Data-Driven Quantitation of Movement Abnormality after Stroke
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Functional Activity
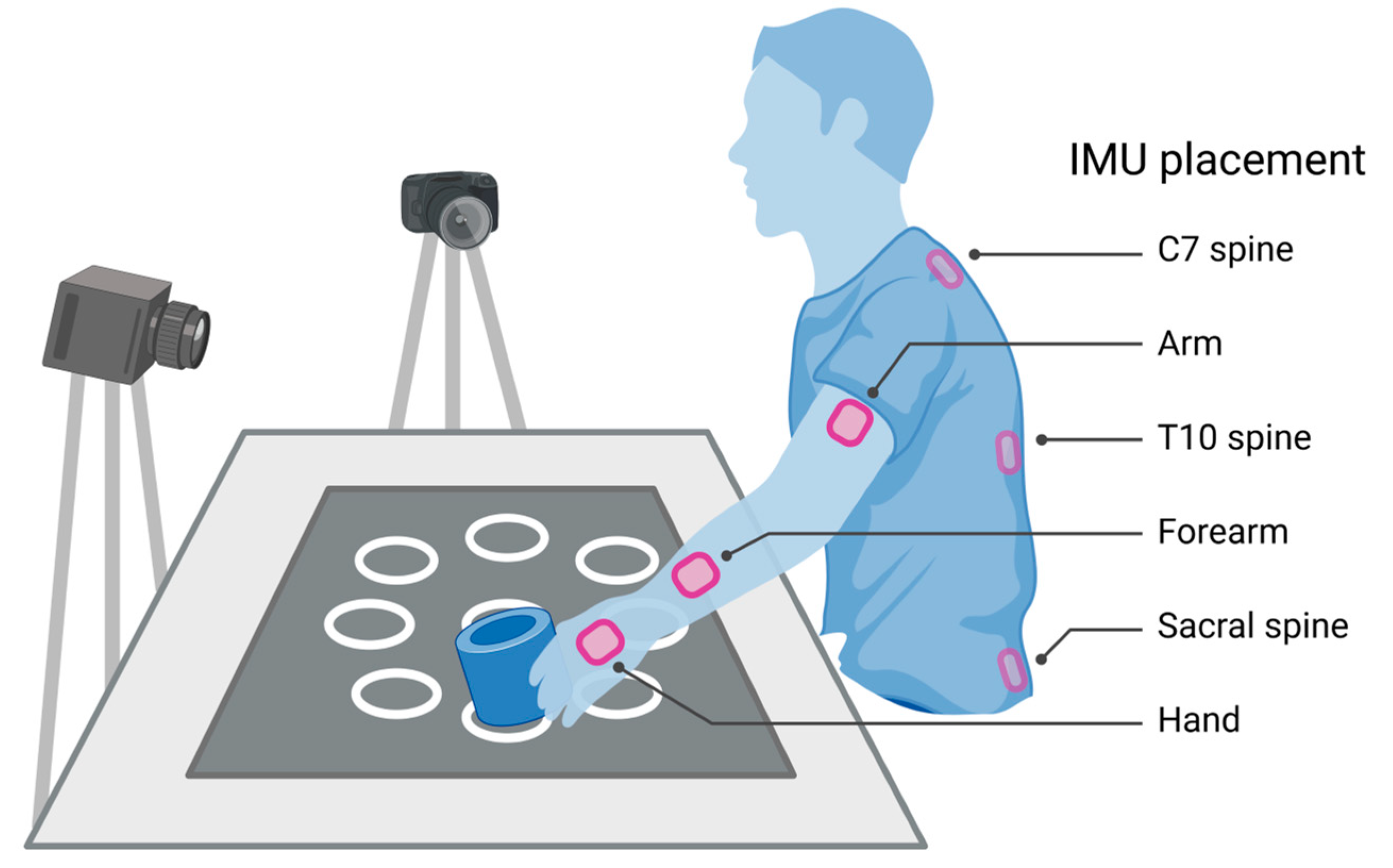
2.3. Motion Capture
2.4. Primitive Classification
2.5. Model Selection
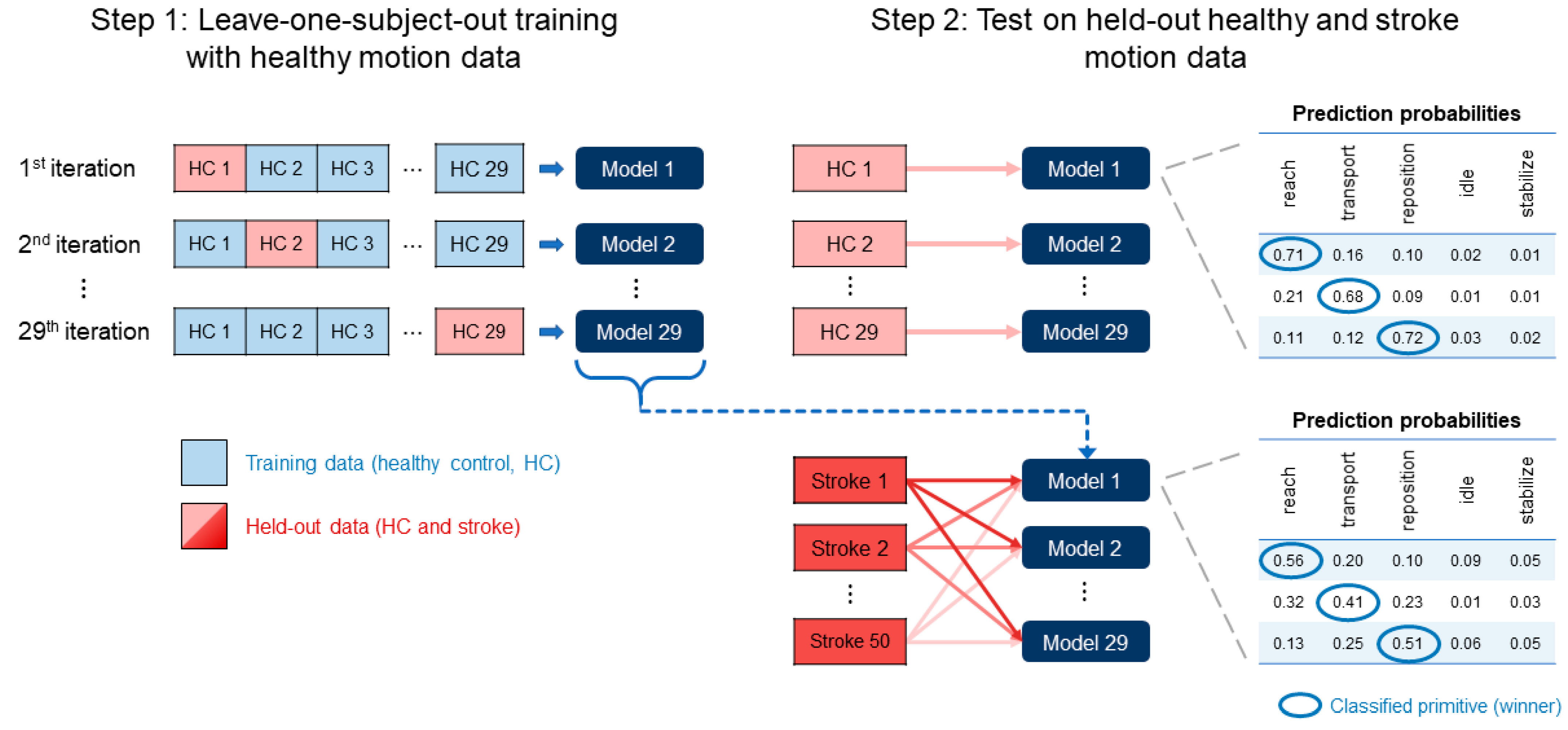
2.6. Model Training and Testing
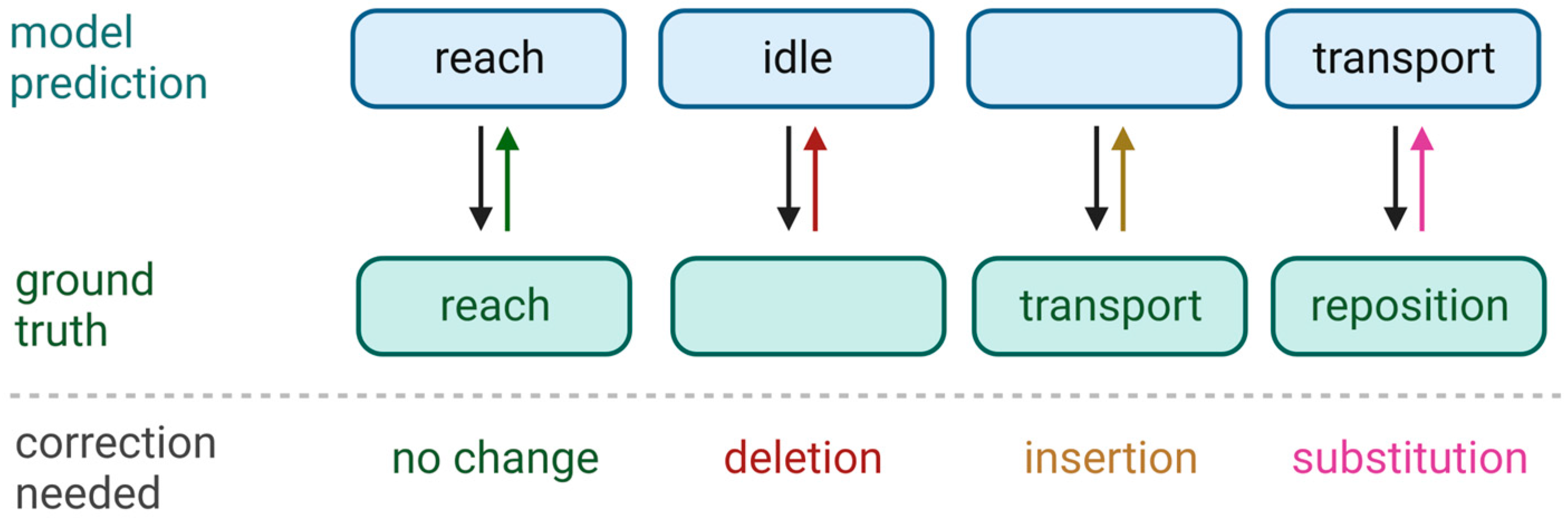
2.7. Assessment of Classification Performance
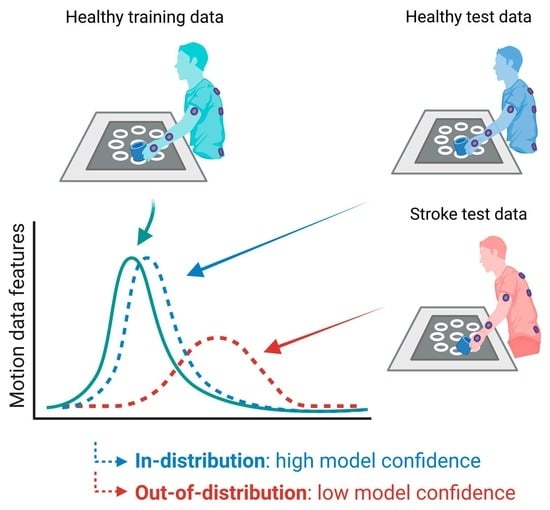
2.8. Assessment of Model Confidence
2.9. Localization of Reduced Confidence
2.10. Analyses
3. Results
3.1. Model Accuracy
3.2. Model Confidence in Healthy and Stroke Groups
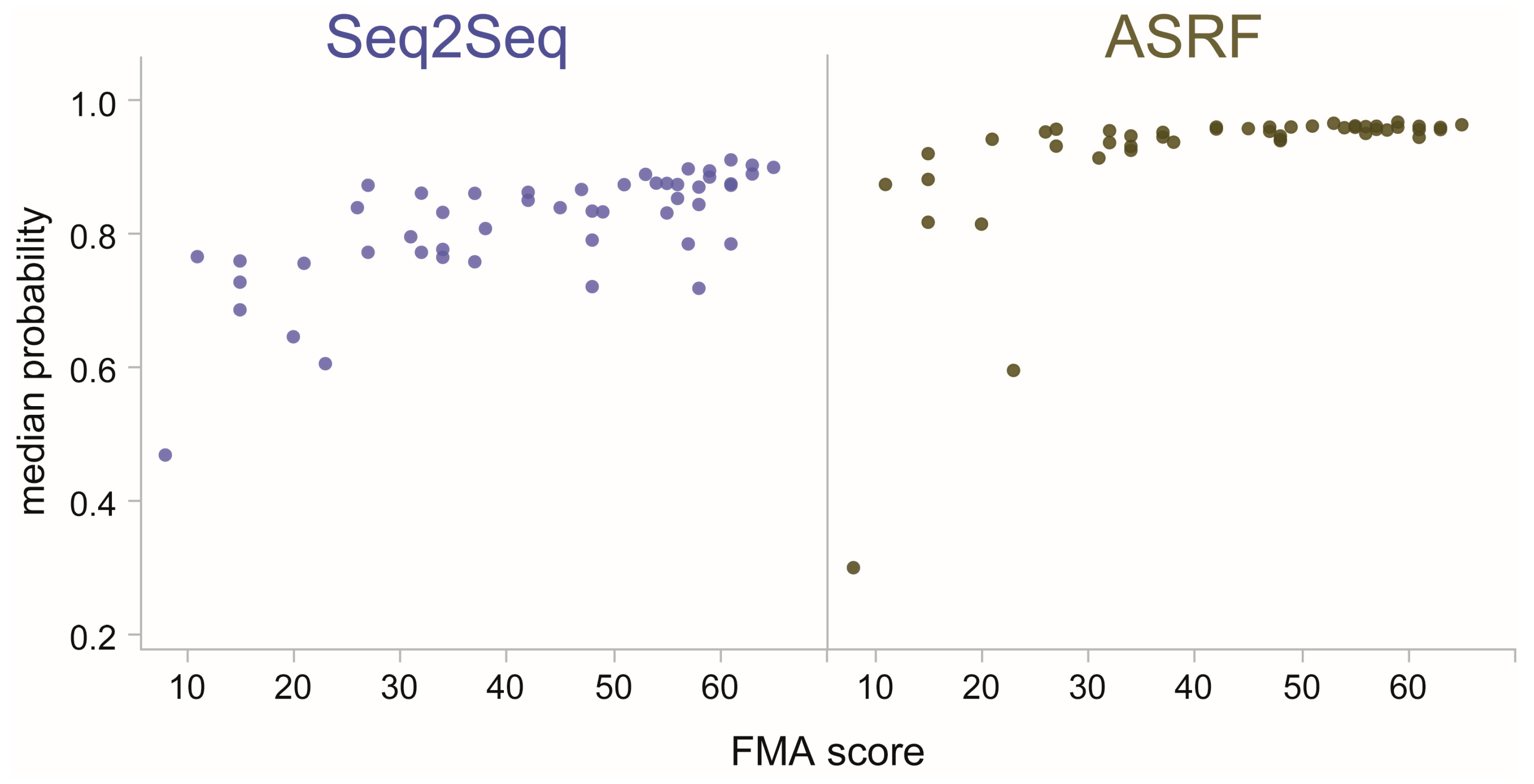
3.3. Model Confidence in Categories of Stroke Impairment
3.4. Model Confidence in Individual Stroke Impairment
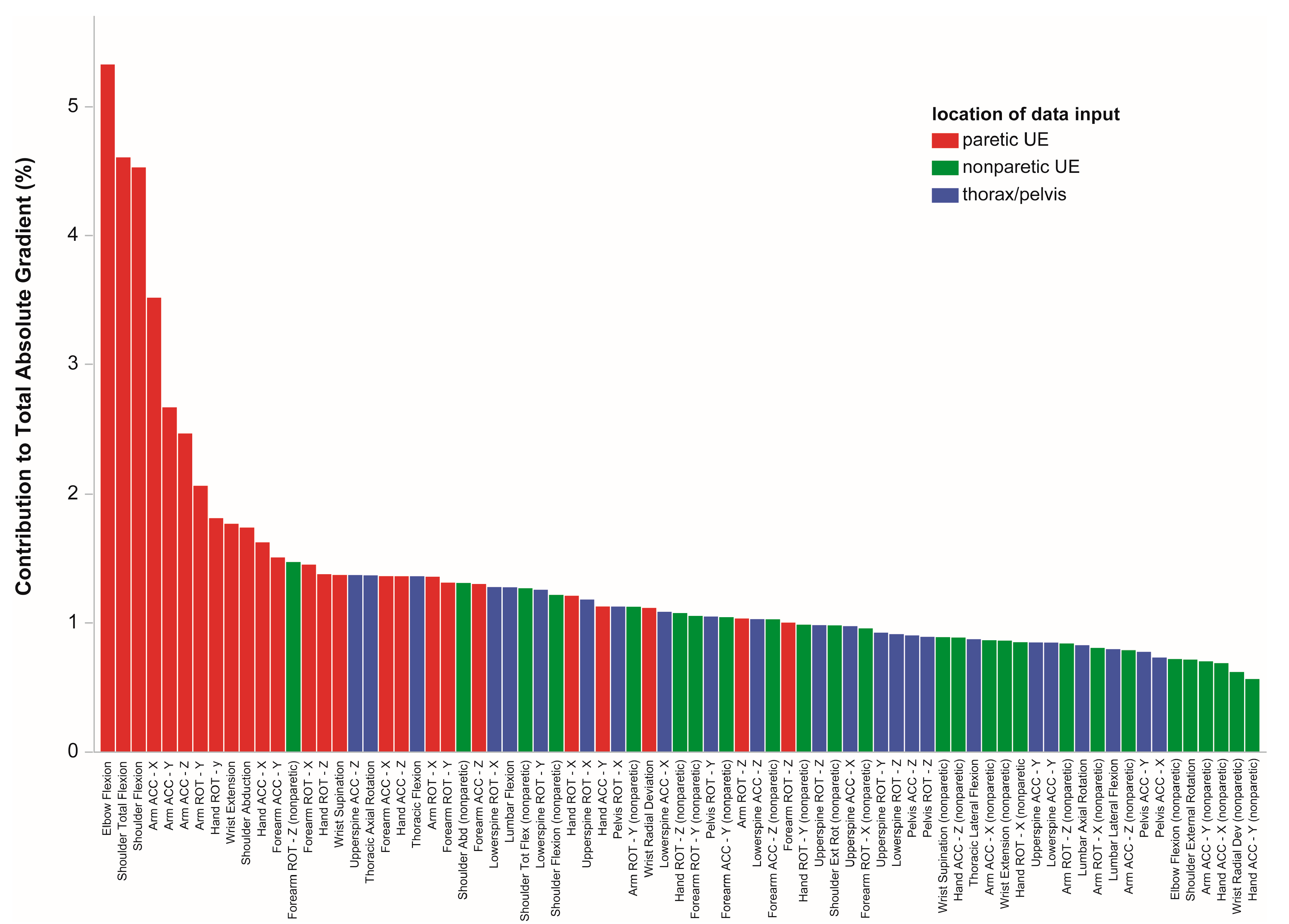
3.5. Locations Driving Model Uncertainty
4. Discussion
4.1. Previous Work
4.2. Practical Considerations
4.3. Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wade, D.T.; Langton-Hewer, R.; Wood, V.A.; Skilbeck, C.E.; Ismail, H.M. The hemiplegic arm after stroke: Measurement and recovery. J. Neurol. Neurosurg. Psychiatry 1983, 46, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Twitchell, T.E. The restoration of motor function following hemiplegia in man. Brain 1951, 74, 443–480. [Google Scholar] [CrossRef]
- Mayo, N.E.; Wood-Dauphinee, S.; Côté, R.; Durcan, L.; Carlton, J. Activity, participation, and quality of life 6 months poststroke. Arch. Phys. Med. Rehabil. 2002, 83, 1035–1042. [Google Scholar] [CrossRef]
- Geller, T. Overcoming the uncanny valley. IEEE Comput. Graph. Appl. 2008, 28, 11–17. [Google Scholar] [CrossRef]
- Fugl-Meyer, A.R.; Jaasko, L.; Leyman, I.; Olsson, S.; Steglind, S. The post-stroke hemiplegic patient. 1. a method for evaluation of physical performance. Scand. J. Rehabil. Med. 1975, 7, 13–31. [Google Scholar] [PubMed]
- Lyle, R.C. A performance test for assessment of upper limb function in physical rehabilitation treatment and research. Int. J. Rehabil. Res. 1981, 4, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.L.; Thompson, P.A.; Morris, D.M.; Rose, D.K.; Winstein, C.J.; Taub, E.; Giuliani, C.; Pearson, S.L. The EXCITE trial: Attributes of the Wolf Motor Function Test in patients with subacute stroke. Neurorehabilit. Neural Repair 2005, 19, 194–205. [Google Scholar] [CrossRef]
- Platz, T.; Pinkowski, C.; van Wijck, F.; Kim, I.H.; di Bella, P.; Johnson, G. Reliability and validity of arm function assessment with standardized guidelines for the Fugl-Meyer Test, Action Research Arm Test and Box and Block Test: A multicentre study. Clin. Rehabil. 2005, 19, 404–411. [Google Scholar] [CrossRef]
- Gladstone, D.J.; Danells, C.J.; Black, S.E. The fugl-meyer assessment of motor recovery after stroke: A critical review of its measurement properties. Neurorehabilit. Neural Repair 2002, 16, 232–240. [Google Scholar] [CrossRef]
- Lin, J.-H.; Hsu, M.-J.; Sheu, C.-F.; Wu, T.-S.; Lin, R.-T.; Chen, C.-H.; Hsieh, C.-L. Psychometric Comparisons of 4 Measures for Assessing Upper-Extremity Function in People With Stroke. Phys. Ther. 2009, 89, 840–850. [Google Scholar] [CrossRef]
- Schwarz, A.; Kanzler, C.M.; Lambercy, O.; Luft, A.R.; Veerbeek, J.M. Systematic Review on Kinematic Assessments of Upper Limb Movements After Stroke. Stroke J. Cereb. Circ. 2019, 50, 718–727. [Google Scholar] [CrossRef] [PubMed]
- Alt Murphy, M.; Häger, C.K. Kinematic analysis of the upper extremity after stroke—How far have we reached and what have we grasped? Phys. Ther. Rev. 2015, 20, 137–155. [Google Scholar] [CrossRef]
- Kwakkel, G.; Lannin, N.A.; Borschmann, K.; English, C.; Ali, M.; Churilov, L.; Saposnik, G.; Winstein, C.; van Wegen, E.E.H.; Wolf, S.L.; et al. Standardized Measurement of Sensorimotor Recovery in Stroke Trials: Consensus-Based Core Recommendations from the Stroke Recovery and Rehabilitation Roundtable. Neurorehabilit. Neural Repair 2017, 31, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Kitago, T.; Goldsmith, J.; Harran, M.; Kane, L.; Berard, J.; Huang, S.; Ryan, S.L.; Mazzoni, P.; Krakauer, J.W.; Huang, V.S. Robotic therapy for chronic stroke: General recovery of impairment or improved task-specific skill? J. Neurophysiol. 2015, 114, 1885–1894. [Google Scholar] [CrossRef]
- Cortes, J.C.; Goldsmith, J.; Harran, M.D.; Xu, J.; Kim, N.; Schambra, H.M.; Luft, A.R.; Celnik, P.; Krakauer, J.W.; Kitago, T. A Short and Distinct Time Window for Recovery of Arm Motor Control Early After Stroke Revealed with a Global Measure of Trajectory Kinematics. Neurorehabilit. Neural Repair 2017, 31, 552–560. [Google Scholar] [CrossRef]
- Przybyla, L.; Good, D.; Sainburg, R. Virtual Reality Arm Supported Training Reduces Motor Impairment In Two Patients with Severe Hemiparesis. J. Neurol. Transl. Neurosci. 2013, 1, 1018. [Google Scholar]
- Wu, C.; Trombly, C.A.; Lin, K.; Tickle-Degnen, L. A kinematic study of contextual effects on reaching performance in persons with and without stroke: Influences of object availability. Arch. Phys. Med. Rehabil. 2000, 81, 95–101. [Google Scholar] [CrossRef]
- Wisneski, K.J.; Johnson, M.J. Quantifying kinematics of purposeful movements to real, imagined, or absent functional objects: Implications for modelling trajectories for robot-assisted ADL tasks. J. Neuroeng. Rehabil. 2007, 4, 7. [Google Scholar] [CrossRef]
- Beer, R.F.; Dewald, J.P.A.; Dawson, M.L.; Rymer, W.Z. Target-dependent differences between free and constrained arm movements in chronic hemiparesis. Exp. Brain Res. 2004, 156, 458–470. [Google Scholar] [CrossRef]
- Schambra, H.M.; Xu, J.; Branscheidt, M.; Lindquist, M.; Uddin, J.; Steiner, L.; Hertler, B.; Kim, N.; Berard, J.; Harran, M.D.; et al. Differential Poststroke Motor Recovery in an Arm Versus Hand Muscle in the Absence of Motor Evoked Potentials. Neurorehabilit. Neural Repair 2019, 33, 568–580. [Google Scholar] [CrossRef]
- van Kordelaar, J.; van Wegen, E.; Kwakkel, G. Impact of time on quality of motor control of the paretic upper limb after stroke. Arch. Phys. Med. Rehabil. 2014, 95, 338–344. [Google Scholar] [CrossRef]
- Lum, P.S.; Mulroy, S.; Amdur, R.L.; Requejo, P.; Prilutsky, B.I.; Dromerick, A.W. Gains in upper extremity function after stroke via recovery or compensation: Potential differential effects on amount of real-world limb use. Top. Stroke Rehabil. 2009, 16, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Olesh, E.V.; Yakovenko, S.; Gritsenko, V. Automated assessment of upper extremity movement impairment due to stroke. PLoS ONE 2014, 9, e104487. [Google Scholar] [CrossRef] [PubMed]
- Arac, A.; Zhao, P.; Dobkin, B.H.; Carmichael, S.T.; Golshani, P. DeepBehavior: A Deep Learning Toolbox for Automated Analysis of Animal and Human Behavior Imaging Data. Front. Syst. Neurosci. 2019, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.; Bhagubai, M.M.C.; Wolterink, G.; Held, J.P.O.; Luft, A.R.; Veltink, P.H. Assessment of Upper Limb Movement Impairments after Stroke Using Wearable Inertial Sensing. Sensors 2020, 20, 4770. [Google Scholar] [CrossRef]
- Murphy, M.A.; Willén, C.; Sunnerhagen, K.S. Kinematic Variables Quantifying Upper-Extremity Performance After Stroke During Reaching and Drinking From a Glass. Neurorehabilit. Neural Repair 2010, 25, 71–80. [Google Scholar] [CrossRef]
- Halilaj, E.; Rajagopal, A.; Fiterau, M.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Machine learning in human movement biomechanics: Best practices, common pitfalls, and new opportunities. J. Biomech. 2018, 81, 1–11. [Google Scholar] [CrossRef]
- Hendrycks, D.; Gimpel, K. A Baseline for Detecting Misclassified and Out-of-Distribution Examples in Neural Networks. arXiv 2016, arXiv:1610.02136. [Google Scholar]
- Yang, J.; Zhou, K.; Li, Y.; Liu, Z. Generalized out-of-distribution detection: A survey. arXiv 2021, arXiv:2110.11334. [Google Scholar]
- DeVries, T.; Taylor, G.W. Learning Confidence for Out-of-Distribution Detection in Neural Networks. arXiv 2018, arXiv:1802.04865. [Google Scholar] [CrossRef]
- van Hespen, K.M.; Zwanenburg, J.J.M.; Dankbaar, J.W.; Geerlings, M.I.; Hendrikse, J.; Kuijf, H.J. An anomaly detection approach to identify chronic brain infarcts on MRI. Sci. Rep. 2021, 11, 7714. [Google Scholar] [CrossRef] [PubMed]
- Linmans, J.; Elfwing, S.; van der Laak, J.; Litjens, G. Predictive uncertainty estimation for out-of-distribution detection in digital pathology. Med. Image Anal. 2023, 83, 102655. [Google Scholar] [CrossRef]
- Zaida, M.; Ali, S.; Ali, M.; Hussein, S.; Saadia, A.; Sultani, W. Out of distribution detection for skin and malaria images. arXiv 2021, arXiv:2111.01505. [Google Scholar] [CrossRef]
- Boyer, P.; Burns, D.; Whyne, C. Out-of-Distribution Detection of Human Activity Recognition with Smartwatch Inertial Sensors. Sensors 2021, 21, 1669. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.; Narayan, S.; Dwivedi, S.K.; Gupta, V.; Ahmed, S.; Khan, F.S.; Shao, L. Out-of-distribution detection for generalized zero-shot action recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 9985–9993. [Google Scholar]
- Schambra, H.M.; Parnandi, A.; Pandit, N.G.; Uddin, J.; Wirtanen, A.; Nilsen, D.M. A Taxonomy of Functional Upper Extremity Motion. Front. Neurol. 2019, 10, 857. [Google Scholar] [CrossRef]
- Saulle, M.F.; Schambra, H.M. Recovery and Rehabilitation after Intracerebral Hemorrhage. Semin. Neurol. 2016, 36, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Parnandi, A.; Kaku, A.; Venkatesan, A.; Pandit, N.; Wirtanen, A.; Rajamohan, H.; Venkataramanan, K.; Nilsen, D.; Fernandez-Granda, C.; Schambra, H. PrimSeq: A deep learning-based pipeline to quantitate rehabilitation training. PLoS Digit. Health 2022, 1, e0000044. [Google Scholar] [CrossRef]
- Kaku, A.; Liu, K.; Parnandi, A.; Rajamohan, H.; Venkataramanan, K.; Venkatesan, A.; Wirtanen, A.; Pandit, N.; Schambra, H.M.; Fernandez-Granda, C. StrokeRehab: A Benchmark Dataset for Sub-second Action Identification. In Proceedings of the Neural Information Processing Systems, New Orleans, LA, USA, 16–19 May 2022. [Google Scholar]
- Kaku, A.; Liu, K.; Parnandi, A.; Rajamohan, H.R.; Venkataramanan, K.; Venkatesan, A.; Wirtanen, A.; Pandit, N.; Schambra, H.; Fernandez-Granda, C. Sequence-to-Sequence Modeling for Action Identification at High Temporal Resolution. arXiv 2021, arXiv:2111.02521. [Google Scholar]
- Levenshtein, V.I. Binary codes capable of correcting deletions, insertions, and reversals. Sov. Phys. Dokl. 1966, 10, 707–710. [Google Scholar]
- Woodbury, M.L.; Velozo, C.A.; Richards, L.G.; Duncan, P.W. Rasch analysis staging methodology to classify upper extremity movement impairment after stroke. Arch. Phys. Med. Rehabil. 2013, 94, 1527–1533. [Google Scholar] [CrossRef]
- Ramdas, A.; Trillos, N.G.; Cuturi, M. On Wasserstein Two-Sample Testing and Related Families of Nonparametric Tests. Entropy 2017, 19, 47. [Google Scholar] [CrossRef]
- Wasserstein, L.N. Markov processes over denumerable products of spaces describing large systems of automata. Probl. Inf. Transm. 1969, 5, 47–52. [Google Scholar]
- Cirstea, M.C.; Levin, M.F. Compensatory strategies for reaching in stroke. Brain 2000, 123 Pt 5, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.F.; Kleim, J.A.; Wolf, S.L. What do motor “recovery” and “compensation” mean in patients following stroke? Neurorehabilit. Neural Repair 2009, 23, 313–319. [Google Scholar] [CrossRef]
- Subramanian, S.K.; Yamanaka, J.; Chilingaryan, G.; Levin, M.F. Validity of movement pattern kinematics as measures of arm motor impairment poststroke. Stroke J. Cereb. Circ. 2010, 41, 2303–2308. [Google Scholar] [CrossRef]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.M.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019, 32, 8024–8035. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Pandas Development Team. Pandas-Dev/Pandas: Pandas (v2.0.1); Zenodo: Meyrin, Switzerland, 2023. [Google Scholar]
- Van Rossum, G. The Python Library Reference, Release 3.8.2; Python Software Foundation: Wilmington DE, USA, 2020. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Robertson, D.; Caldwell, G.; Hamill, J.; Kamen, G.; Whittlesey, S. Research Methods in Biomechanics, 2nd ed.; Human Kinetics: Champaign, IL, USA, 2013. [Google Scholar]
- Snyder, C.; Martínez, A.; Jahnel, R.; Roe, J.; Stöggl, T. Connected Skiing: Motion Quality Quantification in Alpine Skiing. Sensors 2021, 21, 3799. [Google Scholar] [CrossRef] [PubMed]
- Troje, N.F. Decomposing biological motion: A framework for analysis and synthesis of human gait patterns. J. Vis. 2002, 2, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Deluzio, K.J.; Astephen, J.L. Biomechanical features of gait waveform data associated with knee osteoarthritis: An application of principal component analysis. Gait Posture 2007, 25, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Ross, G.B.; Dowling, B.; Troje, N.F.; Fischer, S.L.; Graham, R.B. Objectively Differentiating Movement Patterns between Elite and Novice Athletes. Med. Sci. Sport. Exerc. 2018, 50, 1457–1464. [Google Scholar] [CrossRef]
- Ross, G.B.; Dowling, B.; Troje, N.F.; Fischer, S.L.; Graham, R.B. Classifying Elite From Novice Athletes Using Simulated Wearable Sensor Data. Front. Bioeng. Biotechnol. 2020, 8, 814. [Google Scholar] [CrossRef]
- Otten, P.; Kim, J.; Son, S.H. A Framework to Automate Assessment of Upper-Limb Motor Function Impairment: A Feasibility Study. Sensors 2015, 15, 20097–20114. [Google Scholar] [CrossRef]
- Patel, S.; Hughes, R.; Hester, T.; Stein, J.; Akay, M.; Dy, J.; Bonato, P. Tracking motor recovery in stroke survivors undergoing rehabilitation using wearable technology. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 6858–6861. [Google Scholar]
- Tang, L.; Halloran, S.; Shi, J.Q.; Guan, Y.; Cao, C.; Eyre, J. Evaluating upper limb function after stroke using the free-living accelerometer data. Stat. Methods Med. Res. 2020, 29, 3249–3264. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, B.; Wang, J.; Wu, Q.; Li, X.; Zhou, L.; Xiong, D. Wearable Intelligent Machine Learning Rehabilitation Assessment for Stroke Patients Compared with Clinician Assessment. J. Clin. Med. 2022, 11, 7467. [Google Scholar] [CrossRef]
- Lee, S.-H.; Hwang, Y.-J.; Lee, H.-J.; Kim, Y.-H.; Ogrinc, M.; Burdet, E.; Kim, J.-H. Proof-of-Concept of a Sensor-Based Evaluation Method for Better Sensitivity of Upper-Extremity Motor Function Assessment. Sensors 2021, 21, 5926. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, J.; Ling, Z.; Zhang, B.; Yan, Y.; Xiong, D.; Guo, L. Quantitative Evaluation System of Upper Limb Motor Function of Stroke Patients Based on Desktop Rehabilitation Robot. Sensors 2022, 22, 1170. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, X.; Gong, Y.; Cheng, Y.; Gao, X.; Chen, X. Motor Function Evaluation of Hemiplegic Upper-Extremities Using Data Fusion from Wearable Inertial and Surface EMG Sensors. Sensors 2017, 17, 582. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.; Ross, G.B.; Graham, R.B. Machine Learning and Deep Neural Network Architectures for 3D Motion Capture Datasets. In Proceedings of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 4827–4830. [Google Scholar]
- Zackowski, K.M.; Dromerick, A.W.; Sahrmann, S.A.; Thach, W.T.; Bastian, A.J. How do strength, sensation, spasticity and joint individuation relate to the reaching deficits of people with chronic hemiparesis? Brain 2004, 127, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Côté-Allard, U.; Campbell, E.; Phinyomark, A.; Laviolette, F.; Gosselin, B.; Scheme, E. Interpreting Deep Learning Features for Myoelectric Control: A Comparison With Handcrafted Features. Front. Bioeng. Biotechnol. 2020, 8, 158. [Google Scholar] [CrossRef]
- Schambra, H.M.; Ogden, R.T.; Martinez-Hernandez, I.E.; Lin, X.; Chang, Y.B.; Rahman, A.; Edwards, D.J.; Krakauer, J.W. The reliability of repeated TMS measures in older adults and in patients with subacute and chronic stroke. Front. Cell. Neurosci. 2015, 9, 335. [Google Scholar] [CrossRef]

| Healthy Controls | Stroke Patients | |
|---|---|---|
| n = 29 | n = 50 | |
| Sex 1 | 15 M, 14 F | 23 M, 27 F |
| Age | 62.4 ± 13.1 years | 57.7 ± 14.0 years |
| Race 2 | 11 W, 14 B, 1 A, 1 AI, 2 O | 23 W, 11 B, 8 A, 0 AI, 8 O |
| Paretic side 3 | n/a | 27 L: 23 R |
| Fugl-Meyer score 4 | 65.2 ± 1.0 | 43.1 ± 16.1 |
| Impairment level 5 | n/a | 20 mild, 22 moderate, 8 severe |
| Time since stroke | n/a | 5.4 ± 6.1 years |
| Joint | Anatomical Angles |
|---|---|
| Shoulder | Flexion/extension, internal/external rotation, adduction/abduction, total flexion 1 |
| Elbow | Flexion/extension |
| Wrist | Flexion/extension, pronation/supination, radial/ulnar deviation |
| Thorax 2 | Flexion/extension, axial rotation, lateral flexion/extension |
| Lumbar 3 | Flexion/extension, axial rotation, lateral flexion/extension |
| Seq2Seq | ASRF | |||
|---|---|---|---|---|
| Healthy | Stroke | Healthy | Stroke | |
| True positive rate | 0.842 | 0.912 | 0.868 | 0.929 |
| False discovery rate | 0.115 | 0.272 | 0.159 | 0.312 |
| F1 score | 0.848 | 0.798 | 0.838 | 0.773 |
| Seq2Seq | ASRF | |
|---|---|---|
| Cauchy (median) | 0.701 *** | 0.705 *** |
| Gaussian (mean) | 0.661 *** | 0.619 *** |
| Cauchy PC1 | −0.706 *** | −0.727 *** |
| Gaussian PC1 | −0.660 *** | −0.600 *** |
| Wasserstein | −0.511 ** | 0.372 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parnandi, A.; Kaku, A.; Venkatesan, A.; Pandit, N.; Fokas, E.; Yu, B.; Kim, G.; Nilsen, D.; Fernandez-Granda, C.; Schambra, H. Data-Driven Quantitation of Movement Abnormality after Stroke. Bioengineering 2023, 10, 648. https://doi.org/10.3390/bioengineering10060648
Parnandi A, Kaku A, Venkatesan A, Pandit N, Fokas E, Yu B, Kim G, Nilsen D, Fernandez-Granda C, Schambra H. Data-Driven Quantitation of Movement Abnormality after Stroke. Bioengineering. 2023; 10(6):648. https://doi.org/10.3390/bioengineering10060648
Chicago/Turabian StyleParnandi, Avinash, Aakash Kaku, Anita Venkatesan, Natasha Pandit, Emily Fokas, Boyang Yu, Grace Kim, Dawn Nilsen, Carlos Fernandez-Granda, and Heidi Schambra. 2023. "Data-Driven Quantitation of Movement Abnormality after Stroke" Bioengineering 10, no. 6: 648. https://doi.org/10.3390/bioengineering10060648
APA StyleParnandi, A., Kaku, A., Venkatesan, A., Pandit, N., Fokas, E., Yu, B., Kim, G., Nilsen, D., Fernandez-Granda, C., & Schambra, H. (2023). Data-Driven Quantitation of Movement Abnormality after Stroke. Bioengineering, 10(6), 648. https://doi.org/10.3390/bioengineering10060648