Leveraging Genomic and Bioinformatic Analysis to Enhance Drug Repositioning for Dermatomyositis
Abstract
1. Introduction
2. Results
2.1. Variants Associated with Dermatomyositis from GWAS and PheWAS Catalogs
2.2. Functional Annotation of Dermatomyositis Risk Genes
2.3. Gene Network Expansion through Utilization of the STRING Database
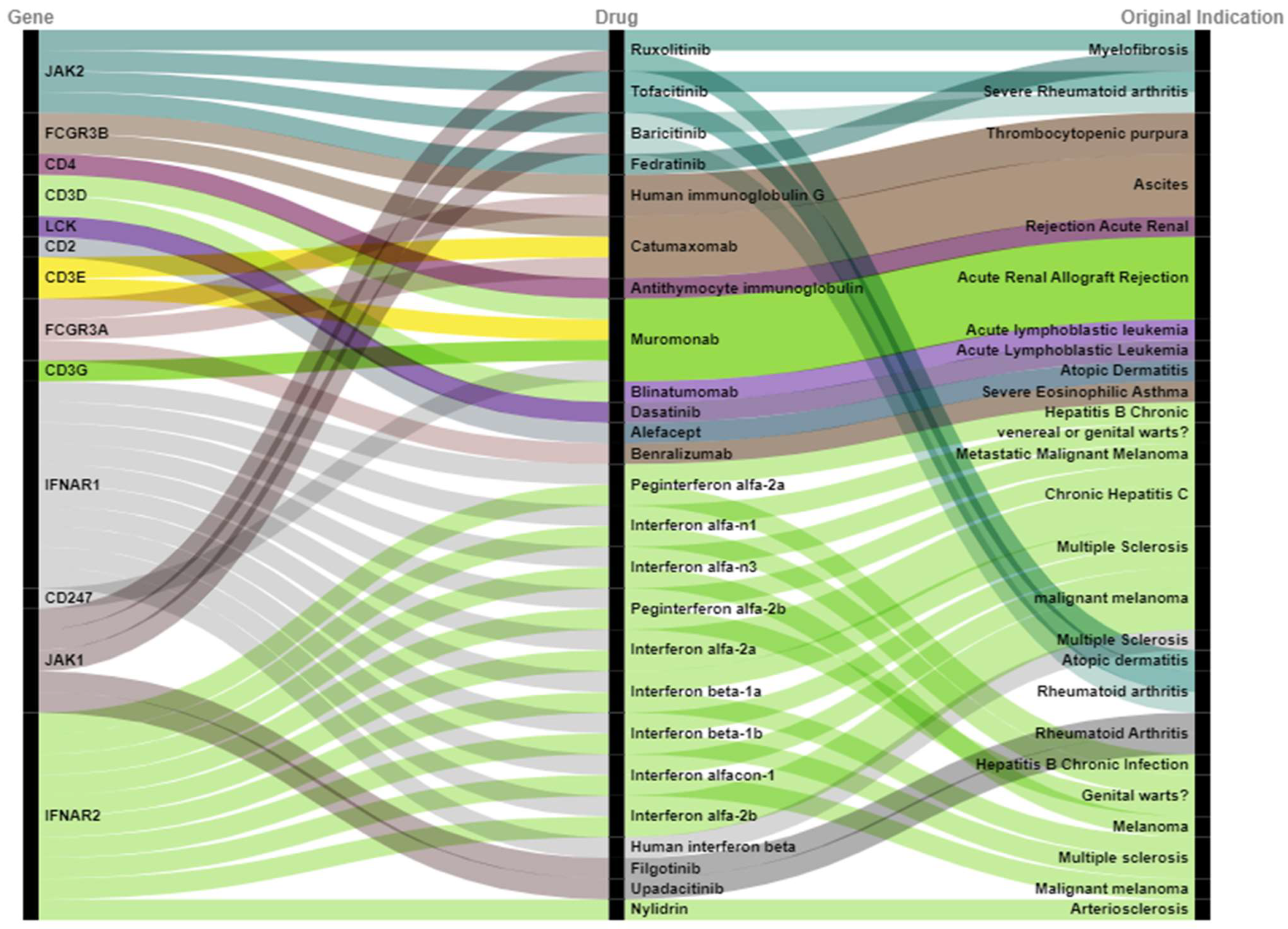
2.4. Prioritization of Drugs Repurposed for Dermatomyositis
3. Discussion
4. Materials and Methods
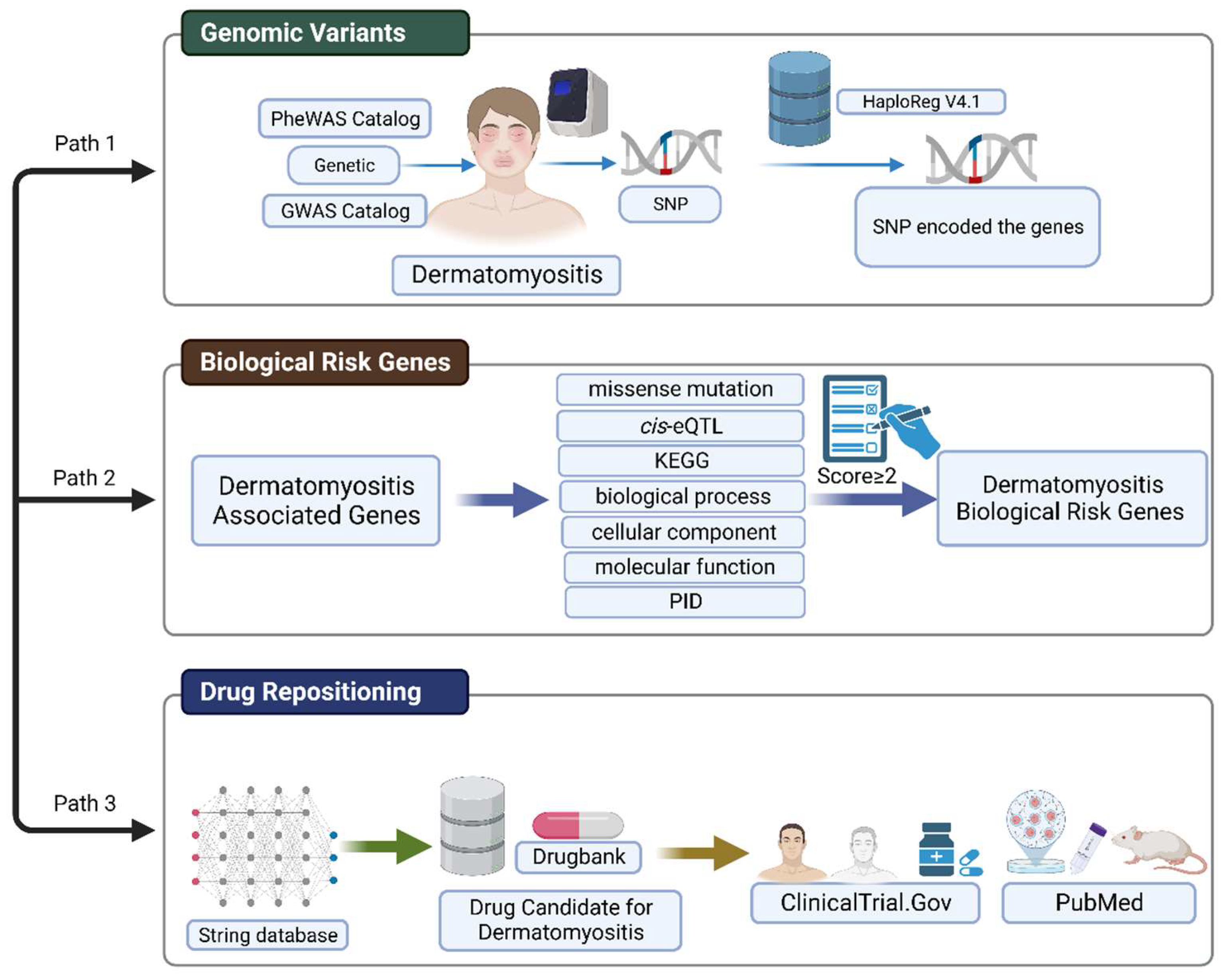
4.1. Workflow for Integrative Analysis of Genomic Variants and Gene Network
4.2. Candidate Risk Genes Associated with Dermatomyositis
4.3. Biological Risk Genes for Dermatomyositis
4.4. Gene Network Expansion by Using STRING Database
4.5. Gene and Drug Overlapping Analysis from Drug Databases
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qudsiya, Z.; Waseem, M. Dermatomyositis. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2022. [Google Scholar]
- Ungprasert, P.; Leeaphorn, N.; Hosiriluck, N.; Chaiwatcharayut, W.; Ammannagari, N.; Raddatz, D.A. Clinical features of inflammatory myopathies and their association with malignancy: A systematic review in asian population. Int. Sch. Res. Not. 2013, 2013, 509354. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.Y.; Fan, Y.; Wang, Y.X.; Zhong, Y.J.; Wang, G.F. Prevalence of interstitial lung disease in polymyositis and dermatomyositis: A meta-analysis from 2000 to 2020. Semin. Arthritis Rheum. 2021, 51, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Bendewald, M.J.; Wetter, D.A.; Li, X.; Davis, M.D. Incidence of dermatomyositis and clinically amyopathic dermatomyositis: A population-based study in Olmsted County, Minnesota. Arch. Dermatol. 2010, 146, 26–30. [Google Scholar] [CrossRef]
- O’Hanlon, T.P.; Carrick, D.M.; Targoff, I.N.; Arnett, F.C.; Reveille, J.D.; Carrington, M.; Gao, X.; Oddis, C.V.; Morel, P.A.; Malley, J.D.; et al. Immunogenetic risk and protective factors for the idiopathic inflammatory myopathies: Distinct HLA-A, -B, -Cw, -DRB1 and -DQA1 allelic profiles and motifs define clinicopathologic groups in caucasians. Medicine 2005, 84, 338–349. [Google Scholar] [CrossRef] [PubMed]
- O’Hanlon, T.P.; Rider, L.G.; Mamyrova, G.; Targoff, I.N.; Arnett, F.C.; Reveille, J.D.; Carrington, M.; Gao, X.; Oddis, C.V.; Morel, P.A.; et al. HLA polymorphisms in African Americans with idiopathic inflammatory myopathy: Allelic profiles distinguish patients with different clinical phenotypes and myositis autoantibodies. Arthritis Rheum. 2006, 54, 3670–3681. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Han, L.; Yuan, L.; Yang, Y.; Gou, G.; Sun, H.; Lu, L.; Bao, L. HLA class II alleles may influence susceptibility to adult dermatomyositis and polymyositis in a Han Chinese population. BMC Dermatol. 2014, 14, 9. [Google Scholar] [CrossRef]
- Krathen, M.S.; Fiorentino, D.; Werth, V.P. Dermatomyositis. Curr. Dir. Autoimmun. 2008, 10, 313–332. [Google Scholar]
- Clemente, G.; Piotto, D.G.P.; Barbosa, C.; Peracchi, O.A.; Len, C.A.; Hilário, M.O.E.; Terreri, M.T.R. High frequency of calcinosis in juvenile dermatomyositis: A risk factor study. Rev. Bras. De Reumatol. 2012, 52, 549–553. [Google Scholar] [CrossRef]
- Dalakas, M.C.; Hohlfeld, R. Polymyositis and dermatomyositis. Lancet 2003, 362, 971–982. [Google Scholar] [CrossRef]
- Okogbaa, J.; Batiste, L. Dermatomyositis: An Acute Flare and Current Treatments. Clin. Med. Insights Case Rep. 2019, 12, 1179547619855370. [Google Scholar] [CrossRef]
- Chyr, J.; Gong, H.; Zhou, X. DOTA: Deep Learning Optimal Transport Approach to Advance Drug Repositioning for Alzheimer’s Disease. Biomolecules 2022, 12, 196. [Google Scholar] [CrossRef] [PubMed]
- Sirota, M.; Dudley, J.T.; Kim, J.; Chiang, A.P.; Morgan, A.A.; Sweet-Cordero, A.; Sage, J.; Butte, A.J. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 2011, 3, 96ra77. [Google Scholar] [CrossRef] [PubMed]
- Fioranelli, M.; Roccia, M.G.; Lotti, T. Treatment of dermatomyositis with ruxolitinib. Dermatol. Ther. 2016, 29, 285. [Google Scholar] [CrossRef] [PubMed]
- Paik, J.J.; Casciola-Rosen, L.; Shin, J.Y.; Albayda, J.; Tiniakou, E.; Leung, D.G.; Gutierrez-Alamillo, L.; Perin, J.; Florea, L.; Antonescu, C.; et al. Study of Tofacitinib in Refractory Dermatomyositis: An Open-Label Pilot Study of Ten Patients. Arthritis Rheumatol. 2021, 73, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Shalabi, M.M.; Garcia, B.; Kendall, C.; Siller, A., Jr.; Austinn, M.; Tyring, S.K. Janus Kinase and Tyrosine Kinase Inhibitors in Dermatology: A Review of Their Utilization, Safety Profile and Future Applications. Skin Therapy Lett. 2022, 27, 4–9. [Google Scholar]
- Fischer, K.; Aringer, M.; Steininger, J.; Heil, J.; Beissert, S.; Abraham, S.; Günther, C. Improvement of cutaneous inflammation and panniculitis in patients with dermatomyositis by the Janus kinase inhibitor baricitinib. Br. J. Dermatol. 2022, 187, 432–435. [Google Scholar] [CrossRef]
- Biggioggero, M.; Becciolini, A.; Crotti, C.; Agape, E.; Favalli, E.G. Upadacitinib and filgotinib: The role of JAK1 selective inhibition in the treatment of rheumatoid arthritis. Drugs Context 2019, 8, 212595. [Google Scholar] [CrossRef]
- Huard, C.; Gullà, S.V.; Bennett, D.V.; Coyle, A.J.; Vleugels, R.A.; Greenberg, S.A. Correlation of cutaneous disease activity with type 1 interferon gene signature and interferon β in dermatomyositis. Br. J. Dermatol. 2017, 176, 1224–1230. [Google Scholar] [CrossRef]
- Shiba, H.; Takeuchi, T.; Isoda, K.; Kokunai, Y.; Wada, Y.; Makino, S.; Hanafusa, T. Dermatomyositis as a complication of interferon-α therapy: A case report and review of the literature. Rheumatol. Int. 2014, 34, 1319–1322. [Google Scholar] [CrossRef]
- Somani, A.K.; Swick, A.R.; Cooper, K.D.; McCormick, T.S. Severe dermatomyositis triggered by interferon beta-1a therapy and associated with enhanced type I interferon signaling. Arch. Dermatol. 2008, 144, 1341–1349. [Google Scholar] [CrossRef]
- Jakobsen, K.R.; Demuth, C.; Sorensen, B.S.; Nielsen, A.L. The role of epithelial to mesenchymal transition in resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Transl. Lung Cancer Res. 2016, 5, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Kingo, K.; Aunin, E.; Karelson, M.; Rätsep, R.; Silm, H.; Vasar, E.; Kõks, S. Expressional changes in the intracellular melanogenesis pathways and their possible role in the pathogenesis of vitiligo. J. Dermatol. Sci. 2008, 52, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Douroudis, K.; Kingo, K.; Traks, T.; Reimann, E.; Raud, K.; Rätsep, R.; Mössner, R.; Silm, H.; Vasar, E.; Kõks, S. Polymorphisms in the ATG16L1 gene are associated with psoriasis vulgaris. Acta Derm. Venereol. 2012, 92, 85–87. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, A.; Zheng, M.; Lyu, L.; Thapa, C.; Gong, S.; Yang, Y.; Lyu, X. JAK-inhibitors for dermatomyositis: A concise literature review. Dermatol. Ther. 2021, 34, e14939. [Google Scholar] [CrossRef] [PubMed]
- Kul Cinar, O.; Papadopoulou, C.; Pilkington, C.A. Treatment of Calcinosis in Juvenile Dermatomyositis. Curr. Rheumatol. Rep. 2021, 23, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tan, J.; Martino, M.M.; Lui, K.O. Regulatory T-Cells: Potential Regulator of Tissue Repair and Regeneration. Front. Immunol. 2018, 9, 585. [Google Scholar] [CrossRef]
- Ladislau, L.; Suárez-Calvet, X.; Toquet, S.; Landon-Cardinal, O.; Amelin, D.; Depp, M.; Rodero, M.P.; Hathazi, D.; Duffy, D.; Bondet, V.; et al. JAK inhibitor improves type I interferon induced damage: Proof of concept in dermatomyositis. Brain 2018, 141, 1609–1621. [Google Scholar] [CrossRef]
- Adu, B.; Dodoo, D.; Adukpo, S.; Hedley, P.L.; Arthur, F.K.; Gerds, T.A.; Larsen, S.O.; Christiansen, M.; Theisen, M. Fc γ receptor IIIB (FcγRIIIB) polymorphisms are associated with clinical malaria in Ghanaian children. PLoS ONE 2012, 7, e46197. [Google Scholar] [CrossRef]
- Lill, M.; Kõks, S.; Soomets, U.; Schalkwyk, L.C.; Fernandes, C.; Lutsar, I.; Taba, P. Peripheral blood RNA gene expression profiling in patients with bacterial meningitis. Front. Neurosci. 2013, 7, 33. [Google Scholar] [CrossRef]
- Reemann, P.; Reimann, E.; Ilmjärv, S.; Porosaar, O.; Silm, H.; Jaks, V.; Vasar, E.; Kingo, K.; Koks, S. Melanocytes in the skin--comparative whole transcriptome analysis of main skin cell types. PLoS ONE 2014, 9, e115717. [Google Scholar] [CrossRef]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef]
- Irham, L.M.; Adikusuma, W.; Perwitasari, D.A.; Dania, H.; Maliza, R.; Faridah, I.N.; Santri, I.N.; Phiri, Y.V.A.; Cheung, R. The use of genomic variants to drive drug repurposing for chronic hepatitis B. Biochem. Biophys. Rep. 2022, 31, 101307. [Google Scholar] [CrossRef] [PubMed]
- Adikusuma, W.; Chou, W.-H.; Lin, M.-R.; Ting, J.; Irham, L.M.; Perwitasari, D.A.; Chang, W.-P.; Chang, W.-C. Identification of Druggable Genes for Asthma by Integrated Genomic Network Analysis. Biomedicines 2022, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Afief, A.R.; Irham, L.M.; Adikusuma, W.; Perwitasari, D.A.; Brahmadhi, A.; Cheung, R. Integration of genomic variants and bioinformatic-based approach to drive drug repurposing for multiple sclerosis. Biochem. Biophys. Rep. 2022, 32, 101337. [Google Scholar] [CrossRef]
- Irham, L.M.; Adikusuma, W.; Perwitasari, D.A. Genomic variants-driven drug repurposing for tuberculosis by utilizing the established bioinformatic-based approach. Biochem. Biophys. Rep. 2022, 32, 101334. [Google Scholar] [CrossRef]
- Puspitaningrum, A.N.; Perwitasari, D.A.; Adikusuma, W.; Djalilah, G.N.; Dania, H.; Maliza, R.; Faridah, I.N.; Sarasmita, M.A.; Rezadhini, M.; Cheung, R.; et al. Integration of genomic databases and bioinformatic approach to identify genomic variants for sjogren’s syndrome on multiple continents. Media Farm. J. Ilmu Farm. (J. Pharm. Sci.) 2022, 19, 71–81. [Google Scholar] [CrossRef]
- Lesmana, M.H.S.; Le, N.Q.K.; Chiu, W.-C.; Chung, K.-H.; Wang, C.-Y.; Irham, L.M.; Chung, M.-H. Genomic-Analysis-Oriented Drug Repurposing in the Search for Novel Antidepressants. Biomedicines 2022, 10, 1947. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
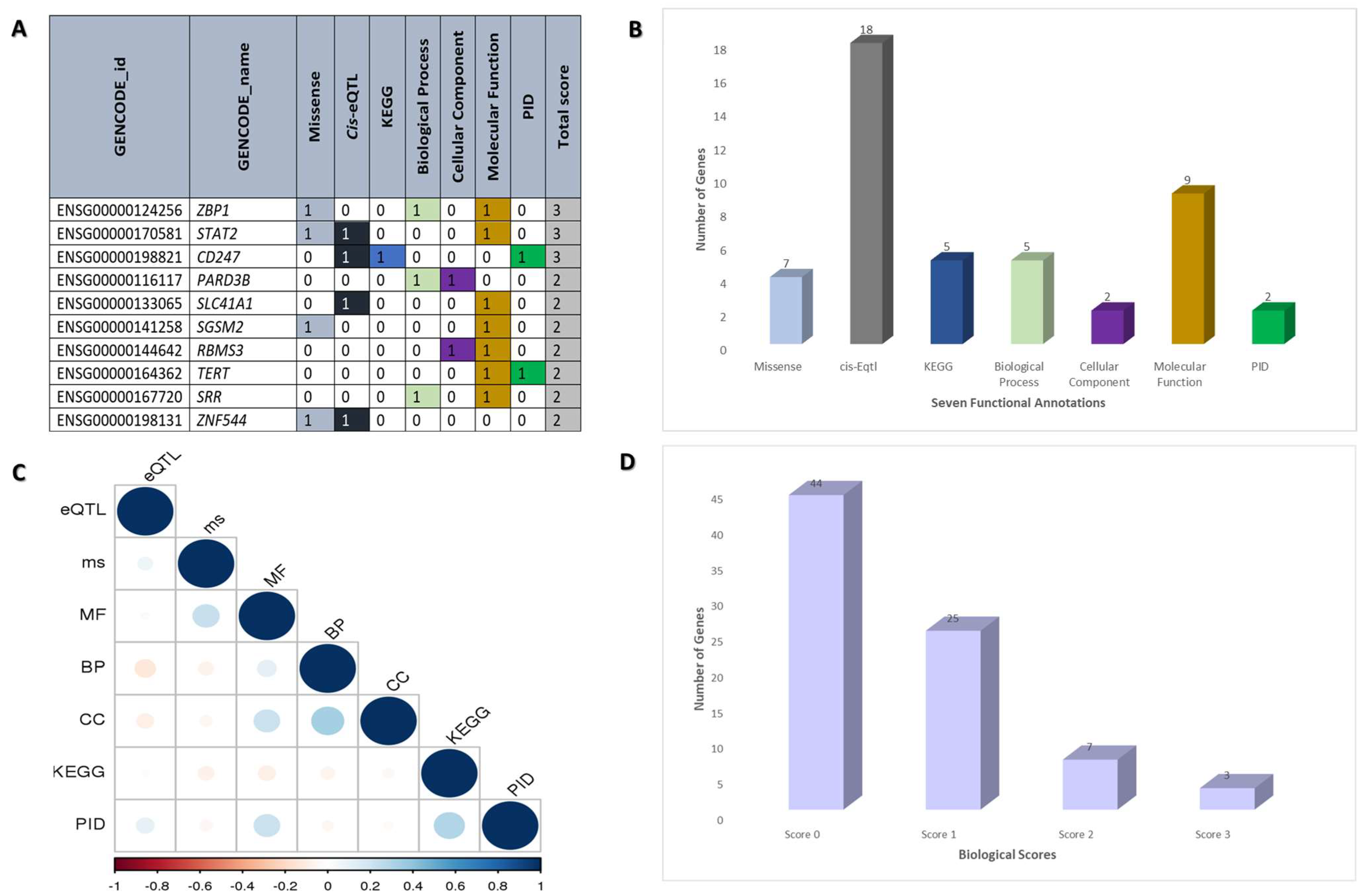
| GENCODE ID | Gene Name (GENCODE) | Missense | Cis-eQTL | KEGG | Biological Process | Cellular Component | Molecular Function | PID | Total Score |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000124256 | ZBP1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 3 |
| ENSG00000170581 | STAT2 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 3 |
| ENSG00000198821 | CD247 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 3 |
| ENSG00000116117 | PARD3B | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 |
| ENSG00000133065 | SLC41A1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 |
| ENSG00000141258 | SGSM2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| ENSG00000144642 | RBMS3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 |
| ENSG00000164362 | TERT | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| ENSG00000167720 | SRR | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 |
| ENSG00000198131 | ZNF544 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| ENSG00000069275 | NUCKS1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000069667 | RORA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000103653 | CSK | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000110944 | IL23A | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000112294 | ALDH5A1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000117280 | RAB7L1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000128815 | WDFY4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000128915 | NARG2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000135469 | COQ10A | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000135823 | STX6 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| ENSG00000135903 | PAX3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| ENSG00000137261 | KIAA0319 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000139540 | SLC39A5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| ENSG00000139645 | ANKRD52 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000144785 | RP11-977G19 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000152595 | MEPE | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| ENSG00000160185 | UBASH3A | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000183354 | KIAA2026 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000204287 | HLA-DRA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000231389 | HLA-DPA1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000237241 | RP11563N6.4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000238809 | snoU13 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000245534 | RP11-219B17 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000259462 | RP11-752G15 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ENSG00000261801 | RP11-941F15 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Gene | Drug | Original Indication | NCT Number |
|---|---|---|---|
| JAK2 | Tofacitinib | Severe Rheumatoid arthritis | NCT03002649 |
| JAK2 | Baricitinib | Severe Rheumatoid arthritis | NCT05361109 |
| FCGR3B | Human immunoglobulin G | Thrombocytopenic purpura | NCT02728752 |
| CD4 | Antithymocyte immunoglobulin | Rejection Acute Renal | NCT00010335 |
| IFNAR1 | Interferon alfa-n1 | Genital warts | NCT00533091 |
| IFNAR1 | Human interferon beta | Multiple Sclerosis | NCT05192200 |
| JAK1 | Tofacitinib | Rheumatoid arthritis | NCT03002649 |
| JAK1 | Baricitinib | Rheumatoid arthritis | NCT05361109 |
| IFNAR2 | Interferon alfa-n1 | Genital warts | NCT00533091 |
| Target Gene | Drug | PMID |
|---|---|---|
| JAK1, JAK2 | Ruxolitinib | 26448614 |
| Tofacitinib | 33258553 | |
| Upadacitinib | 35081305 | |
| Baricitinib | 35318646 | |
| Filgotinib | 32222877 | |
| IFNAR1, IFNAR2 | Human interferon beta | 27564228 |
| Interferon alfa-2a | 24638953 | |
| Interferon beta-1a | 18936398 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irham, L.M.; Adikusuma, W.; La’ah, A.S.; Chong, R.; Septama, A.W.; Angelina, M. Leveraging Genomic and Bioinformatic Analysis to Enhance Drug Repositioning for Dermatomyositis. Bioengineering 2023, 10, 890. https://doi.org/10.3390/bioengineering10080890
Irham LM, Adikusuma W, La’ah AS, Chong R, Septama AW, Angelina M. Leveraging Genomic and Bioinformatic Analysis to Enhance Drug Repositioning for Dermatomyositis. Bioengineering. 2023; 10(8):890. https://doi.org/10.3390/bioengineering10080890
Chicago/Turabian StyleIrham, Lalu Muhammad, Wirawan Adikusuma, Anita Silas La’ah, Rockie Chong, Abdi Wira Septama, and Marissa Angelina. 2023. "Leveraging Genomic and Bioinformatic Analysis to Enhance Drug Repositioning for Dermatomyositis" Bioengineering 10, no. 8: 890. https://doi.org/10.3390/bioengineering10080890
APA StyleIrham, L. M., Adikusuma, W., La’ah, A. S., Chong, R., Septama, A. W., & Angelina, M. (2023). Leveraging Genomic and Bioinformatic Analysis to Enhance Drug Repositioning for Dermatomyositis. Bioengineering, 10(8), 890. https://doi.org/10.3390/bioengineering10080890










