U-Net Convolutional Neural Network for Real-Time Prediction of the Number of Cultured Corneal Endothelial Cells for Cellular Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Cultures
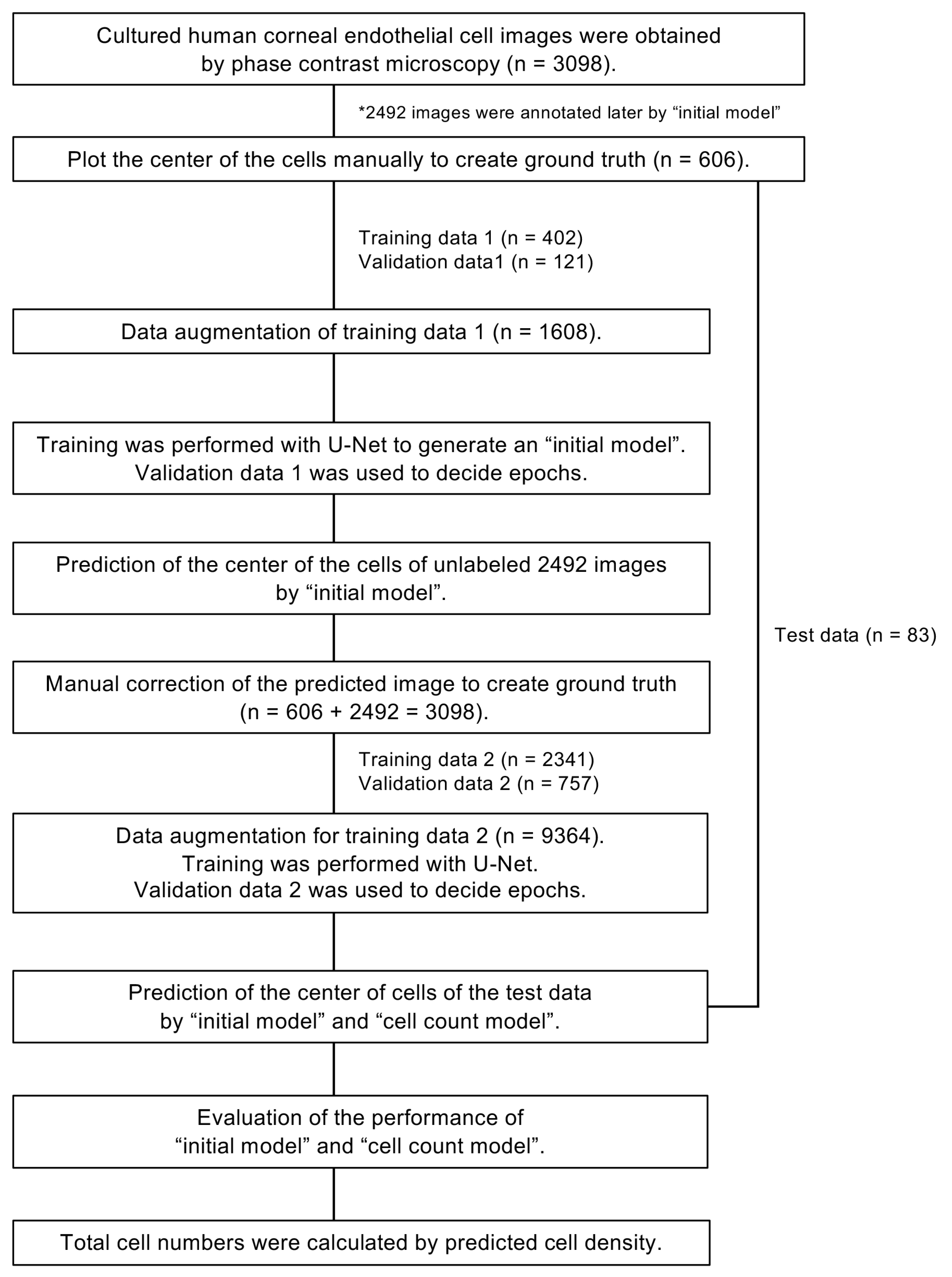
2.3. A Flow Chart for Generating the Models for Predicting Cell Centers
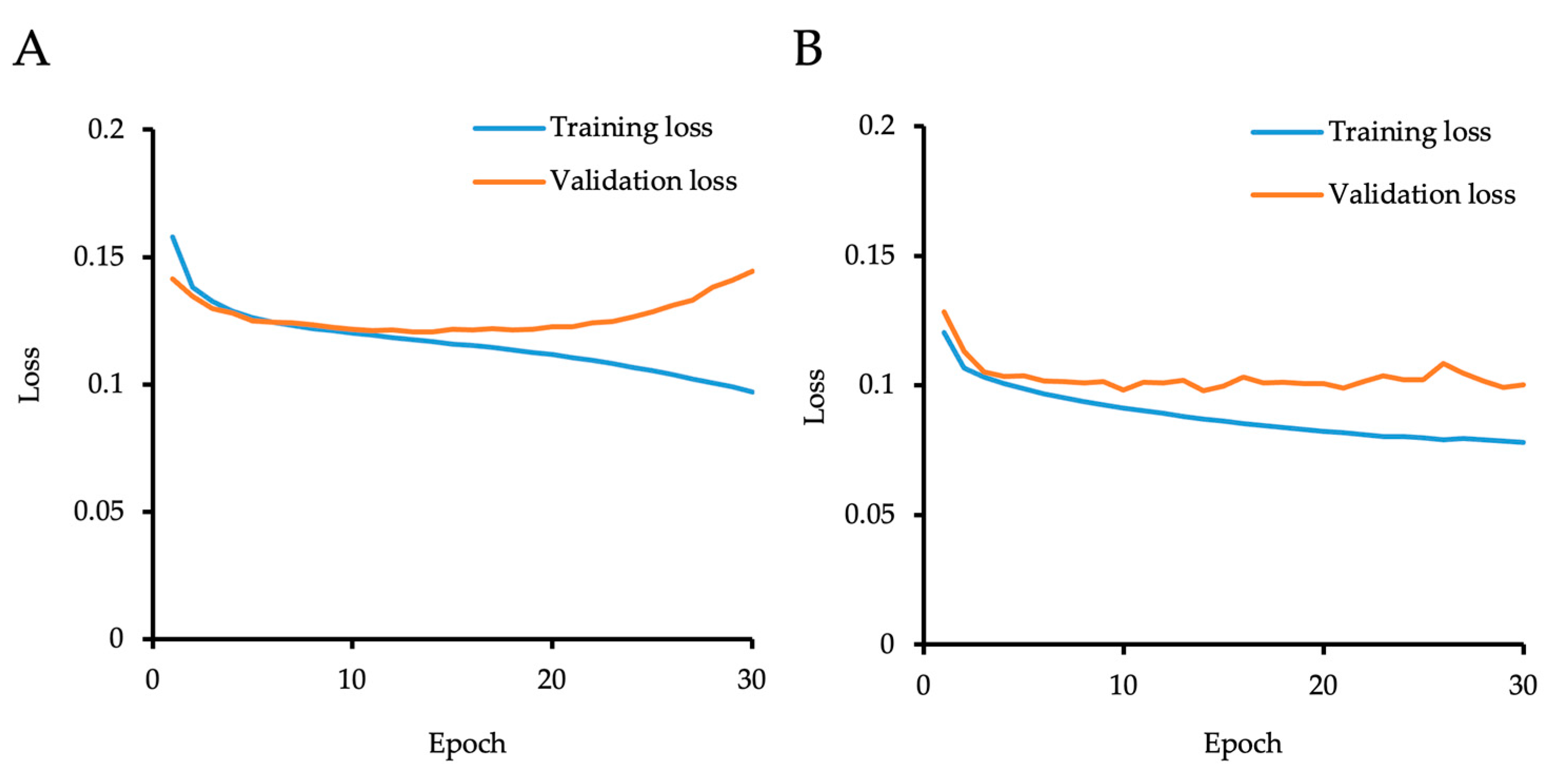
2.4. Generation of the “Initial Model”
2.5. Generation of the “Cell Count Model” Using Semi-Supervised Learning
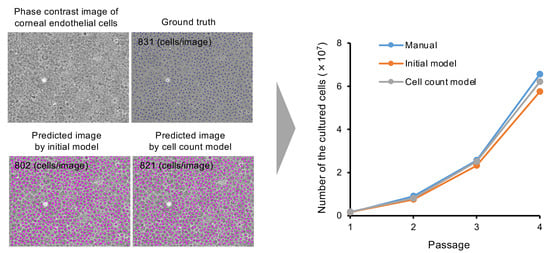
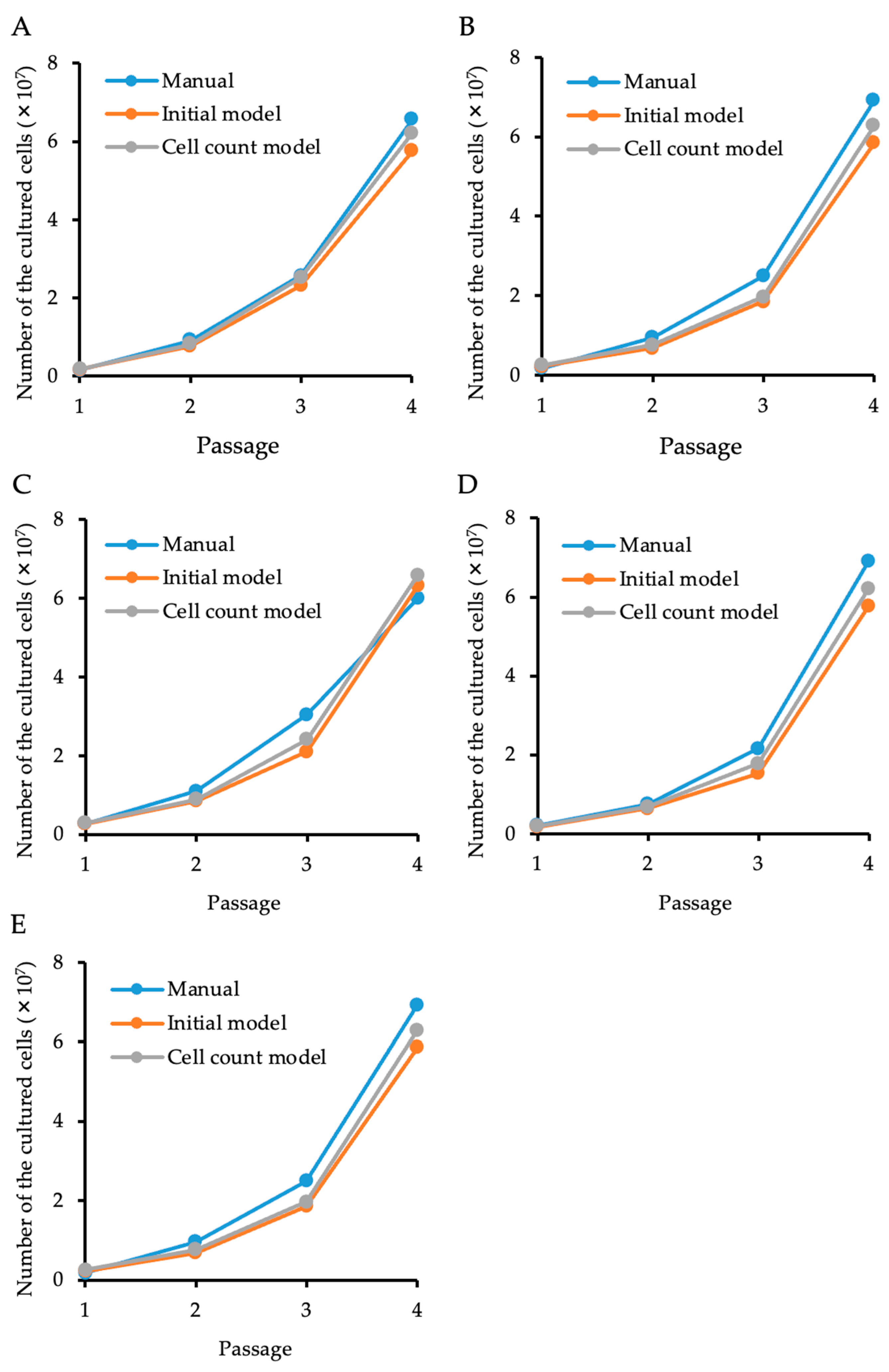
2.6. Feasibility of Using the “Cell Count Model” to Predict Total Cell Numbers in Cultures
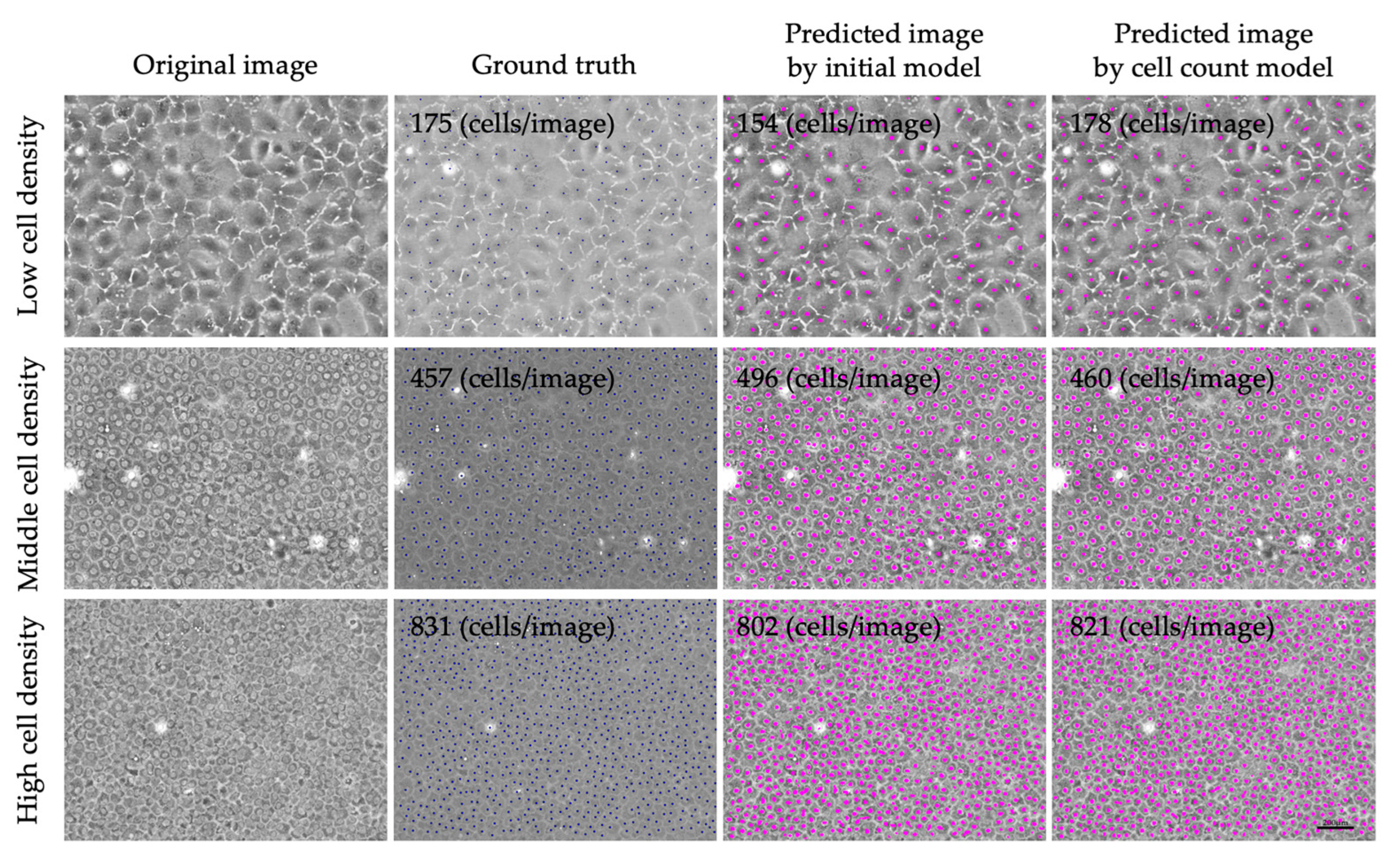
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chaurasia, S.; Mohamed, A.; Garg, P.; Balasubramanian, D.; Rao, G.N. Thirty years of eye bank experience at a single centre in India. Int. Ophthalmol. 2020, 40, 81–88. [Google Scholar] [CrossRef]
- Gao, H.; Huang, T.; Pan, Z.; Wu, J.; Xu, J.; Hong, J.; Chen, W.; Wu, H.; Kang, Q.; Zhu, L.; et al. Survey report on keratoplasty in China: A 5-year review from 2014 to 2018. PLoS ONE 2020, 15, e0239939. [Google Scholar] [CrossRef]
- Eye Bank Association of America. Eye Banking Statistical Report; Eye Bank Association of America: Washington, DC, USA, 2022. [Google Scholar]
- Flockerzi, E.; Turner, C.; Seitz, B.; GeKeR Study Group Collaborators, GeKeR Study Group. Descemet’s membrane endothelial keratoplasty is the predominant keratoplasty procedure in Germany since 2016: A report of the DOG-section cornea and its keratoplasty registry. Br. J. Ophthalmol. 2023, 16, bjo-2022-323162. [Google Scholar] [CrossRef]
- Quilendrino, R.; Rodriguez-Calvo de Mora, M.; Baydoun, L.; Ham, L.; van Dijk, K.; Dapena, I.; Oellerich, S.; Melles, G.R.J. Prevention and management of Descemet membrane endothelial keratoplasty complications. Cornea 2017, 36, 1089–1095. [Google Scholar] [CrossRef]
- Berrospi, R.D.; Galvis, V.; Bhogal, M.; Tello, A. Double- line reflection pattern as a simple method to determine graft orientation of Descemet membrane endothelial keratoplasty. Cornea 2019, 38, 768–771. [Google Scholar] [CrossRef]
- Karadag, R.; Aykut, V.; Esen, F.; Oguz, H.; Demirok, A. Descemet’s membrane endothelial keratoplasty in aphakic and vitrectomized eye. GMS Ophthalmol. Cases 2020, 10, Doc02. [Google Scholar] [CrossRef]
- Aravena, C.; Yu, F.; Deng, S.X. Outcomes of Descemet membrane endothelial keratoplasty in patients with previous glaucoma surgery. Cornea 2017, 36, 284–289. [Google Scholar] [CrossRef]
- Gain, P.; Jullienne, R.; He, Z.; Aldossary, M.; Acquart, S.; Cognasse, F.; Thuret, G. Global survey of corneal transplantation and eye banking. JAMA Ophthalmol. 2016, 134, 167–173. [Google Scholar] [CrossRef]
- Okumura, N.; Koizumi, N. Regeneration of the corneal endothelium. Curr. Eye Res. 2020, 45, 303–312. [Google Scholar] [CrossRef]
- Rodriguez-Fernandez, S.; Pineiro-Ramil, M.; Castro-Vinuelas, R.; Sanjurjo-Rodriguez, C.; Alvarez-Portela, M.; Fuentes-Boquete, I.M.; Rendal-Vazquez, E.; Silvia, M.D.-P. Current development of alternative treatments for endothelial decompensation: Cell-based therapy. Exp. Eye Res. 2021, 207, 108560. [Google Scholar] [CrossRef]
- Soh, Y.Q.; Poh, S.S.J.; Peh, G.S.L.; Mehta, J.S. New therapies for corneal endothelial diseases: 2020 and beyond. Cornea 2021, 40, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Catala, P.; Thuret, G.; Skottman, H.; Mehta, J.S.; Parekh, M.; Ni Dhubhghaill, S.; Collin, R.W.J.; Nuijts, R.; Ferrari, S.; LaPointe, V.L.S.; et al. Approaches for corneal endothelium regenerative medicine. Prog. Retin. Eye Res. 2022, 87, 100987. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.N.; Mehta, J.S. Cell therapy in corneal endothelial disease. Curr. Opin. Ophthalmol. 2022, 33, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Hatou, S.; Shimmura, S. Advances in corneal regenerative medicine with iPS cells. Jpn. J. Ophthalmol. 2023, 67, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, S.; Koizumi, N.; Ueno, M.; Okumura, N.; Imai, K.; Tanaka, H.; Yamamoto, Y.; Nakamura, T.; Inatomi, T.; Bush, J.; et al. Injection of cultured cells with a ROCK inhibitor for bullous keratopathy. N. Engl. J. Med. 2018, 378, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Numa, K.; Imai, K.; Ueno, M.; Kitazawa, K.; Tanaka, H.; Bush, J.D.; Teramukai, S.; Okumura, N.; Koizumi, N.; Hamuro, J.; et al. Five-year follow-up of first 11 patients undergoing injection of cultured corneal endothelial cells for corneal endothelial failure. Ophthalmology 2021, 128, 504–514. [Google Scholar] [CrossRef]
- Nakahara, M.; Okumura, N.; Nakano, S.; Koizumi, N. Effect of a p38 mitogen-activated protein kinase inhibitor on corneal endothelial cell proliferation. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4218–4227. [Google Scholar] [CrossRef]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted Boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Chapelle, O.; Scholkopf, B.; Zien, A. Semi-supervised learning (Chapelle, O. et al., Eds.; 2006) [book reviews]. IEEE Trans. Neural Netw. 2009, 20, 542. [Google Scholar] [CrossRef]
- Eckardt, J.N.; Bornhauser, M.; Wendt, K.; Middeke, J.M. Semi-supervised learning in cancer diagnostics. Front. Oncol. 2022, 12, 960984. [Google Scholar] [CrossRef]
- Mvula, P.K.; Branco, P.; Jourdan, G.V.; Viktor, H.L. A systematic literature review of cyber-security data repositories and performance assessment metrics for semi-supervised learning. Discov. Data 2023, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhang, B. Semi-supervised learning improves gene expression-based prediction of cancer recurrence. Bioinformatics 2011, 27, 3017–3023. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Ahn, J.; Kim, H.; Park, S. Integrative gene network construction to analyze cancer recurrence using semi-supervised learning. PLoS ONE 2014, 9, e86309. [Google Scholar] [CrossRef]
- Hassanzadeh, H.R.; Phan, J.H.; Wang, M.D. A semi-supervised method for predicting cancer survival using incomplete clinical data. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; Volume 2015, pp. 210–213. [Google Scholar] [CrossRef]
- Chai, H.; Li, Z.N.; Meng, D.Y.; Xia, L.Y.; Liang, Y. A new semi-supervised learning model combined with Cox and SP-AFT models in cancer survival analysis. Sci. Rep. 2017, 7, 13053. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Zhang, A. Affinity network fusion and semi-supervised learning for cancer patient clustering. Methods 2018, 145, 16–24. [Google Scholar] [CrossRef]
- Cristovao, F.; Cascianelli, S.; Canakoglu, A.; Carman, M.; Nanni, L.; Pinoli, P.; Masseroli, M. Investigating deep learning based breast cancer subtyping using pan-cancer and multi-omic data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Volume 9351. [Google Scholar] [CrossRef]
- Fabijanska, A. Segmentation of corneal endothelium images using a U-Net-based convolutional neural network. Artif. Intell. Med. 2018, 88, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kolluru, C.; Benetz, B.A.; Joseph, N.; Menegay, H.J.; Lass, J.H.; Wilson, D. Machine learning for segmenting cells in corneal endothelium images. Proc. SPIE Int. Soc. Opt. Eng. 2019, 10950, 1126–1135. [Google Scholar] [CrossRef]
- Daniel, M.C.; Atzrodt, L.; Bucher, F.; Wacker, K.; Böhringer, S.; Reinhard, T.; Böhringer, D. Automated segmentation of the corneal endothelium in a large set of ‘real-world’specular microscopy images using the U-Net architecture. Sci. Rep. 2019, 9, 4752. [Google Scholar] [CrossRef]
- Vigueras-Guillen, J.P.; Sari, B.; Goes, S.F.; Lemij, H.G.; van Rooij, J.; Vermeer, K.A.; van Vliet, L.J. Fully convolutional architecture vs sliding-window CNN for corneal endothelium cell segmentation. BMC Biomed. Eng. 2019, 1, 4. [Google Scholar] [CrossRef]
- Vigueras-Guillén, J.P.; van Rooij, J.; Engel, A.; Lemij, H.G.; van Vliet, L.J.; Vermeer, K.A. Deep learning for assessing the corneal endothelium from specular microscopy images up to 1 year after ultrathin-DSAEK surgery. Transl. Vis. Sci. Technol. 2020, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.-W.; Wu, J. Unet 3+: A full-scale connected U-Net for medical image segmentation. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Wang, J.; Sun, K.; Cheng, T.; Jiang, B.; Deng, C.; Zhao, Y.; Liu, D.; Mu, Y.; Tan, M.; Wang, X.; et al. Deep high-resolution representation learning for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 3349–3364. [Google Scholar] [CrossRef] [PubMed]
- Okumura, N.; Yamada, S.; Nishikawa, T.; Narimoto, K.; Okamura, K.; Izumi, A.; Hiwa, S.; Hiroyasu, T.; Koizumi, N. U-Net convolutional neural network for segmenting the corneal endothelium in a mouse model of Fuchs endothelial corneal dystrophy. Cornea 2022, 41, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Peh, G.S.; Adnan, K.; George, B.L.; Ang, H.P.; Seah, X.Y.; Tan, D.T.; Mehta, J.S. The effects of Rho-associated kinase inhibitor Y-27632 on primary human corneal endothelial cells propagated using a dual media approach. Sci. Rep. 2015, 5, 9167. [Google Scholar] [CrossRef]
- Wieben, E.D.; Aleff, R.A.; Rinkoski, T.A.; Baratz, K.H.; Basu, S.; Patel, S.V.; Maguire, L.J.; Fautsch, M.P. Comparison of TCF4 repeat expansion length in corneal endothelium and leukocytes of patients with Fuchs endothelial corneal dystrophy. PLoS ONE 2021, 16, e0260837. [Google Scholar] [CrossRef]
- Azuma, K. Regulatory landscape of regenerative medicine in Japan. Curr. Stem Cell Rep. 2015, 1, 118–128. [Google Scholar] [CrossRef]
- Qiu, T.; Hanna, E.; Dabbous, M.; Borislav, B.; Toumi, M. Regenerative medicine regulatory policies: A systematic review and international comparison. Health Policy 2020, 124, 701–713. [Google Scholar] [CrossRef]

| Precision (%) (95% CI) | Recall (%) (95% CI) | F-Value (%) (95% CI) | |
|---|---|---|---|
| Initial model | 95.4 (94.4–96.3) | 89.3 (87.5–91.0) | 91.9 (90.9–92.9) |
| Cell count model | 95.1 (94.1–96.1) | 92.3 (90.8–93.8) | 93.4 (92.6–94.2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okumura, N.; Nishikawa, T.; Imafuku, C.; Matsuoka, Y.; Miyawaki, Y.; Kadowaki, S.; Nakahara, M.; Matsuoka, Y.; Koizumi, N. U-Net Convolutional Neural Network for Real-Time Prediction of the Number of Cultured Corneal Endothelial Cells for Cellular Therapy. Bioengineering 2024, 11, 71. https://doi.org/10.3390/bioengineering11010071
Okumura N, Nishikawa T, Imafuku C, Matsuoka Y, Miyawaki Y, Kadowaki S, Nakahara M, Matsuoka Y, Koizumi N. U-Net Convolutional Neural Network for Real-Time Prediction of the Number of Cultured Corneal Endothelial Cells for Cellular Therapy. Bioengineering. 2024; 11(1):71. https://doi.org/10.3390/bioengineering11010071
Chicago/Turabian StyleOkumura, Naoki, Takeru Nishikawa, Chiaki Imafuku, Yuki Matsuoka, Yuna Miyawaki, Shinichi Kadowaki, Makiko Nakahara, Yasushi Matsuoka, and Noriko Koizumi. 2024. "U-Net Convolutional Neural Network for Real-Time Prediction of the Number of Cultured Corneal Endothelial Cells for Cellular Therapy" Bioengineering 11, no. 1: 71. https://doi.org/10.3390/bioengineering11010071
APA StyleOkumura, N., Nishikawa, T., Imafuku, C., Matsuoka, Y., Miyawaki, Y., Kadowaki, S., Nakahara, M., Matsuoka, Y., & Koizumi, N. (2024). U-Net Convolutional Neural Network for Real-Time Prediction of the Number of Cultured Corneal Endothelial Cells for Cellular Therapy. Bioengineering, 11(1), 71. https://doi.org/10.3390/bioengineering11010071