
| CNN-based method [182] | DRIVE and STARE datasets | / | / | / | 0.9716 | 0.8096 | 0.9841 | |
| SA-Unet [183] | DRIVE and CHASE_DB1 datasets | / | / | / | 0.9724 | 0.8573 | 0.9835 | |
| Lightweight Attention CNN [184] | DRIVE, STARE, AND CHASE_DB1 | / | / | / | 0.9627 | 0.8050 | 0.9817 | |
| Pyramid U-Net [185] | DRIVE and CHASE-DB1 datasets | / | / | / | 0.9615 | 0.8213 | 0.9807 | |
| CRAUNet [186] | DRIVE and CHASE_DB1 datasets | / | / | / | 0.9659 | 0.8259 | / | |
| ResDO-U-Net [187] | DRIVE, STARE, and CHASE_DB1 | / | / | / | 0.9623 | 0.8015 | 0.9807 | |
| Wave-Net [188] | DRIVE set, STARE set, and CHASE | / | / | / | 0.9592 | 0.8046 | 0.9798 | |
| Skin lesion | UNet-SCDC [189] | ISIC Skin Lesion Challenge datasets | 0.885 | 0.824 | / | 0.929 | 0.953 | 0.911 |
| IFCN [190] | IEEE ISBI 2017 Challenge and PH2 | 0.9302 | 0.871 | / | 0.9692 | 0.9688 | 0.9531 | |
| CNN [191] | ISBI 2016 and 2017 datasets | 0.862 | 0.783 | / | 0.938 | 0.870 | 0.964 | |
| Res-Unet [192] | ISIC 2017 dataset | 0.924 | 0.854 | / | / | / | / | |
| CNN [193] | HAM10000 dermoscopic image | / | / | 0.8480 | 0.961 | / | / | |
| MSAU-Net [194] | ISIC 2017, ISIC 2018, and PH2 | 0.9377 | 0.9617 | / | 0.9617 | 0.943 | 0.9698 | |
| U-Net [195] | PH2 dataset | / | 0.976 | / | 0.977 | / | / | |
| Ms RED [196] | ISIC 2016, 2017, 2018, and PH2 | 0.8999 | 0.8345 | 0.9049 | 0.9619 | / | / |
| Technique | Advantages | Disadvantages | Suitability for Specific Medical Applications |
|---|---|---|---|
| Thresholding | Simple, fast, computationally efficient [204] | Limited by stray colors and intensity variations, it does not work well for complex images | Works well in high-contrast regions (e.g., bones, lungs) [205,206] |
| Edge-based Segmentation | The effective detection boundary is clear and highly interpretable | Sensitive to noise and struggles with weak or fuzzy edges [207] | Excellent for tumor boundary detection in MRI and CT images with sharp edges [208] |
| Region-based Segmentation | Effectively detects homogeneous regions and excels at capturing local continuity | Dependence on seed point selection makes it difficult to deal with heterogeneous regions [209] | Available for organ and tissue segmentation with minimal intensity variation, such as the liver in CT [210] |
| Clustering-based Segmentation | Can group similar pixels without prior knowledge [211] | Sensitive to noise; may require post-processing | Tumor segmentation where intensity contrasts with surroundings [212] |
| Graph-based Segmentation | Handles global image context well, flexible handling of complex structures [213] | Computationally intensive and sensitive to the quality of graph construction [70] | Complex tissue structures (e.g., brain MRI) [214] |
| CNNs | Learns spatial hierarchies automatically, scalable [215] | Requires large labeled datasets and high computational power [216] | Tumor segmentation, lesion detection (e.g., brain, liver) [217] |
| FCNs | Pixel-level segmentation, preserves spatial resolution [218] | Sensitive to dataset size and variability [219] | Accurate pixel-wise segmentation (e.g., retinal vessels) [220] |
| U-Net | Strong performance with small datasets, effective in medical images [221] | Memory-intensive, requires much tuning for optimal performance [222] | Medical imaging, especially in biomedical fields (e.g., prostate, skin lesions) [195,223] |
| GANs | Generates high-quality, refined segmentations [224] | Challenging to train, requires large datasets, prone to instability in training [225] | Generating realistic medical segmentations (e.g., skin lesions) [189] |
| RNNs | Captures temporal dependencies, can handle sequential imaging data [226] | Difficult to train, prone to vanishing gradient problems, computationally heavy [227] | Volumetric image segmentation (e.g., heart, liver) [228,229] |
| AEs | Excellent for feature learning and dimensionality reduction [230] | Limited for highly complex segmentation tasks, lower accuracy than CNN-based methods | Feature extraction, anomaly detection (e.g., MRI, CT scans) [231] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Quan, R.; Xu, W.; Huang, Y.; Chen, X.; Liu, F. Advances in Medical Image Segmentation: A Comprehensive Review of Traditional, Deep Learning and Hybrid Approaches. Bioengineering 2024, 11, 1034. https://doi.org/10.3390/bioengineering11101034
Xu Y, Quan R, Xu W, Huang Y, Chen X, Liu F. Advances in Medical Image Segmentation: A Comprehensive Review of Traditional, Deep Learning and Hybrid Approaches. Bioengineering. 2024; 11(10):1034. https://doi.org/10.3390/bioengineering11101034
Chicago/Turabian StyleXu, Yan, Rixiang Quan, Weiting Xu, Yi Huang, Xiaolong Chen, and Fengyuan Liu. 2024. "Advances in Medical Image Segmentation: A Comprehensive Review of Traditional, Deep Learning and Hybrid Approaches" Bioengineering 11, no. 10: 1034. https://doi.org/10.3390/bioengineering11101034
APA StyleXu, Y., Quan, R., Xu, W., Huang, Y., Chen, X., & Liu, F. (2024). Advances in Medical Image Segmentation: A Comprehensive Review of Traditional, Deep Learning and Hybrid Approaches. Bioengineering, 11(10), 1034. https://doi.org/10.3390/bioengineering11101034








