An Approach to Binary Classification of Alzheimer’s Disease Using LSTM
Abstract
1. Introduction
- It might be challenging to recall proper words or names.
- Daily work in social settings or an office might be difficult.
- Getting in trouble remembering people’s names while meeting them.
- Having difficulty locating or misplacing a valuable asset.
- Forgetting something you just heard or read.
- It might become much more difficult to plan and manage various daily tasks.
- Objective 1:
- Develop a robust and accurate diagnostic LSTM approach and utilize its capacity for time-series MRI data analysis.
- Objective 2:
- Investigate how well the suggested LSTM-based model performs in categorizing MRI data and making accurate predictions for the early identification of AD.
- Objective 3:
- To offer a thorough examination of the model’s diagnostic skills and evaluate the model’s efficacy using a wide range of measures, such as accuracy, confusion matrix, and AUC ROC.
- Objective 4:
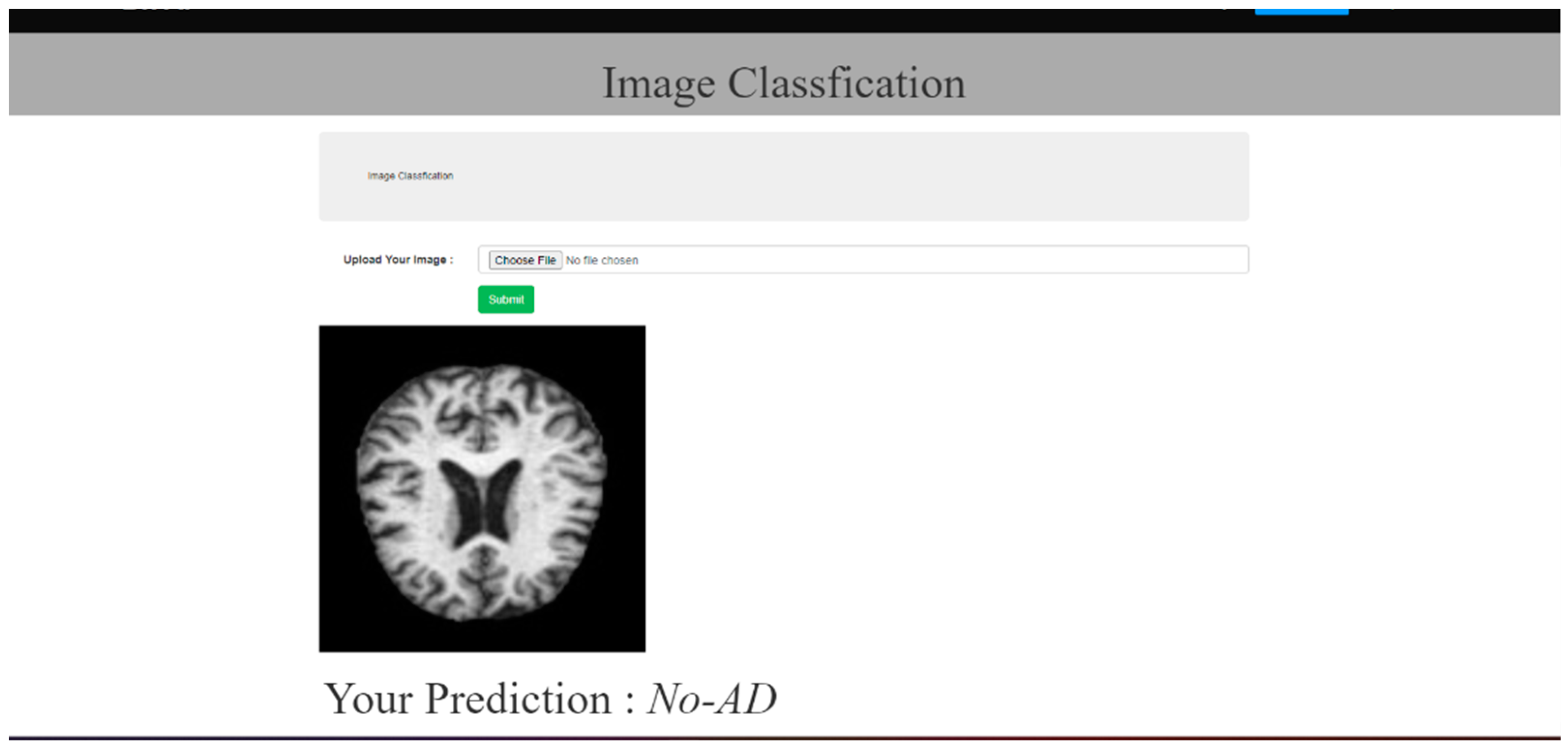
- Create a user-friendly Web-based application to aid in the practical deployment of the generated model.
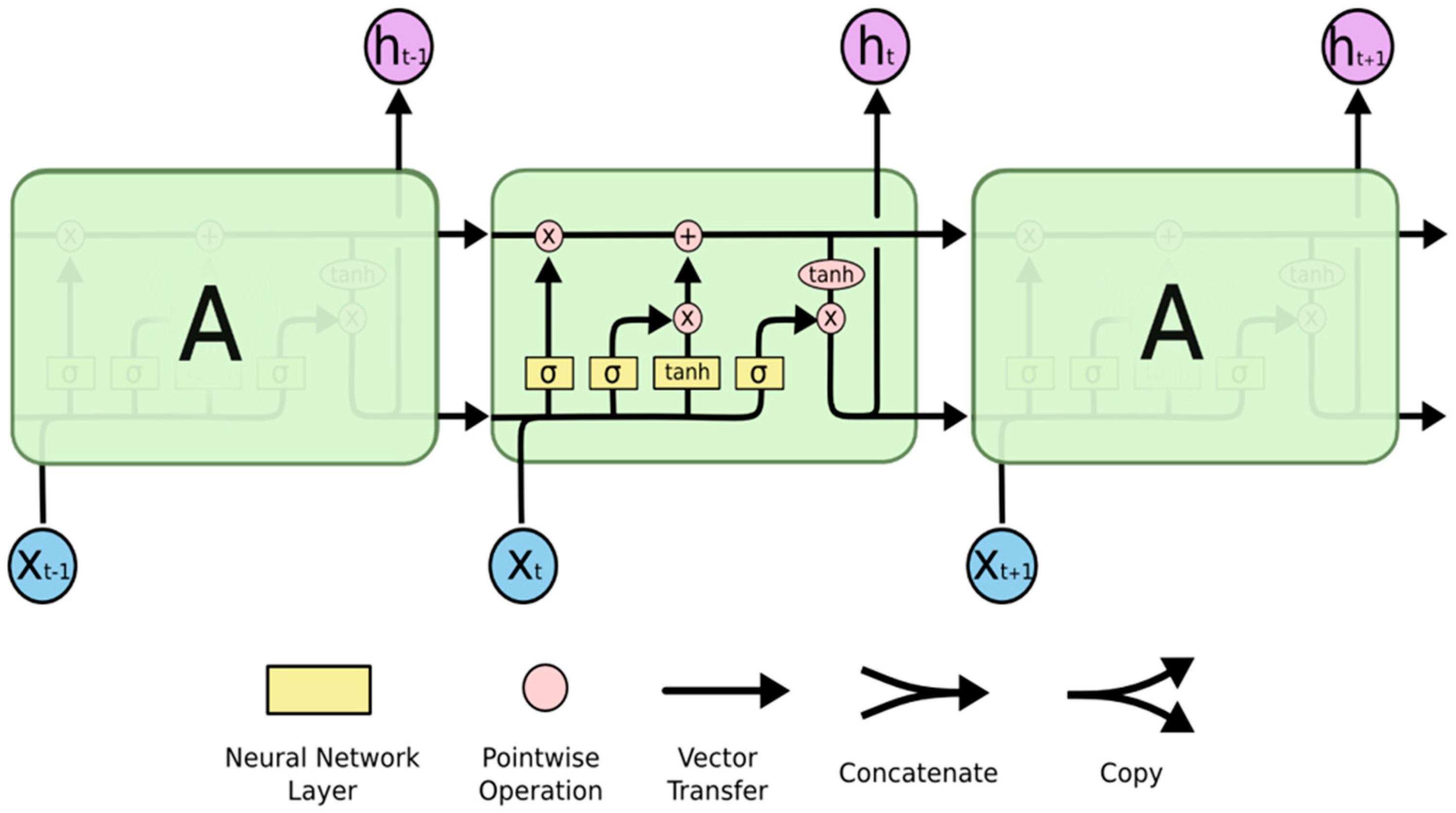
2. Overview of Deep Learning and LSTM Model
- Forget Gate—The input and previous output are combined at the forget gate to produce and generate an output that falls within the range of 0 to 1, and a sigmoid activation function is employed, which indicates how much of the previous timestamp has to be kept or forgotten. The previous state is multiplied by this output after that [22].
- Input Gate—Although the input gate’s output ranges from 0 to 1, it operates with the same signals as the forget gate. The goal here is to choose which new information will be added to the LSTM’s state [18]. The additional values that were to be added to the initial state are then created by multiplying them by the output of the tan h block. The current state is created by adding this gated vector to the previous state.
- Output Gate—The output gate will gate the input and previous state as before to produce an output that ranges from 0 to 1, which is then combined with the output of the tan h block to obtain the current state. The result is then distributed [22]. The LSTM block receives input from both the output and the state.
Importance of Imaging Modality for AD Detection
- Image analysis: AI algorithms can be used to analyze MRI images and detect anomalies or patterns in including. allowing for earlier and more accurate diagnoses.
- Image reconstruction: AI can be used to enhance MR picture quality by reducing noise and boosting resolution.
- Image segmentation: Radiologists can more easily identify particular areas of interest by using AI to segment MRI pictures into various tissues or structures.
- Dose reduction: AI can be used to optimize MRI protocols to reduce the amount of radiation exposure for patients, making the imaging process safer [25].
- Personalization: AI can be used to personalize MRI scans for individual patients based on factors such as body shape, size, and medical history.
3. Related Works
- Temporal information—LSTMs have been used in multiple research projects to incorporate temporal information into the prediction model, which can help us understand Alzheimer’s disease and its progression better. LSTMs are particularly well-suited for studying time-series data [26].
- Improved accuracy—CNNs and transfer learning have been shown in multiple studies to attain a high level of accuracy when it comes to the prediction of Alzheimer’s disease, in contrast to more traditional techniques of machine learning [27]. This could lead to earlier, more accurate diagnoses and better patient outcomes [27].
- Transfer learning—Transfer learning has been utilized in some research to make use of CNN and LSTM models that have already been trained on massive amounts of data and can then be tailored to particular tasks and data sets. This has the benefit of requiring less data and processing during training, which can increase prediction accuracy [28].
- Integration of multi-modal data—Several studies have combined the results of MRI scans with those of other clinical assessments, such as cognitive evaluations or genetic information. This has resulted in a more in-depth and precise understanding of Alzheimer’s disease and how it progresses [29].
4. Materials and Methods
4.1. Data Description
4.2. Proposed Method
- Image Preprocessing—The CV2 library was utilized for image preprocessing. The images were resized, and labels were appended to them.
- Normalization—The image data were normalized by dividing them by 255.0 to scale the pixel values between 0 and 1.
- Categorical Target—The np_utils module from Keras was employed to convert the target labels into categorical format.
- Model Building—The TensorFlow and Keras frameworks were utilized. The necessary modules were imported to construct the model. The model consisted of multiple layers, with the CuDNNLSTM layer serving as the backbone. The layers were added sequentially, employing the he_uniform kernel initializer and setting the return_sequences attribute to true. Dropout layers with a rate of 0.2 were added between each layer to avoid overfitting. The ReLU activation function was applied to the layers. For the final layer, a sigmoid activation function was used since it was a binary classification task.
- Stratified Shuffle-Split Cross-Validation—The Stratified Shuffle-Split Cross-Validation technique was employed with 5 splits and a test size of 0.1. This technique ensured the preservation of the class distribution in the training and testing datasets.
- Model Compilation—The model was compiled with parameters such as a loss function set to binary_crossentropy, metrics for evaluation, and an optimizer. The optimizer chosen was SGD (Stochastic Gradient Descent), and a grid search approach was used to find the best optimal values by trying different learning rates and momentums.
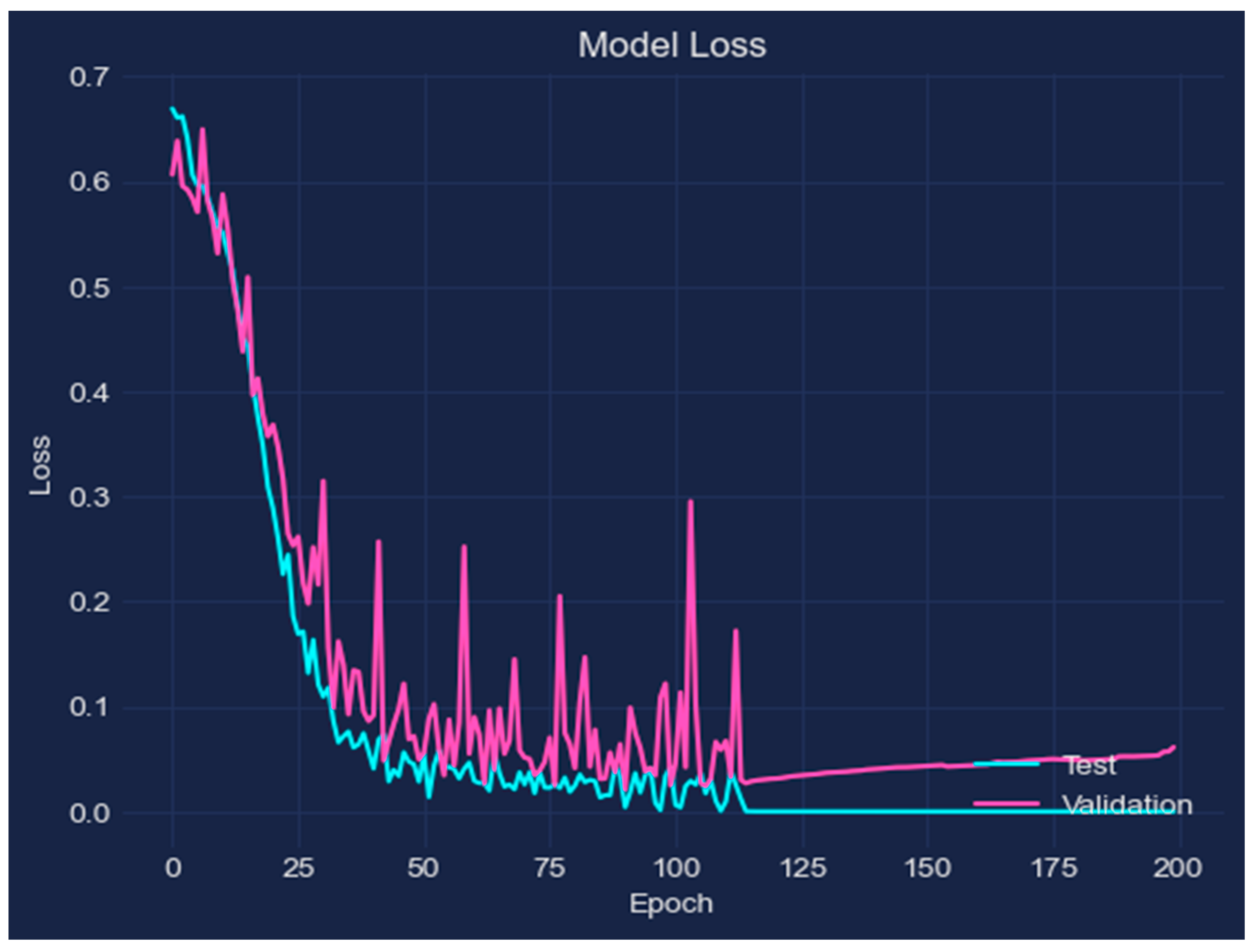
- Early Stopping—Early stopping was put into place to terminate training as soon as the model’s performance on the validation set began to deteriorate to avoid overfitting.
- Best Fold Selection—The best fold (score) was determined using the arg max function, and the corresponding best accuracy was obtained.
5. Experiment and Evaluation
5.1. Setup for Experiment
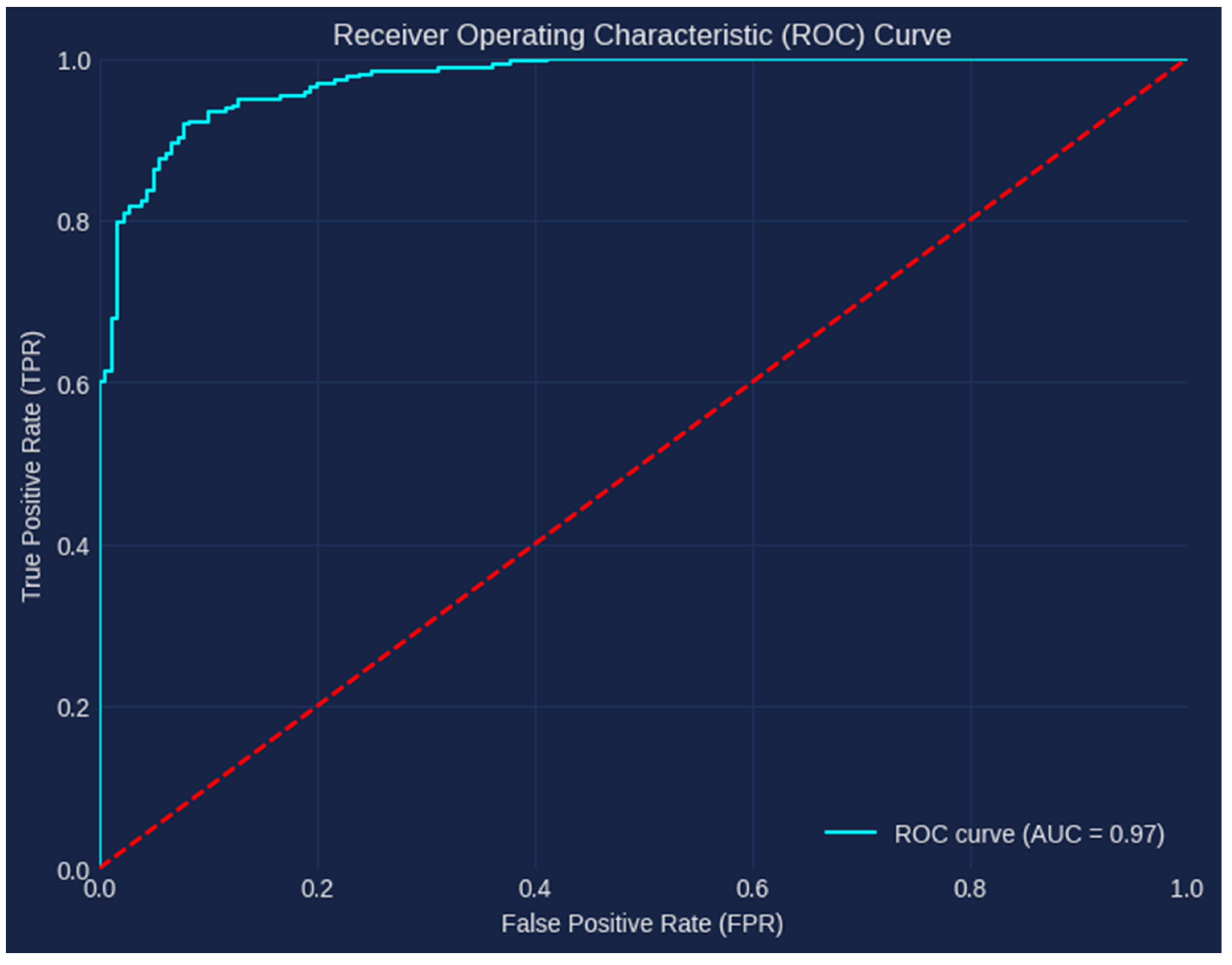
5.2. Results and Discussion
6. Exploratory Data Analysis
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nanni, L.; Salvatore, C.; Cerasa, A.; Castiglioni, I. Combining multiple approaches for the early diagnosis of Alzheimer’s Disease. Pattern Recognit. Lett. 2016, 84, 259–266. [Google Scholar] [CrossRef]
- Alzheimer’s Association. 2016 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2016, 12, 459–509. [Google Scholar] [CrossRef]
- Kavitha, C.; Mani, V.; Srividhya, S.R.; Khalaf, O.I.; Romero, C.A.T. Early-Stage Alzheimer’s Disease Prediction Using Machine Learning Models. Front. Public Health 2022, 10, 853294. [Google Scholar] [CrossRef]
- Zeng, H.; Qi, Y.; Zhang, Z.; Liu, C.; Peng, W.; Zhang, Y. Nanomaterials toward the treatment of Alzheimer’s disease: Recent advances and future trends. Chin. Chem. Lett. 2021, 32, 1857–1868. [Google Scholar] [CrossRef]
- Khan, P.; Kader, F.; Islam, S.M.R.; Rahman, A.B.; Kamal, S.; Toha, M.U.; Kwak, K.-S. Machine Learning and Deep Learning Approaches for Brain Disease Diagnosis: Principles and Recent Advances. IEEE Access 2021, 9, 37622–37655. [Google Scholar] [CrossRef]
- Jack, C.R.; Holtzman, D.M. Biomarker Modeling of Alzheimer’s Disease. Neuron 2013, 80, 1347–1358. [Google Scholar] [CrossRef]
- Frisoni, G.B.; Fox, N.C.; Jack, C.R.; Scheltens, P.; Thompson, P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 67–77. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, L.; Zheng, W.; Yin, L.; Hu, R.; Yang, B. Endoscope image mosaic based on pyramid ORB. Biomed. Signal Process. Control 2022, 71, 103261. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, X.; Yang, B.; Yin, Z.; Liu, S.; Yin, L.; Zheng, W. Three-Dimensional Modeling of Heart Soft Tissue Motion. Appl. Sci. 2023, 13, 2493. [Google Scholar] [CrossRef]
- Cui, R.; Liu, M.; Li, G. Longitudinal analysis for Alzheimer’s disease diagnosis using RNN. In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1398–1401. [Google Scholar]
- Afzal, S.; Maqsood, M.; Nazir, F.; Khan, U.; Aadil, F.; Awan, K.M.; Mehmood, I.; Song, O.-Y. A Data Augmentation-Based Framework to Handle Class Imbalance Problem for Alzheimer’s Stage Detection. IEEE Access 2019, 7, 115528–115539. [Google Scholar] [CrossRef]
- Tomassini, S.; Falcionelli, N.; Sernani, P.; Muller, H.; Dragoni, A.F. An End-to-End 3D ConvLSTM-based Framework for Early Diagnosis of Alzheimer’s Disease from Full-Resolution Whole-Brain sMRI Scans. In Proceedings of the IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 74–78. [Google Scholar]
- Luo, S.; Li, X.; Li, J. Automatic Alzheimer’s Disease Recognition from MRI Data Using Deep Learning Method. J. Appl. Math. Phys. 2017, 5, 1892–1898. [Google Scholar] [CrossRef]
- Islam, Z.; Islam, M.; Asraf, A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform. Med. Unlocked 2020, 20, 100412. [Google Scholar] [CrossRef]
- Sherstinsky, A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) Network. Phys. D Nonlinear Phenom. 2020, 404, 132306. [Google Scholar] [CrossRef]
- Wu, M.; Chen, L. Image recognition based on deep learning. In Proceedings of the 2015 Chinese Automation Congress (CAC), Wuhan, China, 27–29 November 2015; pp. 542–546. [Google Scholar]
- Torfi, A.; Shirvani, R.A.; Keneshloo, Y.; Tavaf, N.; Fox, E.A. Natural Language Processing Advancements by Deep Learning: A Survey. arXiv 2020, arXiv:2003.01200. [Google Scholar]
- Lu, S.; Yang, J.; Yang, B.; Yin, Z.; Liu, M.; Yin, L.; Zheng, W. Analysis and Design of Surgical Instrument Localization Algorithm. Comput. Model. Eng. Sci. 2023, 137, 669–685. [Google Scholar] [CrossRef]
- Wang, F.; Wang, H.; Zhou, X.; Fu, R. A Driving Fatigue Feature Detection Method Based on Multifractal Theory. IEEE Sens. J. 2022, 22, 19046–19059. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Luo, S.; Chiong, R. Deep sequence modelling for Alzheimer’s disease detection using MRI. Comput. Biol. Med. 2021, 134, 104537. [Google Scholar] [CrossRef]
- Understanding LSTM Networks. 2015. Available online: https://colah.github.io/posts/2015-08-Understanding-LSTMs (accessed on 2 February 2023).
- Long Short Term Memory Networks Explanation. 2022. Available online: https://www.geeksforgeeks.org/long-short-term-memory-networks-explanation/ (accessed on 2 March 2023).
- Ekong, F.; Yu, Y.; Patamia, R.A.; Feng, X.; Tang, Q.; Mazumder, P.; Cai, J. Bayesian Depth-Wise Convolutional Neural Network Design for Brain Tumor MRI Classification. Diagnostics 2022, 12, 1657. [Google Scholar] [CrossRef]
- Ebrahimi-Ghahnavieh, A.; Luo, S.; Chiong, R. Transfer Learning for Alzheimer’s Disease Detection on MRI Images. In Proceedings of the IEEE International Conference on Industry 4.0, Artificial Intelligence, and Communications Technology (IAICT), Bali, Indonesia, 1–3 July 2019; pp. 133–138. [Google Scholar]
- Dinesh, T.; Kate, H. Previously Undetected Heart Injury in Patients Recovered from Mild COVID-19. 2020. Available online: https://uhnfoundation.ca/stories/previously-undetected-heart-injury-in-patients-recovered-from-mild-covid-19/ (accessed on 2 June 2023).
- Hong, X.; Lin, R.; Yang, C.; Zeng, N.; Cai, C.; Gou, J.; Yang, J. Predicting Alzheimer’s Disease Using LSTM. IEEE Access 2019, 7, 80893–80901. [Google Scholar] [CrossRef]
- Currie, G.; Hawk, K.E.; Rohren, E.; Vial, A.; Klein, R. Machine Learning and Deep Learning in Medical Imaging: Intelligent Imaging. J. Med. Imaging Radiat. Sci. 2019, 50, 477–487. [Google Scholar] [CrossRef]
- Hon, M.; Khan, N.M. Towards Alzheimer’s disease classification through transfer learning. In Proceeding of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 1166–1169. [Google Scholar]
- Jo, T.; Nho, K.; Saykin, A.J. Deep Learning in Alzheimer’s Disease: Diagnostic Classification and Prognostic Prediction Using Neuroimaging Data. Front. Aging Neurosci. 2019, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Liu, M. RNN-based longitudinal analysis for diagnosis of Alzheimer’s disease. Comput. Med. Imaging Graph. 2019, 73, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dua, M.; Makhija, D.; Manasa, P.Y.L.; Mishra, P. A CNN–RNN–LSTM Based Amalgamation for Alzheimer’s Disease Detection. J. Med. Biol. Eng. 2020, 40, 688–706. [Google Scholar] [CrossRef]
- Aqeel, A.; Hassan, A.; Khan, M.A.; Rehman, S.; Tariq, U.; Kadry, S.; Majumdar, A.; Thinnukool, O. A Long Short-Term Memory Biomarker-Based Prediction Framework for Alzheimer’s Disease. Sensors 2022, 22, 1475. [Google Scholar] [CrossRef] [PubMed]
- Alsaade, F.W.; Alzahrani, M.S. Classification and Detection of Autism Spectrum Disorder Based on Deep Learning Algorithms. Comput. Intell. Neurosci. 2022, 2022, 8709145. [Google Scholar] [CrossRef]
- Ke, F.; Choi, S.J.; Kang, Y.H.; Cheon, K.-A.; Lee, S.W. Exploring the Structural and Strategic Bases of Autism Spectrum Disorders With Deep Learning. IEEE Access 2020, 8, 153341–153352. [Google Scholar] [CrossRef]
- Shoeibi, A.; Ghassemi, N.; Khodatars, M.; Moridian, P.; Khosravi, A.; Zare, A.; Gorriz, J.M.; Chale-Chale, A.H.; Khadem, A.; Acharya, U.R. Automatic diagnosis of schizophrenia and attention deficit hyperactivity disorder in rs-fMRI modality using convolutional autoencoder model and interval type-2 fuzzy regression. Cogn. Neurodyn. 2022, 1–23. [Google Scholar] [CrossRef]
- Shoeibi, A.; Khodatars, M.; Jafari, M.; Ghassemi, N.; Moridian, P.; Alizadehsani, R.; Ling, S.H.; Khosravi, A.; Alinejad-Rokny, H.; Lam, H.; et al. Diagnosis of brain diseases in fusion of neuroimaging modalities using deep learning: A review. Inf. Fusion 2023, 93, 85–117. [Google Scholar] [CrossRef]
- Gunduz, H. Deep Learning-Based Parkinson’s Disease Classification Using Vocal Feature Sets. IEEE Access 2019, 7, 115540–115551. [Google Scholar] [CrossRef]
- Ullah, I.; Hussain, M.; Qazi, E.-U.; Aboalsamh, H. An automated system for epilepsy detection using EEG brain signals based on deep learning approach. Expert Syst. Appl. 2018, 107, 61–71. [Google Scholar] [CrossRef]
- Tandel, G.S.; Biswas, M.; Kakde, O.G.; Tiwari, A.; Suri, H.S.; Turk, M.; Laird, J.R.; Asare, C.K.; Ankrah, A.A.; Khanna, N.N.; et al. A Review on a Deep Learning Perspective in Brain Cancer Classification. Cancers 2019, 11, 111. [Google Scholar] [CrossRef]
- Dipu, N.M.; Alam Shohan, S.; Salam, K.M.A. Deep Learning Based Brain Tumor Detection and Classification. In Proceedings of the 2021 International Conference on Intelligent Technologies (CONIT), Hubli, India, 25–27 June 2021; pp. 1–6. [Google Scholar]
- Sourabh Shastri Sachin Kumar. MRI Preprocessed Dataset. 2022. Available online: https://www.kaggle.com/dsv/3364939 (accessed on 10 March 2023).
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Buda, M.; Saha, A.; Bashir, M.R. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J. Magn. Reson. Imaging 2018, 49, 939–954. [Google Scholar] [CrossRef]
- Judith, A.M.; Priya, S.B.; Mahendran, R.K.; Gadekallu, T.R.; Ambati, L.S. Two-phase classification: ANN and A-SVM classifiers on motor imagery. Asian J. Control 2020. [Google Scholar] [CrossRef]
- Saab, S., Jr.; Fu, Y.; Ray, A.; Hauser, M. A dynamically stabilized recurrent neural network. Neural Process. Lett. 2022, 54, 1195–1209. [Google Scholar] [CrossRef]
- Saab, S., Jr.; Saab, K.; Phoha, S.; Zhu, M.; Ray, A. A multivariate adaptive gradient algorithm with reduced tuning efforts. Neural Netw. 2022, 152, 499–509. [Google Scholar] [CrossRef]
- Badica, C.; Mangioni, G.; Rahimi, S. Intelligent distributed information systems. Inf. Sci. 2010, 180, 1779–1780. [Google Scholar] [CrossRef]
- Lian, Z.; Zeng, Q.; Wang, W.; Gadekallu, T.R.; Su, C. Blockchain-Based Two-Stage Federated Learning With Non-IID Data in IoMT System. IEEE Trans. Comput. Soc. Syst. 2022, 1–10. [Google Scholar] [CrossRef]
- Kumar, A.; Krishnamurthi, R.; Nayyar, A.; Sharma, K.; Grover, V.; Hossain, E. A novel smart healthcare design, simulation, and implementation using healthcare 4.0 processes. IEEE Access 2020, 8, 118433–118471. [Google Scholar] [CrossRef]
- Srikanth, P.; Kumar, A.; Hedabou, M. An Uncertainty Trust Assessment Scheme for Trustworthy Partner Selection in Online Games. IEEE Access 2022, 10, 132232–132249. [Google Scholar] [CrossRef]








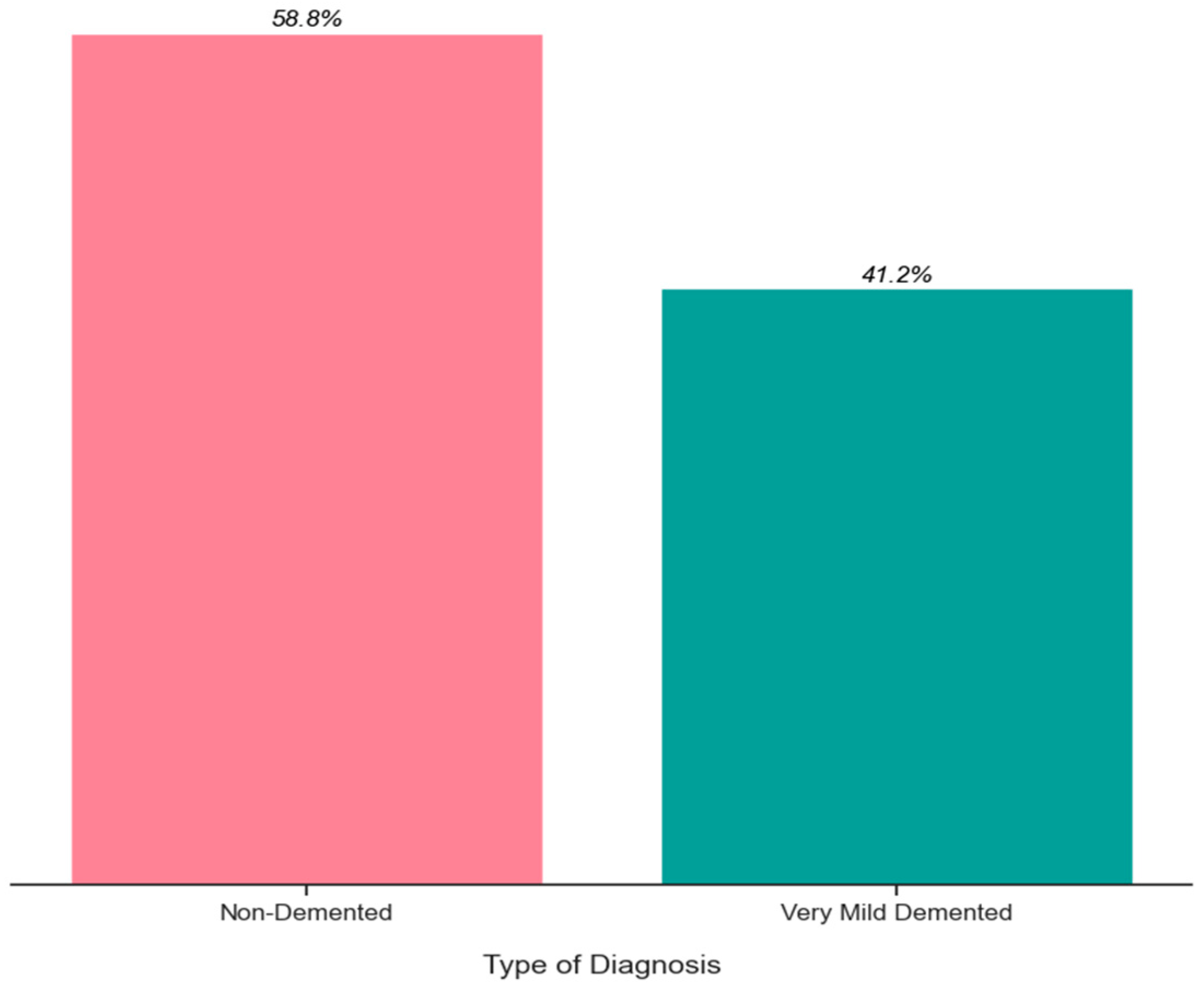

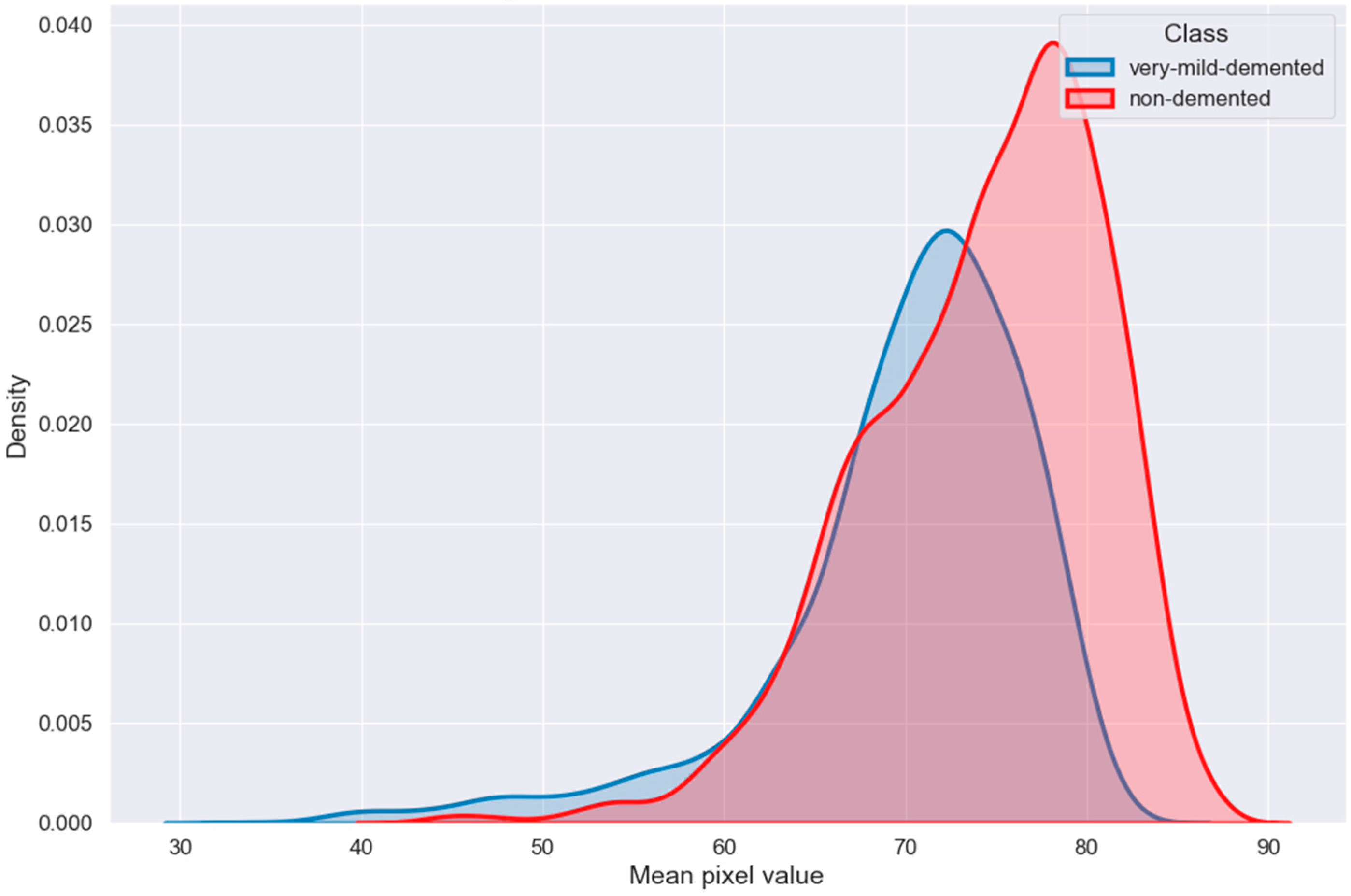

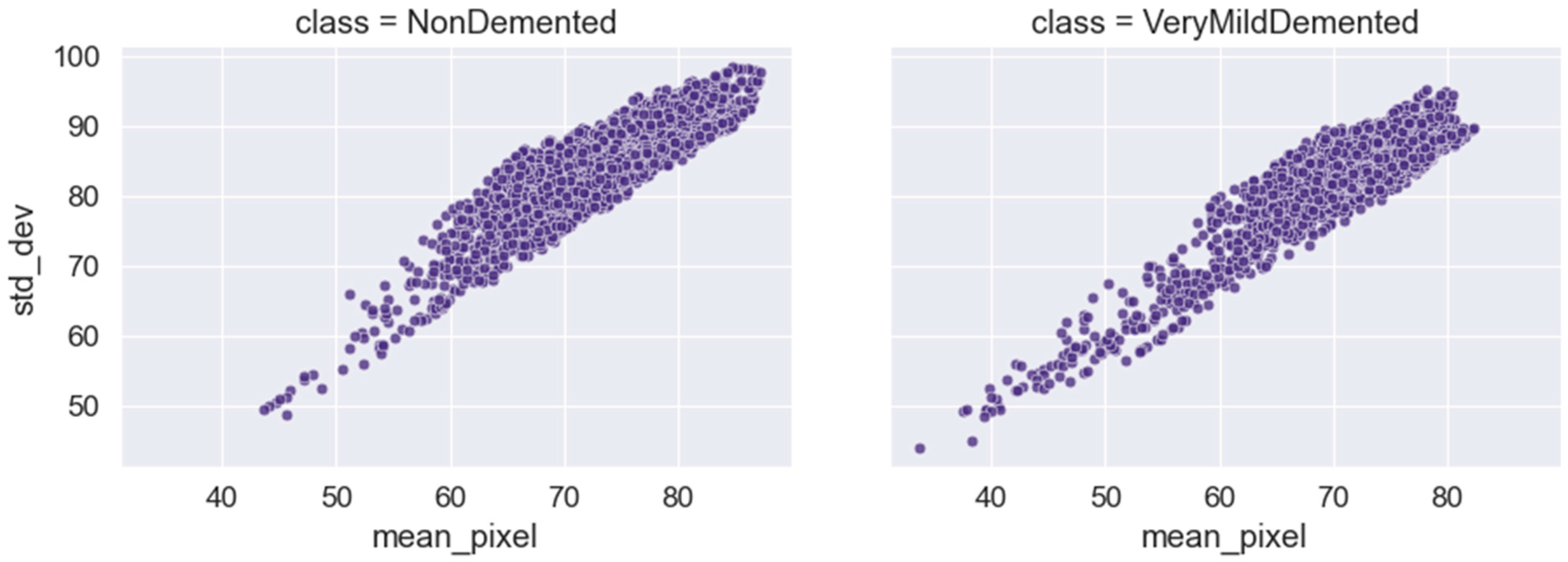
| Ref. | Approach | Methods | Dataset | Result |
|---|---|---|---|---|
| [10] 2018 | Longitudinal analysis for AD diagnosis with RNN | MLP is first developed to learn the spatial characteristics of MR images to classify AD. The MLP outputs are then used to train an RNN with two cascaded (BGRU) layers, which, by extracting longitudinal information from the imaging data at different time intervals, generates a final categorization predicting score. | ADNI, sMRI T1-weighted 428 subjects, 198 AD patients, and 229 NC | Accuracy: 89.7% |
| [26] 2019 | Predicting Alzheimer’s disease using LSTM | LSTM is used to predict the disease’s future state rather than to categorize it in its current state. | 1105 patients are included in the MRI longitudinal time sequence data from the ADNI | AUC of AD vs. NC: 0.935, mAUC of AD vs. MCI 0.798 and mAUC of AD vs. NC vs. MCI 0.777 |
| [30] 2019 | Longitudinal analysis for diagnosis of AD with RNN | The RNN harvests longitudinal data for AD classification using three cascaded BGRU layers, whereas the CNN learns MR image spatial characteristics for classification. | ADNI 198 AD, 229 NC, and 403 MCI | Accuracy 91.33% (AD vs. NC) and 71.71% (pMCI vs. sMCI) |
| [31] 2020 | Hybrid-model-based amalgamation for AD detection | An ensemble approach that uses a weighted average technique which can be employed to merge these models, including CNN, RNN, and LSTM. | Open Access Series of Imaging Studies (OASIS) OASIS dataset-1 for CNN and OASIS dataset-2 for RNN, LSTM | Ensemble of bagged models accuracy 92.22%, Ensemble of primary models 89.75% |
| [12] 2021 | An end-to-end 3D-ConvLSTM for early detection of AD | With the help of high-resolution whole-brain sMRI data, this project seeks to develop or build a comprehensive 3D ConvLSTM-based framework for the early identification of AD. | OASIS-3 and ADNI 1-Screening | Accuracy: 86%, Specificity: 74%, Sensitivity: 96%, F1-score: 88% and AUC of 93% |
| [32] 2022 | An LSTM biomarker-based prediction for AD | After 6, 12, 21, 18, 24, and 36 months, the model can predict the biomarkers (feature vectors) of a patient. These predicted biomarkers will go through layers of a neural network that are all connected. The NN layers will then decide if these biomarkers belong to a person with AD or MCIx. | ADNI, 805 subjects MRI T1-weighted | Accuracy: 88.24% |
| Class | Training | Testing |
|---|---|---|
| NC | 2560 | 640 |
| AD | 1792 | 448 |
| Total | 4352 | 1088 |
| Parameter | Values |
|---|---|
| Dropout rate | 0.2 |
| Batch size | 32 |
| Cross-validation | Stratified Shuffle-Split (5 splits, 0.1 test size) |
| Activation function | Relu and Sigmoid |
| Accuracy | Metric = Accuracy |
| Loss function | binary_crossentropy |
| Optimizer | SGD (Learning Rate with Momentum) |
| Early stopping | Early Stopping with Epoch = 200 |
| Technique | Dataset | Performance |
|---|---|---|
| Longitudinal analysis for AD diagnosis with RNN | ADNI | Accuracy: 89.7% |
| Predicting Alzheimer’s disease using LSTM | ADNI | AUC of AD vs. NC: 0.935 |
| Longitudinal analysis for diagnosis of AD with RNN | ADNI | Accuracy: 91.33% (AD vs. NC) |
| Hybrid model-based amalgamation for AD detection | OASIS | Accuracy: 92.22% |
| A 3D-ConvLSTM end-to-end for AD early detection | OASIS +ADNI | AUC of 93% |
| An LSTM biomarker-based prediction for AD | ADNI | Accuracy: 88.24% |
| Proposed approach | Kaggle Dataset | Accuracy: 98.62% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salehi, W.; Baglat, P.; Gupta, G.; Khan, S.B.; Almusharraf, A.; Alqahtani, A.; Kumar, A. An Approach to Binary Classification of Alzheimer’s Disease Using LSTM. Bioengineering 2023, 10, 950. https://doi.org/10.3390/bioengineering10080950
Salehi W, Baglat P, Gupta G, Khan SB, Almusharraf A, Alqahtani A, Kumar A. An Approach to Binary Classification of Alzheimer’s Disease Using LSTM. Bioengineering. 2023; 10(8):950. https://doi.org/10.3390/bioengineering10080950
Chicago/Turabian StyleSalehi, Waleed, Preety Baglat, Gaurav Gupta, Surbhi Bhatia Khan, Ahlam Almusharraf, Ali Alqahtani, and Adarsh Kumar. 2023. "An Approach to Binary Classification of Alzheimer’s Disease Using LSTM" Bioengineering 10, no. 8: 950. https://doi.org/10.3390/bioengineering10080950
APA StyleSalehi, W., Baglat, P., Gupta, G., Khan, S. B., Almusharraf, A., Alqahtani, A., & Kumar, A. (2023). An Approach to Binary Classification of Alzheimer’s Disease Using LSTM. Bioengineering, 10(8), 950. https://doi.org/10.3390/bioengineering10080950









