Abstract
MicroRNAs (miRNAs) comprise a class of non-coding RNA with extensive regulatory functions within cells. MiR-106a is recognized for its super-regulatory roles in vital processes. Hence, the analysis of its expression in association with diseases has attracted considerable attention for molecular diagnosis and drug development. Numerous studies have investigated miR-106 target genes and shown that this miRNA regulates the expression of some critical cell cycle and apoptosis factors, suggesting miR-106a as an ideal diagnostic and prognostic biomarker with therapeutic potential. Furthermore, the reported correlation between miR-106a expression level and cancer drug resistance has demonstrated the complexity of its functions within different tissues. In this study, we have conducted a comprehensive review on the expression levels of miR-106a in various cancers and other diseases, emphasizing its target genes. The promising findings surrounding miR-106a suggest its potential as a valuable biomolecule. However, further validation assessments and overcoming existing limitations are crucial steps before its clinical implementation can be realized.
1. Introduction
MicroRNAs (miRNAs) are small, endogenous, non-coding RNAs that control gene expression at the translation and even transcription levels. miRNAs are critical regulators of biological processes, including cellular proliferation, differentiation, development, apoptosis, and modulation of the host response to viral infection [1,2]. Moreover, extracellular miRNAs have been widely reported as potential biomarkers for different diseases and disorders while also serving as signaling molecules to mediate cell–cell communications [3,4].
The biogenesis process of these tiny biomolecules generally begins with RNA polymerase II/III activity and can be classified into canonical and noncanonical pathways. While the intron and exon parts of the genome are involved in processing intragenic miRNAs, intergenic miRNAs are believed to be transcribed independently of genes and regulated by their own promoters [5]. miRNAs may be transcribed in the form of long transcripts named clusters. If the clusters share similar seed regions, they are considered to be an miRNA family [4,5].
In this review, we focus on miR-106a, a multi-faceted miRNA, and its regulating role in vital cell processes [6]. miR-106a is a member of the miR-17 family and a part of the miR-106a-363 cluster located on the X chromosome [7]. As listed in Table S1 in Supplementary Materials, there are 1337 predicted targets for hsa-miR-106a-5p in miRDB, with target scores ranging from 100 to 50 (higher miRDB scores generally indicate a stronger likelihood of a functional miRNA-mRNA interaction, suggesting a higher probability that the miRNA can regulate the expression of the targeted mRNA) [8,9]. Considering that the majority of identified target genes are implicated in crucial processes such as cell cycle regulation, drug resistance, tumor progression, and the inflammatory response, it is expected that the dysregulation of miR-106a is associated with a wide range of biological phenomena, diseases, disorders, and malignancies (Figure S1 in Supplementary Materials). In addition, miR-106a regulates several genes involved in cell signaling pathways. However, confirmation of some of these genes still needs further clarification and verification [2,10].
2. mir-106a: Dysregulation in Cancers
2.1. Colorectal Cancer
Colorectal cancer (CRC) is the third most common cancer with the second-highest global mortality rate [11]. Despite progress in its diagnosis and treatment, the overall 5-year survival rate is 40%, and approximately half of the patients die due to the development of distant metastases [12].
More than 200 miRNAs have been found to be dysregulated in CRC, according to the literature and the miRCancer website [12,13]. A significant number of studies have reported the overexpression of miR-106a in plasma, cancer tissues, and stool samples of CRC patients via real-time RT-PCR and microarray analyses [14,15,16,17]; however, the precise role played by miR-106a in CRC is not completely clear. In a study by Feng et al., the authors studied 990 targets of miR-106a predicted by TargetScan and indicated that transforming growth factor-β receptor 2 (TGFBR2) is the direct functional target of miR-106a. As a member of the primary TGF-β signaling molecules, TGFBR2 expression is frequently altered in cancer cell lines through induction of EMT and promotion of tumor cell invasion. The results of this study showed that miR-106a was positively correlated with the migration and invasion potential of CRC cells by downregulating TGFBR2 [2,18,19]. In another study, Hao et al. investigated the correlation between miR-106a and autophagy-related gene 7 (ATG7) in CRC. The obtained results confirmed that miR-106a downregulates ATG7 mRNA, leading to the suppression of tumor cell death [14].
Phosphatase and tensin homolog deleted on chromosome ten (PTEN) is a tumor suppressor downregulated by miR-106a overexpression in CRC. The PTEN/PI3K/AKT signaling pathway (Figure 1) was identified as another target for miR-106a in CRC in research published by Qin et al. [20]. Evidence indicates that the increased activity of PI3K/AKT following PTEN reduction correlates with cell proliferation, migration, invasion, and apoptosis [21,22]. In addition, the inactivation of PTEN results in malignant progression, including advanced stage in CRC [23].
Despite studies emphasizing the overexpression of miR-106a in CRC, Huang and Ma performed an in vitro experiment using cultured CRC cells. Their results showed the tumor-suppressive function of miR-106a in CRC potentially works through regulating E2F1 and caspase-9 expression [24].
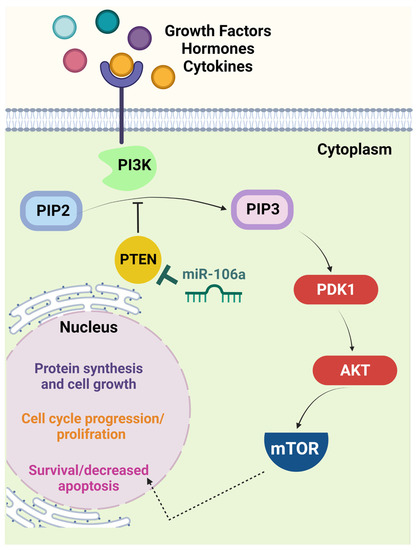
Figure 1.
The PTEN/PI3K/AKT signaling pathway is a crucial target of miR-106a. In the presence of specific growth factors, hormones, or cytokines, interaction with the cell surface receptor activates the receptor complex and PI3K. PI3K then phosphorylates PIP2 to PIP3, which recruits PDK1 to activate AKT. Activated AKT interacts with different molecules, including mTOR. The latter molecule induces cell proliferation and limits apoptosis. PTEN is one of the principal negative regulators of this pathway via reversing the conversion of PIP2 to PIP3. This function makes PTEN a critical tumor suppressor, the expression of which is diminished in many malignancies via different mechanisms, including miR-106a-mediated downregulation. Thus, loss of PTEN correlates with uncontrolled cell cycle progression and probable tumorigenesis [25]. (Created with BioRender.com, accessed on 14 December 2022). AKT, serine/threonine protein kinase B; mTOR, mammalian target of rapamycin; PDK1, phosphoinositide-dependent protein kinase 1; PI3K, phosphatidylinositol 3-kinase; PIP2, phosphatidylinositol diphosphate; PIP3, phosphatidylinositol trisphosphate; PTEN, phosphatase and tensin homolog deleted on chromosome 10.
2.2. Cholangiocarcinoma
Cholangiocarcinoma (CCA) is a malignant tumor originating from the bile duct epithelium with frequent metastasis to the lymph node [26]. However, since its pathogenesis mechanism and provoking factors are still unclear, patients often reach the advanced stages when diagnosed with CCA. For CCA patients, radical resection is almost the only choice, but certainly not a viable one [26,27].
In interesting research by Cheng et al., a pooled analysis was performed to identify a clinically valid miRNA in CCA patients. Their results indicated that the circulating level of miR-106a was significantly lower in CCA patients with a higher susceptibility to lymph node metastasis [26]. However, these findings were not corroborated at the tissue level when assessing miR-106a expression. This paradox may stem from the potential influence of the tumor microenvironment [26]. The origin of circulating miRNAs has been the subject of several studies and remains a topic of debate, and researchers have discussed that serum miRNA levels may not only result from tumors but also from the immune response [26].
Regarding the current data obtained from several studies, the diagnostic value of miR-106a for CCA patients seems clear. Additionally, miR-106a is a possible prognostic biomarker in CCA patients thanks to presenting as a clinically promising indicator for evaluating lymph node metastasis risk [28].
2.3. Ewing Sarcoma
Ewing sarcoma (EWS) is the second most common malignancy related to solid bone and soft tissue in children and young adults [29]. Most EWS tumors harbor t(11,22)(q24:12) chromosomal translocation followed by EWS-FLI1 gene fusion. This fusion acts as an aberrant transcription factor, interacts with RNAs, and dysregulates cellular processes, such as cell cycle progression, and TGF-β signaling [29].
Several studies on the role of miRNAs in EWS have highlighted the upregulation of miR-106a in this malignancy. There is evidence indicating the oncogenic behavior of miR-106a leads to increased expression of Bim (BCL-2 interacting mediator of cell death) and decreased expression of CDK4/CDK6 [29,30,31]. Moreover, the results published by Dylla et al. showed that targeting miR-106a could not effectively inhibit tumor development in responsive EWS cell lines. They suggested that inhibition of the entire miR-106a~363 cluster may be required to obtain significant results [2,30,31].
2.4. Gastric Cancer
Gastric cancer (GC) is the fourth leading cancer and third cause of cancer-related deaths worldwide. Although research on gastric cancer has made significant advancements, the molecular mechanisms underlying cancer invasion and metastasis are still poorly understood [32]. Due to a lack of symptoms in the early stages, most patients with GC are diagnosed with advanced disease or distant metastasis. According to research, early detection in gastric cancer patients leads to a better prognosis and a 5-year survival rate of more than 90% [32,33].
Recently, many ectopically expressed miRNAs have been reported to be involved in the initiation and progression of gastric cancer as novel proto-oncogene and tumor-suppressor genes. The expression level of miR-106a increases and can be detected in tumor tissue, blood, fecal, and gastric fluids [34,35]. Moreover, it has been revealed that miR-106a levels are significantly associated with tumor stage, size, and differentiation, lymphatic and distant metastasis, and invasion [33,36].
Zhu et al. used qualitative, quantitative, and positioning analyses to investigate the biological role of miR-106a in GC. The results demonstrated that the upregulation of miR-106a significantly enhanced GC cell proliferation, migration, and invasion by directly targeting TIMP2. TIMP2, a metallopeptidase inhibitor, plays a crucial role in suppressing endothelial cell proliferation [34]. Moreover, in a study by Wang et al., rescue experiments and examination of caspase-8, PARP, and caspase-3 demonstrated that miR-106a could inhibit gastric cancer cell apoptosis by interfering with the FAS-mediated apoptotic pathway [37]. A significant inverse correlation was also found between miR-106a and FAS expression in gastric cancer cell lines and specimens. Therefore, these findings suggest that ectopically overexpressed miR-106a plays an oncogenic role in gastric carcinogenesis by impairing extrinsic apoptotic pathways via targeting FAS [37,38]. The confirmed target genes of miR-106a in intrinsic and extrinsic apoptotic pathways are shown in Figure 2.
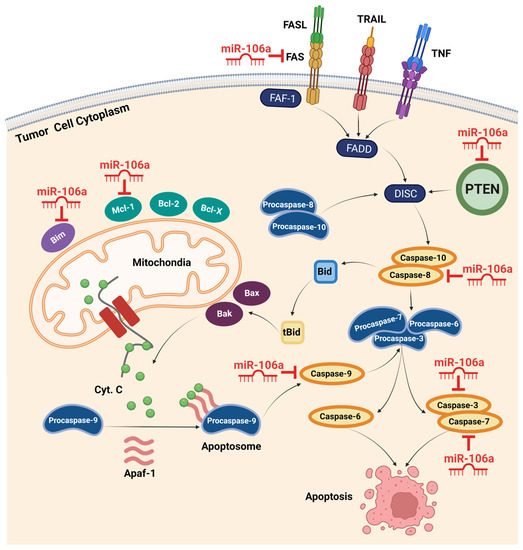
Figure 2.
Intrinsic and extrinsic apoptotic pathways and confirmed target genes of miR-106a. The extrinsic pathway is mediated by death ligands and death receptors, including FASL and FAS. Activation of death receptors initiates a series of reactions with final stimulation of caspases 3, 6, and 7, resulting in apoptosis. miR-106a can hinder apoptosis by suppressing multiple members of this pathway, including FAS, PTEN, and caspase-8, -3, and -7. The intrinsic pathway incorporates mitochondria in promoting apoptosis. Activating this pathway in response to different stresses, such as oncogenic activity, stimulates mitochondrial membrane proteins to release cytochrome C into the cytoplasm. Recruiting Apaf-1 and cytochrome C, procaspase-9 forms the apoptosome complex and converts to active caspase-9. By activating other apoptotic caspases, the cell consequently heads to apoptosis. miR-106a overexpression inhibits this pathway via targeting Bim, Mcl-1, and caspase-9 in different cancers. (Created with BioRender.com, accessed on 14 December 2022). Apaf-1, apoptotic peptidase activating factor 1; Bak, Bcl-2-antagonist killer; Bax, Bcl-2-associated x protein; Bcl-2, B-cell lymphoma 2; Bcl-x, B-cell lymphoma-extra-large; Bid, BH3 interacting-domain death agonist; Bim, Bcl-2 interacting mediator of cell death; DISC, death-inducing signaling complex; FADD, FAS-associated death domain; FAF-1, FAS-associated factor 1; FASL, FAS ligand; Mcl-1, myeloid cell leukemia 1; PTEN, phosphatase and tensin homolog deleted on chromosome 10; TNF, tumor necrosis factor; TRAIL, TNF-related apoptosis-inducing ligand.
Recent research has revealed that certain long noncoding RNAs (lncRNAs) function as competing endogenous RNAs (ceRNAs) to regulate gene expression [39]. These lncRNAs, along with other RNAs, act as miRNA sponges by sharing miRNA response elements (MREs), thereby modulating intracellular miRNA function [32]. Given that noncoding RNAs are predominantly untranslated, they are presumed to be highly effective ceRNAs. One such lncRNA, FER1L4 (long noncoding RNA Fer-1 protein 4), has been shown to play crucial regulatory roles in tumor progression [40]. FER1L4 functions as a ceRNA by sequestering miR-106a-5p in gastric cancer, thereby impacting tumor suppression. Consequently, this ceRNA pathway can ultimately regulate the expression levels of several critical genes, including PTEN, RB1, RUNX1, VEGFA, CDKN1A, E2F1, HIPK3, IL-10, and PAK7, through lncRNA-FER1L4 [40,41].
The development of multidrug resistance (MDR) in gastric cancer poses significant challenges in effective treatment strategies. Dysregulation of key molecular pathways and the involvement of specific microRNAs have been implicated in MDR mechanisms. In this context, the role of miR-106a and its impact on the PTEN/Akt pathway and downstream targets have gained considerable attention.
PTEN, encoding a phosphatase involved in the negative regulation of the Akt pathway by dephosphorylating PIP3 and counteracting PI3K activity, has emerged as one of the main targets of miR-106a, particularly in MDR GC [41,42]. Studies have reported that miR-106a overexpression leads to PTEN knockdown, resulting in dysregulation of the PTEN/Akt pathway. This dysregulation has been associated with the development of MDR in cisplatin-treated GC patients [41,42].
Further investigations have shed light on additional mechanisms through which miR-106a induces MDR in GC patients. It has been observed that miR-106a targets P-glycoprotein (P-gp), an ATP-binding cassette transporter responsible for drug efflux [42]. Moreover, miR-106a overexpression correlates with the downregulation of runt-related transcription factor 3 (RUNX3), a tumor suppressor known to inhibit cell progression and tumorigenesis (Guo et al., 2005). According to Guo et al., RUNX3 prevents the expression of MDR-1, Bcl-2, and multidrug resistance protein-1, while enhancing the sensitivity of GC cells to chemotherapeutic drugs [43]. The dysregulation caused by miR-106a actively promotes MDR in GC cells by desensitizing tumor cells to anticancer drugs, increasing the efflux of chemotherapeutic agents, and suppressing GC cell apoptosis through the dysregulation of tumor suppressor genes [42,43].
2.5. Esophageal Carcinoma
Esophageal cancer is among the malignancies with high mortality worldwide [44]. Esophageal cancer is mainly classified as esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC). ESCC is responsible for >90% of esophageal cancer and is the most aggressive carcinoma of the gastrointestinal tract [45,46].
Since the first study on the levels of serum miRNAs in ESCC patients was published [47], the differential expression of circulating miRNAs and the potential application of biomarkers in ESCC have gained considerable attention in the cancer research field [44,46]. One study analyzed the expression level of miR-106a in the tissues of 21 ESCC patients and reported its reduction in patients that developed recurrent disease or had a tumor-related death [48]. However, because of the small number of examined samples, these findings were not reliable.
In a study conducted by Zhou et al., an miRNA signature consisting of five upregulated miRNAs (miR-106a, miR-18a, miR-20b, miR-486-5p, and miR-584) and one down-regulated miRNA (miR-223-3p) was identified as a potential biomarker for the diagnosis of ESCC. The study utilized PCR assays to demonstrate that the levels of miR-106a and miR-584 were consistently elevated in both the plasma and tissue samples of ESCC patients. These findings suggest that the plasma levels of these two miRNAs hold promise as potential screening biomarkers for ESCC [44]. However, further investigations are necessary to identify the specific target genes or functional pathways associated with miR-106a in this particular cancer in order to establish the clinical validity of the reported results.
2.6. Renal Carcinoma
Renal cell carcinoma (RCC) is a highly lethal form of genitourinary cancer, comprising approximately 90% of all renal malignancies, with an annual increase in the incidence of 2%–3% [49]. The major problems of RCC are the unsatisfactory postoperative survival rate and poor responsiveness to radiotherapy and chemotherapy [49].
The regulatory role of miRNAs and their dysregulation in RCC have been investigated in numerous studies and miR-106a has been identified as a tumor suppressor with dramatically decreased levels in the plasma and tissue of RCC patients. It was shown that increased miR-106a levels inhibited the proliferation of RCC cells and prevented S/G2 transition [50].
It was also found that miR-106a expression was inversely correlated with the expression of insulin receptor substrate 2 (IRS-2) [51]. Insulin receptor substrates (IRSs), comprising six members (IRS-1-6) [52,53], are signaling adaptor proteins. IRSs are immediate downstream effectors of insulin-like growth factor-1 (IGF-1), insulin receptors, prolactin, growth hormone, cytokine, VEGF receptors, and integrin receptor family members [50,51]. The results reported by Ma et al. indicated that IRS-2 was the functional and direct target of miR-106a. Therefore, downregulation of miR-106a in RCC activates IRS-2, which is involved in the PI3K/Akt signaling pathway in different cancer model systems. This signaling pathway has been implicated in promoting tumor cell proliferation, migration, invasion, and survival [51].
PAK5 is a serine/threonine kinase downstream of Rho GTPases. Many studies have found that PAK5 plays a crucial role in promoting cell migration and invasion in tumorigenesis [54,55,56]. Due to harboring a highly conserved p21-GTPase-binding domain, PAK5 may easily interact with a Rho GTPase (GTP-binding Cdc42) and other Rho GTPases like Rho and Rac that affect the cytoskeleton, cell dynamics, and cell morphology. In addition, PAK5-mediated signaling pathways such as PAK5-Egr1-MMP2 are involved in tumor cell migration and invasion [54,56]. Pan et al. identified PAK5 as a target of miR-106a, which promotes the malignancy process in RCC via reduction of its miRNA levels [49].
Furthermore, it was reported that miR-106a-5p directly targets the 3′-UTR of VEGFA mRNA and decreases VEGFA expression, followed by degrading effects on tumor cell growth and colony formation [57]. Therefore, the low level of miR-106a in RCC tissues was correlated with the upregulation of VEGFA, which contributes to angiogenesis and tumor development [57,58].
2.7. Lung Cancer
Non-small cell lung cancer (NSCLC), which represents about 80% of all lung cancer cases, is a leading cause of cancer death in the developed world. Less than 15% of NSCLC patients live more than five years after diagnosis [59].
To study the principal miRNAs involved in NSCLC, Heegaard et al. examined serum and plasma samples from 220 patients with early-stage NSCLC and 220 matched controls using qRT-PCR. They found that the expression levels of miR-146b, miR-221, let-7a, miR-155, miR-17-5p, miR-27a, and miR-106a were significantly reduced in the serum of NSCLC cases while that of miR-29c was considerably increased. However, no significant differences were observed in the plasma of patients compared to controls, and expression levels in serum and plasma did not correlate well [60].
By studying the miRNAs dysregulated in NSCLC tissues and cell lines, Xie et al. confirmed miR-106a upregulation and identified PTEN as a target for this miRNA. This study suggested that miR-106a inhibited the growth and metastasis of NSCLC cells by decreasing PTEN expression [61].
Adenosine triphosphatase-binding cassette A1 (ABCA1) is a member of the ABC transporter family and is fundamental to cellular metabolism. ABCA1 is also one of the main players connected to miR-106a-induced drug resistance in NSCLC. Lee et al. [62] discovered that cancer-specific hypermethylation and downregulated expression of ABCA1 contributed to an elevated level of intracellular cholesterol, followed by enhanced vulnerability to tumor progression. These findings indicated the anticancer role of cholesterol exporter ABCA1 and its impact on tumor development, which were supported by the results of Smith and Land [63]. Their research provided evidence suggesting that the restoration of ABCA1 expression could inhibit tumor development [63]. In this regard, Ma et al. reported that the upregulation of miR-106a expression levels was associated with cisplatin (DDP) resistance by targeting ABCA1. It was also confirmed that the modulation of miR-106a in DDP-treated cells re-sensitized them to DDP [64].
2.8. Pancreatic Cancer
Pancreatic cancer (PC) is one of the leading causes of cancer-related mortality due to poor early diagnosis and imprecise prognosis [65]. The lack of warning signs at the initial stages of PC leads to late and commonly inefficient interventions, followed by a low survival rate [65]. Studies, however, indicate that detection of PC before reaching the ultimate stages or metastatic level, followed by surgical resection and chemotherapy, might notably increase the survival rate of patients (Rawat et al., 2019).
The potential of miRNAs as PC biomarkers has been broadly investigated (Vieira et al., 2021; Yan et al., 2020). miRNA dysregulation patterns can be practically evaluated to differentiate PC from other pancreatic diseases, such as pancreatitis and benign pancreatic masses, compared to healthy samples (Vieira et al., 2021; Yan et al., 2020).
Studies investigating the molecular mechanisms underlying the impact of miR-106a in pancreatic cancer have revealed additional key insights [65,66]. It has been observed that miR-106a promotes pancreatic tumorigenesis by directly targeting multiple genes involved in tumor suppression and cellular proliferation pathways [65,66]. For instance, miR-106a was been found to target retinoblastoma protein (RB1), a critical tumor suppressor involved in cell cycle regulation. By inhibiting RB1 expression, miR-106a disrupts cell cycle control and promotes uncontrolled cell growth in pancreatic cancer cells [66]. Moreover, miR-106a overexpression in PC is correlated with pancreatic tumorigenesis by inducing cancer cell proliferation, epithelial–mesenchymal transition, and invasion by targeting tissue inhibitor of metalloproteinase-2 (TIMP-2) [66]. These molecular interactions highlight the intricate role of miR-106a in driving pancreatic cancer progression through the modulation of key regulatory pathways.
2.9. Ovarian Cancer
Ovarian cancer (OC) is the most lethal gynecological malignancy commonly diagnosed at an advanced stage with extensive peritoneal metastases due to the absence of warning symptoms and lack of effective screening methods at the early stage [67]. Investigating the miRNAs dysregulated in OC led to the identification of miR-106a as an oncomiR that is overexpressed in tumor tissue samples and cell lines and facilitates cell growth and metastasis by reducing PTEN expression [68]. Chen et al. also indicated that IL-6 is a regulator of miR-106a and inhibits miR-106a expression by activating STAT3 [68]. In a series of experiments using the OncomiR database, western blotting, and luciferase reporter assays, Chao et al. validated Rho GTPase-activating protein 24 (ARHGAP24) as a direct target of miR-106a. The expression of ARHGAP24 protein was suppressed by the overexpression of miR-106a-5p [67].
In a study by Liu et al., miR-106a upregulation was indicated in high-grade serous ovarian carcinomas (HGSOC) based on the real-time reverse transcriptase PCR (qRT-PCR) results and miRNA in situ hybridization in a large cohort study of HGSOC samples. They additionally showed the correlation of miR-106a with HGSOC tumor growth and differentiation both in vivo and in vitro, confirming p130 (RBL2), a retinoblastoma (Rb) tumor suppressor family member, as a specific target of miR-106a [69].
The role of miR-106a in the drug resistance behavior of OC-involved tissue has also been explored, revealing that miR-106a-mediated resistance of ovarian cancer cell line A2780 to the chemotherapeutic agent cisplatin (DDP) was accomplished through targeting Mcl-1 [70]. Reports showed that the Mcl-1 gene was involved in cisplatin and paclitaxel resistance in OC. Mcl-1, a member of the Bcl-2 family, contributes to the ability of cells to survive and evade the toxic effects of the drug [70,71,72].
The association between PDCD4 (programmed cell death 4) protein expression level and miR-106a upregulation in the ovarian cancer OVCAR3 cell line and the cisplatin (DDP)-resistant ovarian cancer OVCAR3/CIS cell line was studied using stem-loop qPCR [73]. PDCD4, a tumor suppressor, reportedly inhibited tumorous transformation, progression, and translation. By interacting with such factors as RNA helicase eIF4A and scaffold protein eIF4G, PDCD4 inhibits protein synthesis by suppressing translation initiation [2]. The results demonstrated that PDCD4 is a target of miR-106a and played a role in DDP resistance in the OVCR3 cell line. Furthermore, the knockdown of PDCD4 significantly increased the cell survival rate and had an overall effect similar to miR-106a overexpression [73].
Moreover, through immunoblotting and luciferase assays, Huh et al. identified BCL10 and caspase-7 as direct target genes of miR-106a downregulation in OC. The authors suggested that the chemoresistance associated with miR-106a in the OC cell line may be induced by direct regulation of BCL10 and/or caspase-7 [71].
Guo et al. conducted investigations on the lncRNA X inactive specific transcript (XIST) and observed its correlation with OC and miR-106a expression [74]. Although the XIST mechanism of action in OC still requires validation and in vivo proof, it is clear that its expression declines as OC progresses. Since the interaction between miR-106a and XIST has been shown using various techniques, such as dual-luciferase reporter and RNA pull-down assays, XIST is considered another target for miR-106a in OC. Further in vitro and in vivo results have also demonstrated the probable therapeutic potential of XIST in restoring tumor cell apoptosis via sponging miR-106a [74].
2.10. Brain Tumors
Tumors that originate in the brain are classified as primary brain tumors, which can be either benign or malignant. Approximately half of all primary brain tumors arise from glial cells, collectively named gliomas. The World Health Organization (WHO) has divided gliomas into two categories: low-grade (WHO grades I and II: pilocytic astrocytoma and diffuse astrocytoma, respectively) and high-grade (WHO grades III and IV: anaplastic astrocytoma and glioblastoma multiforms (GBMs), respectively) [75,76].
Other types of brain tumors include choroids plexus tumors, primitive neuroectodermal tumors (PNET, e.g., medulloblastoma, neuroblastoma, retinoblastoma, pineoblastoma), tumors originating from neuronal cells (gangliocytoma, central neurocytoma), mixed glioneuronal tumors (tumors displaying a neuronal as well as a glial component, such as ganglioglioma), and dysembryoplastic neuroepithelial tumors (DNET) [76].
The role of miRNAs and their dysregulation in brain tumors have been extensively investigated in separate studies and miR-106a has an undeniable but unclear effect on brain tumor development and progression [75,76,77] Although different researchers have reported the overexpression of miR-106a in brain tumors [7,77,78,79], there are studies paradoxically indicating the tumor suppression behavior of this miRNA in GMB and astrocytoma [80,81,82,83].
According to published data of glioblastoma cell cultures, it is currently believed that miR-106a promotes tumor cell invasion via activation of the WNT signaling pathway and altering β-catenin cellular localization [78]. Binding β-catenin to the promoter regions of a series of oncogenes, including MMP2, NANOG, MYC, RUNX2, CD44, SOX9, and OCT4, is functionally related to cell invasion [78]. Wang et al. have also explored two functionally related microRNAs, miR-20a and miR-106a, in human glioma stem cells (GSCs) and found that these two molecules were significantly overexpressed in related tissues and enhanced the invasiveness of CD133 + GSCs by directly targeting TIMP-2 [79].
On the other hand, one of the first studies reporting the tumor-suppressive effect of miR-106a in GBM demonstrated that the low expression of miR-106a in human glioma specimens is associated with the accumulation of E2F1 protein and high-grade glioma [81]. The authors identified E2F1 as a direct functional target of miR-106a, suggesting that the effect of miR-106a on the glioma suppressive effect may result from the inhibition of E2F1 via posttranscriptional regulation. In addition, their results revealed that miR-106a could increase p53 expression via E2F1 inhibition, whereas the effect of miR-106a on the proliferation of glioma cells was independent of p53 status [81]. Furthermore, Dai et al. reported the tumor suppressor role of miR-106a and its involvement in GBM cell proliferation and even glucose uptake by targeting SLC2A3. In addition, they showed that the downregulation of miR-106a in GBM tissues leads to poor survival in GBM patients [80].
Significant decreases in miR-106a levels have also been reported in brain tumor types other than GBM. It was revealed that miR-106a targets FASTK in astrocytoma cells and inhibits their proliferation and migration while inducing apoptosis [83]. Jones et al. reported the upregulation of miR-106a in PA and suggested its effect via targeting the MAPK and NF-κB pathways [84]. Some studies demonstrated that overexpression of miR-92, miR-106a, miR-17-5p, and miR-93 in neuroblastoma was functionally linked to MYCN amplification. These miRNAs were correlated with MYCN amplification and activation [85].
To explore the effects and mechanisms of miR-106a on MDR reversal in human glioma cells, Wang et al. knocked down miR-106a in the cisplatin-resistant U87 (U87/DDP) and gefitinib-resistant U251 (U251/G) glioma cell lines and evaluated their sensitivity to drugs, apoptosis rate, and rhodamine 123 content. In addition, they detected decreased expression levels of P-gp, MDR1, MRP1, GST-π, CDX2, ERCC1, RhoE, Bcl-2, Survivin, and Topo-II, as well as reduced production of IL-6, IL-8, and TGF-β in these cell lines. Furthermore, they found decreased expression of p-AKT and transcriptional activation of NF-κB, Twist, AP-1, and Snail in these cell lines. These results suggest that miR-106a is a promising therapeutic target for treating human multidrug-resistant glioma [86].
2.11. Breast Cancer
Breast cancer (BC) is the most common cancer among women worldwide, accounting for 25%–30% of all newly diagnosed cancer and exhibiting the second highest mortality among all cancer types in women [87]. Despite advances in early detection and diagnosis, breast cancer’s overall prevalence and survival rate have only marginally improved. Developed countries have maintained a high incidence of breast cancer, whereas the incidence of breast cancer in the developing world is also increasing due to lifestyle changes and longer life expectancies [87,88].
There is not much information published regarding the role of miR-106a in BC. However, recent reports have shown that miRNA-106a promotes breast cancer cell proliferation, clonogenicity, migration, and invasion by inhibiting apoptosis and chemosensitivity [88,89,90]. To identify candidate miRNAs as BC diagnostic biomarkers, Li et al. accomplished a three-phase study to evaluate the expression of 12 miRNAs from the miR-106a-363 cluster in plasma and serum samples. The candidate miRNAs identified utilizing qRT-RCR were subjected to further analysis in breast tissue, plasma exosomes, and serum exosomes. The results showed the significant elevation of miR-106a expression was associated with specific clinical parameters in plasma and serum, indicating the potential roles of this miRNA in BC pathogenesis [89]. MiR-106a overexpression additionally upregulated the levels of Bcl-2 and ABCG2 and downregulated the expression of P53, Bax, and RUNX3 [88]. In an interesting study by Yang et al., miR-106a was among the miRNAs that target ZBTB4a, a transcriptional repressor that inactivates genes through binding their respective GC-rich promoters. These results suggested miR-106a indirectly regulates the enhancer of zeste homolog 2 (EZH2) and its overexpression is inversely correlated with ZBTB4, as has been observed in multiple cancers [91]. Recently, Liu et al. identified DAX-1 as a direct target of miR-106a in BC. DAX-1 is a member of the atypical nuclear receptor family that is downregulated in BC. The authors showed that miR-106a was responsible for promoting the invasion, migration, and proliferation of BC cells by targeting DAX-1. Therefore, the expression level of this miRNA is closely correlated to BC stages, distant metastasis, lymph node metastasis, and poor prognosis [90].
2.12. Endometrial Adenocarcinoma
Endometrial cancer (EMC) is a gynecological malignancy with increasing incidence because of today’s lifestyle, particularly in developed countries [92,93].
The evaluation of miRNA expression patterns in EMC cells compared to normal tissue has indicated significant alterations and identified multiple miRNA candidates as diagnostic and prognostic biomarkers [92,94]. In a differential analysis-based study by Wang et al., miR-106a was reported among the five specific prognostic miRNA markers involved in constructing a prognostic model using machine learning [95].
In addition, it was reported that BCL2L11 is one of the main miR-106a targets in EMC. BCL2L11 is a known regulator in the cell cycle and apoptosis. Therefore, miR-106a overexpression correlated with tumor growth, migration, and invasion as the result of BCL2L11 downregulation in EMC tumor cells [96]. Other reports also validated these findings, where knocking down miR-106a notably attenuated EMC cell proliferation and tumor invasiveness while inducing apoptosis [93,95].
Furthermore, a study by Oplawski et al. provided evidence of miR-106a’s involvement in the epithelial–mesenchymal transition (EMT) in endometrial cancer. The researchers not only identified BCL2L as a target of miR-106a but also highlighted its association with EMT [97].
2.13. Cervical Cancer
Despite widespread screening for cervical cancer (CC) and significant advances in the development of vaccines, cervical cancer frequently occurs in women worldwide, with high mortality in developing countries [98,99]. Currently, there is no effective treatment for advanced-stage or recurrent cervical cancer. Thus, the search for therapeutically functional molecules or effective targets is critical. Pathogenesis studies have provided reliable evidence for promoting the role of human papillomavirus (HPV) infection in the tumorigenesis of CC [100]. Additional factors, such as changes in levels of cellular proteins that are crucial regulators in certain molecular signaling pathways, are involved in the progression of HPV-infected lesions to cancer [100,101].
There are few reports concerning the dysregulation of miR-106a in CC. However, published results indicate the overexpression of miR-106a in CC cell lines and tissues [98,101,102]. It was revealed that the upregulation of miR-106a was associated with increases in cell migration, invasion, and invasion-related gene expression [98,102]. In addition, miR-106a regulates TIMP-2 by directly binding to its 3’-UTR and inducing the expression of matrix metalloproteinases (MMPs) [102]. On the other hand, the results of clinical sample analysis showed an inverse correlation between miR-106a and TIMP-2 expression since re-expression of TIMP-2 led to the restriction of tumor cell migration, invasion, and MMP expression in CC cells [102].
In an interesting study by Cui et al., the role of miR-106a was studied in HPV-16-positive CC patients. The results showed the overexpression of miR-106a in both HPV-16-positive CC tissues and cell lines [101]. Liver kinase B (LKB1) was also reported as a newly discovered target of miR-106a. MiR-106a appears to play an oncogenic role by targeting the AMPK-mTOR pathway via silencing LKB1. Furthermore, HPV-16 E7 was found to upregulate miR-106a in both E7-expressing and cervical cancer cells [101].
2.14. Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is one of the most diagnosed malignancies and represents the second leading cause of cancer-related deaths [103,104]. Despite improvements in surgery and other treatments, the overall 5-year survival rate for HCC patients remains extremely low, mainly caused by the tumor’s late detection. Another important reason for poor outcomes is tumor metastasis and recurrence [105,106,107].
As Yuan et al. reported, the overexpression of miR-106a by its promoter hypomethylation contributes to the progression of HCC. The observations confirmed the downregulation of TIMP-2, TP53INP1, and CDKN1A expression in HCC tissues, and these genes were considered as possible miR-106a direct targets [104].
It is known that chemotherapeutic drug gemcitabine-resistant (GR) HCC cells acquire EMT characteristics. EMT, which stands for epithelial–mesenchymal transition, is a phenomenon in which epithelial cells convert into mesenchymal cells through losing epithelial cell–cell junction and epithelial markers (E-cadherin and γ-catenin) while inducing mesenchymal markers (Twist, Vimentin, Snail, and Slug) and is associated with facilitating migration and invasion of HCC tumor cells [108]. Some studies confirmed the drug-resistance role of miR-106a in HCC [105,108,109].
FBXW7 (F-box and WD repeat domain containing 7) is a protein downregulated in some malignancies, including HCC. The published findings indicated the involvement of FBXW7 in suppressing tumor cell migration, invasion, and MDR in cancers [109]. The correlation between FBXW7 and miR-106a in HCC has been assessed by adopting various molecular and cellular methods, such as qRT-PCR, western blotting, luciferase activity, and RNA immunoprecipitation analyses. The results confirmed the upregulation of miR-106a and validated FBXW7 as one of its targets. As shown in one study, decreasing the miR-106a level via elimination by FBXW7 restored the normal features of liver cells [109].
Bioinformatic analyses demonstrated the inverse correlation between long noncoding RNA T cell leukemia/lymphoma 6 (IncTCL6) and miR-106a in HCC [103]. In a study conducted by Luo et al., the functional mechanism of miR-106a and IncTCL6 dysregulation in HCC was investigated, revealing that IncTCL6 directly binds to miR-106a and reduces its expression, resulting in suppressed tumor progression through the PI3K/AKT signaling pathway and restored PTEN expression [103].
In a recent study by Liang et al., downregulation of protein tyrosine phosphatase non-receptor type 12 (PTPN12) in HCC was linked to miR-106a overexpression followed by a significant increase in the proliferation, invasion, and migration of tumor cells [110].
On the contrary, in a study by Wang et al., it was demonstrated that platelet-derived growth factor-D (PDGF-D) mediated EMT through inhibition of miR-106a and subsequent upregulation of Twist1 in HCC GR cells [108]. In line with the role of miRNAs in regulating Twist, this study revealed that miR-106a inhibited cell invasion through the downregulation of Twist1 in HCC GR cells. Since the downregulation of Twist1 reversed EMT to MET (mesenchymal–epithelial transition) in GR cells, miR-106a suppressed EMT partly due to targeting Twist1 [108]. This research group additionally confirmed the tumor suppression behavior of miR-106a in HCC by indicating the role of Fer-1-like family member 4 (FER1L4) in tumor progression via reducing miR-106a and miR-372 levels in the liver. These two miRNAs are believed to inhibit HCC by targeting E2F1 as an NF-kB gene activator at the transcription level [105].
2.15. Melanoma
Melanoma is the most dangerous form of skin cancer and accounts for more than 70% of skin tumor-related deaths [111]. A number of studies reviewed miRNA expression patterns as potential biomarkers for screening, early detection, diagnosis, and prognosis of melanoma. Along with other candidates, miR-106a is among those that regulate the highest number of transcription factors in melanoma [112,113,114].
Connexin 43 (Cx43) is a vital gap junction protein in the tumor microenvironment (TME). As Wang et al. reported, the expression of Cx43 is lower in melanoma cells than in human epidermal melanocytes (HEMn) [111]. The lower expression of Cx43 is significantly associated with increased malignancy in multiple cancers [115]. Bioinformatic analyses and luciferase reporter assays confirmed Cx43 as the main target of miR-106a in melanoma cells. The in vitro experiments with melanoma cells also showed a direct correlation between Cx43 overexpression and decreased cell proliferation as well as colony formation ability [111].
On the other hand, in another study by Luan et al., the downregulation of miR-106a was reported. The authors indicated that the overexpression of lncRNA H19 in melanoma was correlated with poor prognosis in patients [116]. According to the tumor-suppressing activity of miR-106a by targeting E2F3 (a member of the E2F transcription factor family), it was demonstrated that lncRNA H19 may function as a sponge of miR-106a-5p to upregulate E2F3 expression and consequently promote glucose metabolism and growth in melanoma cells. Thus, lncRNA H19 may serve as a survival indicator and potential therapeutic target for melanoma patients [116].
2.16. Osteosarcoma
Osteosarcoma (OS) is one of the most common types of malignant bone tumors with high mortality rates in children and adolescents, and despite medical interventions such as neoadjuvant chemotherapy and surgery, the survival rate remains low, which suggests poor prognosis [117,118].
In genome-wide miRNA profiling and a cohort study using serum and plasma samples from OS patients and healthy persons, it was discovered that a total of 56 miRNAs were upregulated and 164 miRNAs were downregulated [119]. Subsequently, the researchers validated three miRNAs, miR-21, miR-106a, and miR-221, with elevated levels in the peripheral blood samples of OS patients as compared to healthy controls. In contrast, they did not find that miR-106a was significantly upregulated or downregulated in OS tumors compared to normal bone controls. The increase in miR-106a levels in the plasma of OS patients may be derived from other tissues in the body rather than the tumor itself [119].
In addition, Chen et al. performed a study that indicated the regulating role of miR-106a in the proliferation and invasion of human osteosarcoma cells by targeting vascular non-inflammatory molecule 2 (VNN2) [117]. VNN2 protein is a novel glycosylphosphatidyl inositol-anchored protein member of the VNN family that serves an essential role in the transendothelial migration of cells [120]. The VNN family includes membrane-associated proteins reported to be associated with regulating neutrophil trafficking and adherence. Moreover, the VNN family belongs to a broader pantetheinase family with a role in redox regulation, which may be associated with tumor progression in vitro. The results of this study showed that miR-106a is upregulated in human osteosarcoma cells and the knockdown of miR-106a mediated tumor progression at least partially by targeting VNN2 in osteosarcoma [117].
The upregulation of miR-106a in OS was also confirmed by qRT-PCR and statistical analysis in a study by Zhao et al. The authors demonstrated that the expression of miR-106a was higher in lower OS grades, and this miRNA’s increased level was detected in OS with lung metastasis. Thus, miR-106a was identified as a potential biomarker for early diagnosis and targeted therapy for OS [121].
Furthermore, it was reported that some lncRNAs, including lncRNA HOTAIR (HOX antisense intergenic RNA), are dysregulated in cancer conditions [118]. As Guo et al. reported, the increased level of lncRNA HOTAIR in drug-resistant OS was associated with higher tumor cell proliferation and invasion. Interestingly, they found that lncRNA HOTAIR binds to miR-106a, which is downregulated in DDP-resistant OS. According to the confirmed attenuation of STAT3 by miR-106a, it was concluded that lncRNA HOTAIR overexpression results in the upregulation of STAT3 via inhibiting miR-106a [118].
3. Cancer Drug Resistance: The Role of miR-106a
Although the advancement of chemotherapeutic agents in terms of personalized medicine and high-tech development has empowered oncologists to fight cancers, drug resistance still seems to be one of the main challenges in cancer treatment, being responsible for more than 90% of cancer-related mortalities [122,123].
As a complex phenomenon, drug resistance is affected by numerous factors and has cancer-specific features. The mechanisms of miRNA involvement in promoting or suppressing drug resistance are different for each miRNA and each cancer. However, they mainly include alterations in cell cycle checkpoints and drug efflux functions, affecting apoptosis and autophagy, stimulating aberrant DNA damage repair, and inducing abnormal metabolism [122,124,125]. The proven roles of miR-106a in promoting drug resistance in cancers include its involvement in increasing drug clearance or accumulation of oncogenic factors by targeting membrane protein genes. Additionally, miR-106a disruption of apoptosis via targeting key genes and pathways is the critical reason underlying drug resistance in cisplatin-treated patients. Furthermore, it is indicated that miR-106a can modulate drug resistance in colorectal tumors by regulating the properties of cancer stem cells via suppressing tumor suppressor genes [2,42,71,124]. It should be noted that the expression level of miR-106a and its impact on the sensitivity of tumor cells to therapeutics may vary depending on the stage of chemotherapy [122,124] and the mechanism of action of the drug [126].
According to the importance of miRNAs in drug resistance, particularly in cancer chemotherapy, novel approaches have recently emerged for developing medications designed as a combination of chemotherapeutic agents and miRNA-based treatments. Such synergistic strategies for simultaneously targeting tumorigenic pathways and controlling critical miRNA functions may be more effective in improving the clinical outcomes of chemotherapy and prolonging the survival of cancer patients [127,128].
4. mir-106a: Dysregulation in Non-Cancer Diseases
4.1. Hepatitis B
The Hepatitis B virus (HBV) was one of the first viruses demonstrated to be associated with cancer [129]. Although HBV is well known today as the cause of acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma, it does not directly cause malignancy [130]. It is believed that HBV infection makes the immune system release complex and abnormal responses that are the cause of liver destruction. Thus, exploring the mechanisms of the immune response in chronic hepatitis B (CHB) patients has been the focus of intense research [130,131].
Continuous release of cytokines by immune cells in response to virus infections frequently results in harmful impacts due to recruiting inflammatory cells, constraining virus replication and spread, and inducing adaptive immunity [132]. In the case of HBV infection, IL-8 is one of the main involved inflammatory cytokines that can stimulate immune responses via granulocytes, NK cells, and T-cell chemotaxis [133].
In a study by Hong et al., the qRT-PCR results suggested that the miR-106a level of peripheral blood mononuclear cells was decreased in CHB patients [134]. Using luciferase activity assays, the authors indicated that IL-8 levels were inversely correlated with miR-106a levels in CHB and they identified IL-8 as a target of miR-106a. Moreover, it was confirmed that IL-8 overexpression in CHB was attenuated by inducing miR-106a to reverse the damage. These results have represented miR-106a as an important player to be considered in the molecular research of CHB [134].
4.2. Multiple Sclerosis
Multiple sclerosis (MS) is an autoimmune disease that impacts the brain’s white matter and spinal cord (central nervous system). The rate of MS is higher in women than in men, with a ratio of three to one [135,136]. There are four major types of MS, including primary progressive multiple sclerosis (PPMS), relapsing-remitting multiple sclerosis (RRMS), secondary progressive multiple sclerosis (SPMS), and progressive relapsing multiple sclerosis (PRMS) [136].
With evidence regarding the involvement of miRNAs in modulating the immune response, inflammatory diseases, and autoimmunity [137,138,139], Rahimirad et al. performed a multi-stage experimental study to identify MS-associated miRNAs and their target genes. The qRT-PCR results indicated that miR-106a levels tended to decrease in the blood samples of MS patients. A gene target interaction network of hsa-miR-106a-5p was constructed using the miRTarBase file deposited in Cytoscape and 28 targets with strong support interactions were identified. RBL2, APP, CYP19A1, and BMP2 were upregulated during MS progression and reasonably reported as the potential targets of miR-106a. Hence, miR-106a may be therapeutically valuable in MS condition attenuation [136].
4.3. Myasthenia Gravis
Myasthenia gravis (MG) is an antibody-mediated and T cell-dependent autoimmune disease of the neuromuscular junction (NMJ), the pathogenesis of which is poorly understood. About 50%-85% of ocular myasthenia gravis (OMG) patients progress to generalized myasthenia gravis (GMG), and approximately 15%-20% of patients with MG will experience potentially fatal myasthenic crises due to respiratory muscle weakness [140]. In recent years, many studies have revealed that miRNAs are key regulators of MG pathogenesis [140,141].
In a study by Xu et al., the expression of plasma exosomal miRNAs in MG and their involvement in MG pathogenesis were investigated [142]. The authors used deep sequencing and qRT-PCR analyses to reveal that exosomal miR-106a-5p was significantly downregulated in patients with OMG and GMG compared to healthy control subjects. They additionally indicated that levels of this miRNA were notably reduced in patients with GMG compared to OMG [142].
4.4. Cardiac Hypertrophy
It is now believed that cardiac hypertrophy is a pathological process, as it can lead to heart failure, arrhythmia, and even sudden death [143]. Cardiac hypertrophy is a condition associated with growth of cardiomyocyte size and the overexpression of many fetal genes, causing left ventricular (LV) wall thickness and diminished normal cardiac function [144].
The reports on miRNA involvement in the regulatory network of cardiovascular disease have shown the upregulation of miR-106a in cardiac hypertrophy, and mitofusin 2 (Mfn2) was identified as the potential target gene for this miRNA. Mfn2, a key member of the mitofusin protein family, is located in the outer membrane of mitochondria and acts to maintain this organelle’s structure [145,146]. Mfn2 is known as a cell proliferation suppressor gene, probably by inhibiting the MAPK/ERK pathway. Recent studies indicate that Mfn2 deficiency is involved in cardiovascular disease [144,146,147].
Based on these facts, a study was conducted by Guan et al. to investigate the role of miR-106a in hypertrophic growth of the heart by targeting Mfn2 using a mouse model of cardiac hypertrophy and a cellular model of cardiomyocyte hypertrophy [144]. They showed that miR-106a overexpression was sufficient to induce hypertrophic growth via directly targeting Mfn2. It was confirmed that the knockdown of miR-106a could inhibit the damage while reversing the hypertrophic alterations. Thus, miR-106a may be a promising molecular target in treating pathological hypertrophy and other cardiac disorders [144,148].
5. The Role in Spermatogenesis
Spermatogenesis in male mammals has three phases: mitotic proliferation of spermatogonia, meiosis of spermatocytes, and haploid differentiation of spermatids. The genome of male germ cells is actively transcribed to generate a complex transcriptome with a specific expression pattern in every spermatogenesis phase [149]. According to the importance of the precise regulation needed for successful sperm production, the role of miRNAs has been highlighted particularly for their post-translation effects on stable mRNA expression [150]. Kotaja has comprehensively reviewed spermatogenesis-engaged miRNAs regarding their specific roles in the phases and also their confirmed or potential targets [151].
In a study by He et al. using genetically modified mouse models and various molecular and cellular methodologies, including real-time PCR, microarray, in situ hybridization, immunohistochemistry, etc., the impact of the miRNA expression profile in spermatogenesis was investigated. Additional evidence obtained from clinical evaluation of human spermatozoa or seminal plasma not only confirmed the regulatory role of miRNAs in spermatogenesis but also specified them as potential biomarkers for male factor infertility [152]. Through conducting in vitro and in vivo analyses on mouse spermatogonial stem cells (SSCs), the authors reported that miRNA-20 and miRNA-106a are preferentially expressed in SSCs. They further demonstrated that these two miRNAs promoted post-transcriptional regulation via targeting STAT3 and Ccnd1 and that knockdown of STAT3, Fos, and Ccnd1 resulted in the regeneration of SSCs [152].
6. mir-106a and Human Aging
Aging may be the most apparent but still mysterious process in nature. It is accepted that both genetic and non-genetic factors are correlated with the deterioration of body functions due to attenuation of the repair system’s abilities. With all the phenotypic and genotypic alterations, there are always unknown niches to be studied about this condition [153,154].
Many studies have addressed the genes, proteins, and mechanisms involved in this multifactorial process; however, the confirmation of miRNA roles in regulating cellular functions opened new doors for scientists to perform innovative research identifying the miRNA candidates engaged in aging. Although the number of aging-related miRNAs is increasing, their mechanisms of action and specific targets have remained elusive [154,155,156,157].
To address this question, Hackl et al. performed an interesting study (Figure S2 in Supplementary Materials) [153]. They selected four replicative cell aging models, which included endothelial cells, replicated CD8+ T cells, renal proximal tubular epithelial cells, and skin fibroblasts. In addition, they included three organismal aging models, namely foreskin cells, mesenchymal stem cells, and CD8+ T cell populations from old and young donors. Through the utilization of miRNA microarrays using locked nucleic acid technology, a total of four commonly regulated miRNAs were identified. These miRNAs included miR-17, which was downregulated in all seven models; miR-19b and miR-20a, which showed downregulation in six models; and miR-106a, which was downregulated in five models. The reduction in levels of these miRNAs was correlated with an increase in transcript levels of specific target genes, particularly CDK inhibitor p21/CDKN1A. These findings established miRNAs as novel indicators of cell aging in humans [153].
Furthermore, in a miRNA analysis of human longevity at a genome-wide scale, a panel of 863 miRNAs was examined to identify significant differences in miRNA regulation between 15 long-lived individuals (LLI), consisting of centenarians and nonagenarians, and 55 younger individuals [158]. The study design encompassed two distinct experimental approaches: microarrays for initial screening and qRT-PCR for technical validation and replication [158]. The results of this screening study confirmed downregulation of miR-17, miR-106a, and miR-20a in accordance with Hackl et al.’s report [153,158]. Moreover, it was found that the downregulated miRNAs were significantly enriched in disease-associated pathways and had a higher number of reported associations with diseases compared to the upregulated miRNAs. The clustering of these downregulated miRNAs suggested that they may work together in a coordinated manner rather than functioning independently. This finding supports the hypothesis that miRNA dysregulation during aging likely occurs in groups or “packs” rather than as isolated entities [158]. KEGG mapping highlighting miR-106a as a target in the longevity regulating pathway is presented in Figure S3 in Supplementary Materials.
7. Perspectives and Conclusions
Since their discovery, miRNAs have remained among hot biology topics and an increasing number of studies have been undertaken to elucidate more about these biomolecules. As presented in this review exploring the dysregulation of miR-106a, this miRNA can be selected for further investigation and validation for application as a biomarker and/or therapeutic target (Table S2 in Supplementary Materials). It cannot be denied that the multi-aspect roles of this miRNA in critical cellular processes make it a great candidate for medical and biomedical applications [159].
Many published reports have indicated that most miRNAs hardly pass the validation evaluation to be applied as a biomarker. A wide variety of analysis methods with different levels of sensitivity, lack of standard normalization methods, and challenges in reliable real sample investigation may be the main reasons why miRNAs have not yet entered the market as clinically approved biomarkers [160]. Table 1 lists the current kits and platforms developed for miRNA-based diagnosis.

Table 1.
miRNA-based diagnostics in the market.
In the case of miR-106a, determining its precise healthy and non-healthy expression levels in blood and related tissue samples appears to be a primary challenge. Being a circulating miRNA, miR-106a originates from extracellular vesicles released into the blood by various cell types [161]. Despite its remarkable stability in blood, extracting and analyzing miR-106a from these vesicles may present difficulties due to potential false results caused by release from blood cells in response to different conditions [161,162]. Moreover, the levels of circulating miRNAs can be influenced by numerous factors, such as age, gender, ethnicity, medication, and smoking, further complicating the assessment [161,163].
Additionally, as a potential diagnostic tool in clinical settings, miR-106a faces another limitation. It has been detected in patients with different tumor types; however, the results are inconsistent, even among closely related studies of the same diseases. These inconsistencies raise doubts about its reliability and applicability as a universal biomarker in clinical practice [163].
Despite about ten miRNA-based drugs having reached clinical trials, none are in phase III yet, while the number of discontinued studies appears to be increasing (Table 2). In addition to delivery system limitations, dosage concerns, and potential silent side effects, the current regulatory gap covering essential safety and quality attributes for advanced therapies, including miRNA-based therapies, is a bottleneck in translating miRNAs from bench to clinic [163,164].

Table 2.
miRNA-based therapeutics in pre-clinical and clinical phases.
In an intriguing study, Zhang et al. conducted a review of the challenges in miRNA therapeutics, focusing on the available resources from a drug target perspective [165]. Although miRNA therapeutics showed promise at the outset, the researchers highlighted a phenomenon they termed “too many targets for miRNA effect” (TMTME). This refers to the fact that most miRNAs target tens to hundreds of genes [165]. While miRNAs like miR-106 that participate in multiple regulatory networks and target essential genes in crucial biological pathways seem like attractive therapeutic candidates, developing drugs based on them often proves difficult and complex due to the multiple consequences of altering their levels in the body.
An analysis of unsuccessful miRNA therapeutic candidates revealed that unforeseen side effects emerged during phases I and II of clinical studies, leading to the discontinuation of drug development. These unexpected side effects hindered the advancement of miRNA-based therapies and underscored the challenges in translating miRNA research into successful clinical applications [165].
Therefore, despite the abundance of evidence showcasing the significant regulatory roles of miR-106a in various diseases, including cancers, it is imperative to emphasize the crucial step of validation, particularly through the comprehensive analysis of real samples, before considering the inclusion of this miRNA in pre-clinical or clinical studies. The data on the confirmed target genes of miR-106a, especially those altered in cancer resistance conditions, may be a potential niche that promotes miR-106a to be upgraded in clinical-oriented studies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bioengineering10080892/s1, Figure S1: KEGG Maps highlighting miR-106a targets in cancer pathways. The target genes are highlighted in yellow. The graph is rendered by miRPath v4.0.; Figure S2: Experimental design of differential miRNA analysis in Hackl et al. study on alteration in miRNAs’ expression in human aging. Reproduced from [153].; Figure S3: KEGG Maps highlighting miR-106a targets in longevity regulating pathway. The target genes are highlighted in yellow. The graph is rendered by miRPath v4.0.; Table S1: miRDB results for has-miR-106a-5p. There are 1337 predicted targets for hsa-miR-106a-5p in miRDB; Table S2: miR-106a, its regulation alterations, and its target genes by condition type (cancers, diseases, spermatogenesis, and aging).
Author Contributions
Conceptualization, M.D. and A.G.-D.; writing—original draft preparation, M.D. and A.G.-D.; writing—review and editing, M.D.; visualization, M.D. and A.G.-D.; supervision, M.D.; project administration, M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Not Applicable.
Acknowledgments
This study is respectfully dedicated to Massoud Saidijam, a great human being and fabulous professor. After a period of endurance, he passed away last year due to leukemia.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ABCA1 | Adenosine triphosphatase-binding cassette A1 |
| ABCG2 | Adenosine triphosphatase-binding cassette G2 |
| AKT (PKB) | Serine/threonine protein kinase B |
| AMPK-mTOR | AMP-activated protein kinase-mammalian target of the rapamycin |
| AP-1 | Activator protein 1 |
| Apaf-1 | Apoptotic peptidase activating factor 1 |
| APP | Amyloid precursor protein |
| ARHGAP24 | Rho GTPase-activating protein 24 |
| ATG7 | Autophagy-related gene 7 |
| Bak | Bcl-2-antagonist killer |
| Bax | Bcl-2-associated x protein |
| BC | Breast cancer |
| Bcl-1 | B-cell lymphoma 1 |
| Bcl-2 | B-cell lymphoma 2 |
| Bcl-x | B-cell lymphoma-extra-large |
| BCL2L11 | Bcl-2-like protein 11 |
| Bid | BH3 interacting-domain death agonist |
| Bim | Bcl-2 interacting mediator of cell death |
| BMP2 | Bone morphogenetic protein 2 |
| CC | Cervical cancer |
| CCA | Cholangiocarcinoma |
| Ccnd1 | Cyclin D1 |
| CDK | Cyclin-dependent kinase |
| CDKN1A | Cyclin dependent kinase inhibitor 1A |
| CDX2 | Caudal-type homeobox transcription factor 2 |
| ceRNA | Competing endogenous RNA |
| CHB | Chronic hepatitis B |
| CRC | Colorectal cancer |
| Cx43 | Connexin 43 |
| CYP19A1 | Cytochrome P450 family 19 subfamily A |
| DAX-1 | Dosage sensitive sex reversal, adrenal hypoplasia, critical region on the X chromosome, gene 1 |
| DDP | Diamminedichloroplatinum |
| DISC | Death inducing signaling complex |
| E2F1 | E2 promoter binding factor 1 (Transcription factor E2F1) |
| eIF4A | Eukaryotic initiation factor 4A-I |
| EMC | Endometrial cancer |
| EMT | Epithelial–mesenchymal transition |
| ERCC1 | Excision repair cross complementation group 1 |
| ESCC | Esophageal squamous cell carcinoma |
| EWS | Ewing sarcoma |
| EZH2 | Enhancer of zeste homolog 2 |
| FADD | Fas-associated death domain |
| FAF-1 | FAS-associated factor 1 |
| FASL | FAS ligand |
| FASTK | Fas-activated serine/threonine kinase |
| FBXW7 | F-box and WD repeat domain containing 7 |
| FER1L4 | Long non-coding RNA Fer-1 protein 4 |
| FER1L4 | Fer-1 like family member 4 |
| GBMs | Glioblastoma multiforms |
| GC | Gastric cancer |
| GMG | Generalized myasthenia gravis |
| GR | Gemcitabine-resistant |
| GST-π | Glutathione S-transferase π |
| HBV | Hepatitis B virus |
| HCC | Hepatocellular carcinoma |
| HEMn | Human epidermal melanocytes |
| HGSOC | High-grade serous ovarian carcinomas |
| HIPK3 | Homeodomain-interacting protein kinase 3 |
| HOTAIR | HOX antisense intergenic RNA |
| HPV | Human papillomavirus |
| IGF-1 | Insulin-like growth factor-1 |
| IL | Interleukin |
| IncTCL6 | Long noncoding RNA T cell leukemia/lymphoma 6 |
| IRS | Insulin receptor substrates |
| lncRNA | Long noncoding RNA |
| lncRNA | Long noncoding RNA |
| lncRNA XIST, XIST | Long noncoding RNA X inactive specific transcript |
| MAPK | Mitogen-activated protein kinase |
| Mcl-1 | Myeloid cell leukemia 1 |
| MDR | Multidrug resistance |
| MDR1, MRP1 | Multidrug resistance protein 1 |
| MET | Mesenchymal-epithelial transition |
| mfn2 | Mitofusin 2 |
| MG | Myasthenia gravis |
| miRNA | Microrna |
| MMP | Matrix metalloproteinase |
| MMP2 | Mmp2 |
| MRE | Mirna response element |
| MS | Multiple sclerosis |
| MYCN | MYC expressed in neuroblastoma |
| NF-ᴋB | Nuclear factor kappa B |
| NMJ | Neuromuscular junction |
| NSCLC | Non-small cell lung cancer |
| OC | Ovarian cancer |
| 4-Oct | Octamer-binding transcription factor 4 |
| OMG | Ocular myasthenia gravis |
| OS | Osteosarcoma |
| PA | Pilocytic astrocytomas |
| PAK7 | P21-activated kinase 7 |
| PARP | Poly-ADP ribose polymerase |
| PC | Pancreatic cancer |
| PDCD4 | Programmed cell death 4 |
| PDGF-D | Platelet-derived growth factor-D |
| PDK1 | Phosphoinositide-dependent protein kinase 1 |
| P-gp | P-glycoprotein |
| PI3K | Phosphatidylinositol 3-kinase |
| PIP2 | Phosphatidylinositol diphosphate |
| PIP3 | Phosphatidylinositol trisphosphate |
| PNET | Primitive neuroectodermal tumor |
| PPMS | Primary progressive multiple sclerosis |
| PRMS | Progressive relapsing multiple sclerosis |
| PTEN | Phosphatase and tensin homolog deleted on chromosome 10 |
| PTPN12 | Protein tyrosine phosphatase non-receptor type 12 |
| RB1 | Retinoblastoma protein 1 |
| RBL2 | Retinoblastoma 2 |
| RCC | Renal cell carcinoma |
| RNA | Ribonucleic acid |
| RRMS | Relapsing-remitting multiple sclerosis |
| RT-PCR | Real-time reverse transcriptase PCR |
| RUNX1 | Runt-related transcription factor 1 |
| RUNX2 | Runt-related transcription factor 2 |
| RUNX3 | Runt-related transcription factor 3 |
| SLC2A3, GLUT3 | Solute carrier family 2 member 3 |
| SOX9 | Sry-box transcription factor 9 |
| SPMS | Secondary progressive multiple sclerosis |
| SSCs | Spermatogonial stem cells |
| STAT3 | Signal transducers and activators of transcription 3 |
| TGFBR2 | Transforming growth factor-b receptor 2 |
| TGF-β | Transforming growth factor-β |
| TIMP2 | Tissue inhibitors of metalloproteinase 2 |
| TME | Tumor microenvironment |
| TNF | Tumor necrosis factor |
| Topo-II | DNA-topoisomerase II |
| TP53INP1 | Tumor protein p53-inducible nuclear protein 1 |
| TRAIL | TNF-related apoptosis-inducing ligand |
| VEGF | Vascular endothelial growth factor |
| VEGFA | Endogenous vascular endothelial growth factor-a |
| VNN2 | Vascular non-inflammatory molecule 2 |
| WHO | World Health Organization |
| WNT | Wingless-related integration site |
References
- Leitao, A.L.; Enguita, F.J. A Structural View of miRNA Biogenesis and Function. Non-Coding RNA 2022, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Zhuang, Y.; Zheng, J.N.; Pei, D.S. MiR-106a: Promising biomarker for cancer. Bioorganic Med. Chem. Lett. 2016, 26, 5373–5377. [Google Scholar] [CrossRef] [PubMed]
- Kennel, P.J.; Schulze, P.C. A Review on the Evolving Roles of MiRNA-Based Technologies in Diagnosing and Treating Heart Failure. Cells 2021, 10, 3191. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Azmi, A.S.; Moore, A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip. Rev. RNA 2021, 12, e1662. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Daneshpour, M.; Omidfar, K.; Ghanbarian, H. A novel electrochemical nanobiosensor for the ultrasensitive and specific detection of femtomolar-level gastric cancer biomarker miRNA-106a. Beilstein J. Nanotechnol. 2016, 7, 2023–2036. [Google Scholar] [CrossRef]
- Gruszka, R.; Zakrzewska, M. The Oncogenic Relevance of miR-17-92 Cluster and Its Paralogous miR-106b-25 and miR-106a-363 Clusters in Brain Tumors. Int. J. Mol. Sci. 2018, 19, 879. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Kaur, P.; Kotru, S.; Singh, S.; Munshi, A. Role of miRNAs in diabetic neuropathy: Mechanisms and possible interventions. Mol. Neurobiol. 2022, 59, 1836–1849. [Google Scholar] [CrossRef]
- Kudelova, E.; Holubekova, V.; Grendar, M.; Kolkova, Z.; Samec, M.; Vanova, B.; Mikolajcik, P.; Smolar, M.; Kudela, E.; Laca, L.; et al. Circulating miRNA expression over the course of colorectal cancer treatment. Oncol. Lett. 2022, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- Ak, S.; Tunca, B.; Tezcan, G.; Cecener, G.; Egeli, U.; Yilmazlar, T.; Ozturk, E.; Yerci, O. MicroRNA expression patterns of tumors in early-onset colorectal cancer patients. J. Surg. Res. 2014, 191, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Yang, W.; Duan, L.; Wang, X.; Li, Y.; Xu, C.; Liu, C.; Zhang, Y.; Zhou, W.; Liu, J.; et al. Biological Implications and Clinical Potential of Metastasis-Related miRNA in Colorectal Cancer. Mol. Ther. Nucleic Acids 2021, 23, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Xia, G.; Wang, C.; Zhong, F.; Liu, L.; Zhang, D. miR-106a suppresses tumor cells death in colorectal cancer through targeting ATG7. Med. Mol. Morphol. 2017, 50, 76–85. [Google Scholar] [CrossRef]
- He, Y.; Wang, G.; Zhang, L.; Zhai, C.; Zhang, J.; Zhao, X.; Jiang, X.; Zhao, Z. Biological effects and clinical characteristics of microRNA-106a in human colorectal cancer. Oncol. Lett. 2017, 14, 830–836. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, P.; Yang, J.; Liu, Z.; Yang, Z.; Qin, H. Candidate microRNA biomarkers in human colorectal cancer: Systematic review profiling studies and experimental validation. Int. J. Cancer 2012, 130, 2077–2087. [Google Scholar] [CrossRef]
- Zhu, G.F.; Xu, Y.W.; Li, J.; Niu, H.L.; Ma, W.X.; Xu, J.; Zhou, P.R.; Liu, X.; Ye, D.L.; Liu, X.R.; et al. Mir20a/106a-WTX axis regulates RhoGDIa/CDC42 signaling and colon cancer progression. Nat. Commun. 2019, 10, 112. [Google Scholar] [CrossRef]
- Feng, B.; Dong, T.T.; Wang, L.L.; Zhou, H.M.; Zhao, H.C.; Dong, F.; Zheng, M.H. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS ONE 2012, 7, e43452. [Google Scholar] [CrossRef]
- Liu, J.; Huang, Y.; Wang, H.; Wu, D. MiR-106a-5p promotes 5-FU resistance and the metastasis of colorectal cancer by targeting TGFbetaR2. Int. J. Clin. Exp. Pathol. 2018, 11, 5622–5634. [Google Scholar]
- Qin, Y.; Huo, Z.; Song, X.; Chen, X.; Tian, X.; Wang, X. mir-106a regulates cell proliferation and apoptosis of colon cancer cells through targeting the PTEN/PI3K/AKT signaling pathway. Oncol. Lett. 2018, 15, 3197–3201. [Google Scholar] [CrossRef] [PubMed]
- Bermudez Brito, M.; Goulielmaki, E.; Papakonstanti, E.A. Focus on PTEN Regulation. Front. Oncol. 2015, 5, 166. [Google Scholar] [CrossRef]
- Miao, Y.; Zheng, W.; Li, N.; Su, Z.; Zhao, L.; Zhou, H.; Jia, L. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci. Rep. 2017, 7, 41942. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ke, F.; Chen, T.; Zhou, Q.; Weng, L.; Tan, J.; Shen, W.; Li, L.; Zhou, J.; Xu, C.; et al. MicroRNAs that regulate PTEN as potential biomarkers in colorectal cancer: A systematic review. J. Cancer Res. Clin. Oncol. 2020, 146, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Ma, Q. MicroRNA-106a inhibits cell proliferation and induces apoptosis in colorectal cancer cells. Oncol. Lett. 2018, 15, 8941–8944. [Google Scholar] [CrossRef]
- Phin, S.; Moore, M.W.; Cotter, P.D. Genomic Rearrangements of PTEN in Prostate Cancer. Front. Oncol. 2013, 3, 240. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Feng, F.; Zhu, L.; Zheng, Y.; Luo, X.; Liu, C.; Yi, B.; Jiang, X. Circulating miR-106a is a Novel Prognostic and Lymph Node Metastasis Indicator for Cholangiocarcinoma. Sci. Rep. 2015, 5, 16103. [Google Scholar] [CrossRef] [PubMed]
- Barbato, A.; Piscopo, F.; Salati, M.; Reggiani-Bonetti, L.; Franco, B.; Carotenuto, P. Micro-RNA in Cholangiocarcinoma: Implications for Diagnosis, Prognosis, and Therapy. J. Mol. Pathol. 2022, 3, 88–103. [Google Scholar] [CrossRef]
- Sun, C.; Zhu, J.; Wu, B.; Chen, J.; Zhu, Z.; Cai, P.; Guo, W.; Gu, Z.; Wang, J.; Huang, S. Diagnostic and prognostic value of microRNAs in cholangiocarcinoma: A systematic review and meta-analysis. Cancer Manag. Res. 2018, 10, 2125–2139. [Google Scholar] [CrossRef]
- Dylla, L.; Moore, C.; Jedlicka, P. MicroRNAs in Ewing Sarcoma. Front. Oncol. 2013, 3, 65. [Google Scholar] [CrossRef]
- Drobna, M.; Szarzynska, B.; Jaksik, R.; Sedek, L.; Kuchmiy, A.; Taghon, T.; Van Vlierberghe, P.; Szczepanski, T.; Witt, M.; Dawidowska, M. hsa-miR-20b-5p and hsa-miR-363-3p Affect Expression of PTEN and BIM Tumor Suppressor Genes and Modulate Survival of T-ALL Cells In Vitro. Cells 2020, 9, 1137. [Google Scholar] [CrossRef]
- Dylla, L.; Jedlicka, P. Growth-promoting role of the miR-106a~363 cluster in Ewing sarcoma. PLoS ONE 2013, 8, e63032. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Z.; Cheng, T.T.; He, Q.J.; Lei, Z.Y.; Chi, J.; Tang, Z.; Liao, Q.X.; Zhang, H.; Zeng, L.S.; Cui, S.Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol. Cancer 2018, 17, 126. [Google Scholar] [CrossRef]
- Ahadi, A. A systematic review of microRNAs as potential biomarkers for diagnosis and prognosis of gastric cancer. Immunogenetics 2021, 73, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, N.; He, S.; Lui, Y.; Lu, G.; Zhao, L. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastric cancer. FEBS Lett. 2014, 588, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Daneshpour, M.; Karimi, B.; Omidfar, K. Simultaneous detection of gastric cancer-involved miR-106a and let-7a through a dual-signal-marked electrochemical nanobiosensor. Biosens. Bioelectron. 2018, 109, 197–205. [Google Scholar] [CrossRef]
- Zhou, H.; Guo, J.M.; Lou, Y.R.; Zhang, X.J.; Zhong, F.D.; Jiang, Z.; Cheng, J.; Xiao, B.X. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J. Mol. Med. 2010, 88, 709–717. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, M.; Zhu, H.; Zhang, W.; He, S.; Hu, C.; Quan, L.; Bai, J.; Xu, N. miR-106a is frequently upregulated in gastric cancer and inhibits the extrinsic apoptotic pathway by targeting FAS. Mol. Carcinog. 2013, 52, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ma, R.; Yi, B.; Riker, A.I.; Xi, Y. MicroRNAs are involved in the development and progression of gastric cancer. Acta Pharmacol. Sin. 2021, 42, 1018–1026. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C. Identification and validation of potential mRNA- microRNA- long-noncoding RNA (mRNA-miRNA-lncRNA) prognostic signature for cervical cancer. Bioengineered 2021, 12, 898–913. [Google Scholar] [CrossRef]
- Xia, T.; Chen, S.; Jiang, Z.; Shao, Y.; Jiang, X.; Li, P.; Xiao, B.; Guo, J. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci. Rep. 2015, 5, 13445. [Google Scholar] [CrossRef]
- Rhode, P.; Mehdorn, M.; Lyros, O.; Kahlert, C.; Kurth, T.; Venus, T.; Schierle, K.; Estrela-Lopis, I.; Jansen-Winkeln, B.; Lordick, F.; et al. Characterization of Total RNA, CD44, FASN, and PTEN mRNAs from Extracellular Vesicles as Biomarkers in Gastric Cancer Patients. Cancers 2021, 13, 5975. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, Q.; Cai, X. MicroRNA-106a induces multidrug resistance in gastric cancer by targeting RUNX3. FEBS Lett. 2013, 587, 3069–3075. [Google Scholar] [CrossRef]
- Guo, C.; Ding, J.; Yao, L.; Sun, L.; Lin, T.; Song, Y.; Sun, L.; Fan, D. Tumor suppressor gene Runx3 sensitizes gastric cancer cells to chemotherapeutic drugs by downregulating Bcl-2, MDR-1 and MRP-1. Int. J. Cancer 2005, 116, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wen, W.; Zhu, J.; Huang, Z.; Zhang, L.; Zhang, H.; Qi, L.W.; Shan, X.; Wang, T.; Cheng, W.; et al. A six-microRNA signature in plasma was identified as a potential biomarker in diagnosis of esophageal squamous cell carcinoma. Oncotarget 2017, 8, 34468–34480. [Google Scholar] [CrossRef] [PubMed]
- Hiyama, T.; Yoshihara, M.; Tanaka, S.; Chayama, K. Genetic polymorphisms and esophageal cancer risk. Int. J. Cancer 2007, 121, 1643–1658. [Google Scholar] [CrossRef] [PubMed]
- Jamali, L.; Tofigh, R.; Tutunchi, S.; Panahi, G.; Borhani, F.; Akhavan, S.; Nourmohammadi, P.; Ghaderian, S.M.H.; Rasouli, M.; Mirzaei, H. Circulating microRNAs as diagnostic and therapeutic biomarkers in gastric and esophageal cancers. J. Cell Physiol. 2018, 233, 8538–8550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, C.; Chen, X.; Yang, C.; Li, K.; Wang, J.; Dai, J.; Hu, Z.; Zhou, X.; Chen, L.; et al. Expression profile of microRNAs in serum: A fingerprint for esophageal squamous cell carcinoma. Clin. Chem. 2010, 56, 1871–1879. [Google Scholar] [CrossRef] [PubMed]
- Hummel, R.; Hussey, D.J.; Michael, M.Z.; Haier, J.; Bruewer, M.; Senninger, N.; Watson, D.I. MiRNAs and their association with locoregional staging and survival following surgery for esophageal carcinoma. Ann. Surg. Oncol. 2011, 18, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Wei, L.L.; Wu, X.J.; Huo, F.C.; Mou, J.; Pei, D.S. MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis. 2017, 8, e3155. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Abak, A.; Tondro Anamag, F.; Shoorei, H.; Majidpoor, J.; Taheri, M. The emerging role of non-coding RNAs in the regulation of PI3K/AKT pathway in the carcinogenesis process. Biomed. Pharmacother. 2021, 137, 111279. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, H.; He, X.; Song, H.; Qiang, Y.; Li, Y.; Gao, J.; Wang, Z. miR-106a* inhibits the proliferation of renal carcinoma cells by targeting IRS-2. Tumour Biol. 2015, 36, 8389–8398. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Dhe-Paganon, S.; Melendez, P.A.; Lee, J.; Shoelson, S.E. Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J. Biol. Chem. 2003, 278, 25323–25330. [Google Scholar] [CrossRef]
- Lee, Y.H.; White, M.F. Insulin receptor substrate proteins and diabetes. Arch. Pharm. Res. 2004, 27, 361–370. [Google Scholar] [CrossRef]
- Huo, F.C.; Zhu, Z.M.; Du, Q.Y.; Pei, D.S. PAK5-stabilized Smuc confers renal cell carcinoma metastasis. Clin. Transl. Med. 2021, 11, e559. [Google Scholar] [CrossRef]
- Li, Y.K.; Zou, J.; Ye, D.M.; Zeng, Y.; Chen, C.Y.; Luo, G.F.; Zeng, X. Human p21-activated kinase 5 (PAK5) expression and potential mechanisms in relevant cancers: Basic and clinical perspectives for molecular cancer therapeutics. Life Sci. 2020, 241, 117113. [Google Scholar] [CrossRef]
- Yang, L.; Zou, X.; Zou, J.; Zhang, G. A Review of Recent Research on the Role of MicroRNAs in Renal Cancer. Med. Sci. Monit. 2021, 27, e930639. [Google Scholar] [CrossRef]
- Ma, J.; Wang, W.; Azhati, B.; Wang, Y.; Tusong, H. miR-106a-5p Functions as a Tumor Suppressor by Targeting VEGFA in Renal Cell Carcinoma. Dis. Markers 2020, 2020, 8837941. [Google Scholar] [CrossRef]
- Situ, Y.; Xu, Q.; Deng, L.; Zhu, Y.; Gao, R.; Lei, L.; Shao, Z. System analysis of VEGFA in renal cell carcinoma: The expression, prognosis, gene regulation network and regulation targets. Int. J. Biol. Markers 2022, 37, 90–101. [Google Scholar] [CrossRef]
- Tian, Y.; Sun, C.; Zhang, L.; Pan, Y. Clinical significance of miRNA-106a in non-small cell lung cancer patients who received cisplatin combined with gemcitabine chemotherapy. Cancer Biol. Med. 2018, 15, 157–164. [Google Scholar] [CrossRef]
- Heegaard, N.H.; Schetter, A.J.; Welsh, J.A.; Yoneda, M.; Bowman, E.D.; Harris, C.C. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int. J. Cancer 2012, 130, 1378–1386. [Google Scholar] [CrossRef]
- Xie, X.; Liu, H.T.; Mei, J.; Ding, F.B.; Xiao, H.B.; Hu, F.Q.; Hu, R.; Wang, M.S. miR-106a promotes growth and metastasis of non-small cell lung cancer by targeting PTEN. Int. J. Clin. Exp. Pathol. 2015, 8, 3827–3834. [Google Scholar]
- Lee, B.H.; Taylor, M.G.; Robinet, P.; Smith, J.D.; Schweitzer, J.; Sehayek, E.; Falzarano, S.M.; Magi-Galluzzi, C.; Klein, E.A.; Ting, A.H. Dysregulation of cholesterol homeostasis in human prostate cancer through loss of ABCA1. Cancer Res. 2013, 73, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.; Land, H. Anticancer activity of the cholesterol exporter ABCA1 gene. Cell Rep. 2012, 2, 580–590. [Google Scholar] [CrossRef]
- Ma, Y.; Li, X.; Cheng, S.; Wei, W.; Li, Y. MicroRNA-106a confers cisplatin resistance in non-small cell lung cancer A549 cells by targeting adenosine triphosphatase-binding cassette A1. Mol. Med. Rep. 2015, 11, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Shams, R.; Saberi, S.; Zali, M.; Sadeghi, A.; Ghafouri-Fard, S.; Aghdaei, H.A. Identification of potential microRNA panels for pancreatic cancer diagnosis using microarray datasets and bioinformatics methods. Sci. Rep. 2020, 10, 7559. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xu, Q.; Zhang, D.; Li, X.; Han, L.; Lei, J.; Duan, W.; Ma, Q.; Wu, Z.; Wang, Z. Upregulated miR-106a plays an oncogenic role in pancreatic cancer. FEBS Lett. 2014, 588, 705–712. [Google Scholar] [CrossRef]
- Chao, H.; Zhang, M.; Hou, H.; Zhang, Z.; Li, N. HOTAIRM1 suppresses cell proliferation and invasion in ovarian cancer through facilitating ARHGAP24 expression by sponging miR-106a-5p. Life Sci. 2020, 243, 117296. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, F.; Sheng, X.G.; Zhang, S.Q.; Chen, Y.T.; Liu, B.W. MicroRNA-106a regulates phosphatase and tensin homologue expression and promotes the proliferation and invasion of ovarian cancer cells. Oncol. Rep. 2016, 36, 2135–2141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Z.; Gersbach, E.; Zhang, X.; Xu, X.; Dong, R.; Lee, P.; Liu, J.; Kong, B.; Shao, C.; Wei, J.J. miR-106a represses the Rb tumor suppressor p130 to regulate cellular proliferation and differentiation in high-grade serous ovarian carcinoma. Mol. Cancer Res. 2013, 11, 1314–1325. [Google Scholar] [CrossRef]
- Rao, Y.M.; Shi, H.R.; Ji, M.; Chen, C.H. MiR-106a targets Mcl-1 to suppress cisplatin resistance of ovarian cancer A2780 cells. J. Huazhong Univ. Sci. Technol. Med. Sci. 2013, 33, 567–572. [Google Scholar] [CrossRef]
- Huh, J.H.; Kim, T.H.; Kim, K.; Song, J.A.; Jung, Y.J.; Jeong, J.Y.; Lee, M.J.; Kim, Y.K.; Lee, D.H.; An, H.J. Dysregulation of miR-106a and miR-591 confers paclitaxel resistance to ovarian cancer. Br. J. Cancer 2013, 109, 452–461. [Google Scholar] [CrossRef]
- Kim, Y.W.; Kim, E.Y.; Jeon, D.; Liu, J.L.; Kim, H.S.; Choi, J.W.; Ahn, W.S. Differential microRNA expression signatures and cell type-specific association with Taxol resistance in ovarian cancer cells. Drug Des. Dev. Ther. 2014, 8, 293–314. [Google Scholar] [CrossRef]
- Li, H.; Xu, H.; Shen, H.; Li, H. microRNA-106a modulates cisplatin sensitivity by targeting PDCD4 in human ovarian cancer cells. Oncol. Lett. 2014, 7, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Yuan, D.; Zhang, W.; Zhu, D.; Xiao, A.; Mao, G.; Jiang, W.; Lin, M.; Wang, J. Upregulation of long noncoding RNA XIST has anticancer effects on ovarian cancer through sponging miR-106a. Hum. Cell 2021, 34, 579–587. [Google Scholar] [CrossRef]
- Buruiana, A.; Florian, S.I.; Florian, A.I.; Timis, T.L.; Mihu, C.M.; Miclaus, M.; Osan, S.; Hrapsa, I.; Cataniciu, R.C.; Farcas, M.; et al. The Roles of miRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations. Int. J. Mol. Sci. 2020, 21, 1950. [Google Scholar] [CrossRef] [PubMed]
- Mathupala, S.P.; Mittal, S.; Guthikonda, M.; Sloan, A.E. MicroRNA and brain tumors: A cause and a cure? DNA Cell Biol. 2007, 26, 301–310. [Google Scholar] [CrossRef]
- Petrescu, G.E.D.; Sabo, A.A.; Torsin, L.I.; Calin, G.A.; Dragomir, M.P. MicroRNA based theranostics for brain cancer: Basic principles. J. Exp. Clin. Cancer Res. 2019, 38, 231. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Z.; Chen, Z.; Lin, L.; Wang, Y.; Sailike, D.; Luo, K.; Du, G.; Xiang, X.; Jiafu, G.D. MicroRNA-106a-5p facilitates human glioblastoma cell proliferation and invasion by targeting adenomatosis polyposis coli protein. Biochem. Biophys. Res. Commun. 2016, 481, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, B.; Shi, Y.; Xu, C.; Xiao, H.L.; Ma, L.N.; Xu, S.L.; Yang, L.; Wang, Q.L.; Dang, W.Q.; et al. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene 2015, 34, 1407–1419. [Google Scholar] [CrossRef]
- Dai, D.W.; Lu, Q.; Wang, L.X.; Zhao, W.Y.; Cao, Y.Q.; Li, Y.N.; Han, G.S.; Liu, J.M.; Yue, Z.J. Decreased miR-106a inhibits glioma cell glucose uptake and proliferation by targeting SLC2A3 in GBM. BMC Cancer 2013, 13, 478. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, R.; Chen, X.; Mu, Y.; Ai, J.; Shi, C.; Liu, Y.; Shi, C.; Sun, L.; Rainov, N.G.; et al. MiR-106a inhibits glioma cell growth by targeting E2F1 independent of p53 status. J. Mol. Med. 2011, 89, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Yang, G.; Mu, Y.; Han, D.; Shi, C.; Chen, X.; Deng, Y.; Zhang, D.; Wang, L.; Liu, Y.; et al. MiR-106a is an independent prognostic marker in patients with glioblastoma. Neuro Oncol. 2013, 15, 707–717. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhi, F.; Zhou, G.; Shao, N.; Xia, X.; Shi, Y.; Wang, Q.; Zhang, Y.; Wang, R.; Xue, L.; Wang, S.; et al. miR-106a-5p inhibits the proliferation and migration of astrocytoma cells and promotes apoptosis by targeting FASTK. PLoS ONE 2013, 8, e72390. [Google Scholar] [CrossRef]
- Jones, T.A.; Jeyapalan, J.N.; Forshew, T.; Tatevossian, R.G.; Lawson, A.R.; Patel, S.N.; Doctor, G.T.; Mumin, M.A.; Picker, S.R.; Phipps, K.P.; et al. Molecular analysis of pediatric brain tumors identifies microRNAs in pilocytic astrocytomas that target the MAPK and NF-kappaB pathways. Acta Neuropathol. Commun. 2015, 3, 86. [Google Scholar] [CrossRef]
- Schulte, J.H.; Horn, S.; Otto, T.; Samans, B.; Heukamp, L.C.; Eilers, U.C.; Krause, M.; Astrahantseff, K.; Klein-Hitpass, L.; Buettner, R.; et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int. J. Cancer 2008, 122, 699–704. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Z.; Chu, L.; Li, X.; Kan, P.; Xin, X.; Zhu, Y.; Yang, P. The Effects and Molecular Mechanisms of MiR-106a in Multidrug Resistance Reversal in Human Glioma U87/DDP and U251/G Cell Lines. PLoS ONE 2015, 10, e0125473. [Google Scholar] [CrossRef]
- Davey, M.G.; Davies, M.; Lowery, A.J.; Miller, N.; Kerin, M.J. The Role of MicroRNA as Clinical Biomarkers for Breast Cancer Surgery and Treatment. Int. J. Mol. Sci. 2021, 22, 8290. [Google Scholar] [CrossRef]
- You, F.; Luan, H.; Sun, D.; Cui, T.; Ding, P.; Tang, H.; Sun, D. miRNA-106a Promotes Breast Cancer Cell Proliferation, Clonogenicity, Migration, and Invasion Through Inhibiting Apoptosis and Chemosensitivity. DNA Cell Biol. 2019, 38, 198–207. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef]
- Liu, C.; Song, Y.H.; Mao, Y.; Wang, H.B.; Nie, G. MiRNA-106a promotes breast cancer progression by regulating DAX-1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1574–1583. [Google Scholar] [CrossRef]
- Yang, W.S.; Chadalapaka, G.; Cho, S.G.; Lee, S.O.; Jin, U.H.; Jutooru, I.; Choi, K.; Leung, Y.K.; Ho, S.M.; Safe, S.; et al. The transcriptional repressor ZBTB4 regulates EZH2 through a MicroRNA-ZBTB4-specificity protein signaling axis. Neoplasia 2014, 16, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Favier, A.; Rocher, G.; Larsen, A.K.; Delangle, R.; Uzan, C.; Sabbah, M.; Castela, M.; Duval, A.; Mehats, C.; Canlorbe, G. MicroRNA as Epigenetic Modifiers in Endometrial Cancer: A Systematic Review. Cancers 2021, 13, 1137. [Google Scholar] [CrossRef] [PubMed]
- Ravegnini, G.; Gorini, F.; De Crescenzo, E.; De Leo, A.; De Biase, D.; Di Stanislao, M.; Hrelia, P.; Angelini, S.; De Iaco, P.; Perrone, A.M. Can miRNAs be useful biomarkers in improving prognostic stratification in endometrial cancer patients? An update review. Int. J. Cancer 2022, 150, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.N.; Bai, J.X.; Zhou, Q.; Yan, B.; Qin, W.W.; Jia, L.T.; Meng, Y.L.; Jin, B.Q.; Yao, L.B.; Wang, T.; et al. TSA suppresses miR-106b-93-25 cluster expression through downregulation of MYC and inhibits proliferation and induces apoptosis in human EMC. PLoS ONE 2012, 7, e45133. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, K.; Tong, Y.; Dai, X.; Xu, T.; He, D.; Ying, J. Novel miRNA markers for the diagnosis and prognosis of endometrial cancer. J. Cell. Mol. Med. 2020, 24, 4533–4546. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Li, J.; Liu, H.; Zhou, F.; Liu, M. MiR-106a promotes tumor growth, migration, and invasion by targeting BCL2L11 in human endometrial adenocarcinoma. Am. J. Transl. Res. 2017, 9, 4984–4993. [Google Scholar]
- Oplawski, M.; Nowakowski, R.; Srednicka, A.; Ochnik, D.; Grabarek, B.O.; Boron, D. Molecular Landscape of the Epithelial-Mesenchymal Transition in Endometrioid Endometrial Cancer. J. Clin. Med. 2021, 10, 1520. [Google Scholar] [CrossRef]
- Lai, Y.; Zhou, B.; Tan, Q.; Xu, J.; Wan, T.; Zhang, L. LINC00116 enhances cervical cancer tumorigenesis through miR-106a/c-Jun pathway. J. Cell. Biochem. 2020, 121, 2247–2257. [Google Scholar] [CrossRef]
- Mitra, T.; Elangovan, S. Cervical cancer development, chemoresistance, and therapy: A snapshot of involvement of microRNA. Mol. Cell. Biochem. 2021, 476, 4363–4385. [Google Scholar] [CrossRef]
- Sadri Nahand, J.; Moghoofei, M.; Salmaninejad, A.; Bahmanpour, Z.; Karimzadeh, M.; Nasiri, M.; Mirzaei, H.R.; Pourhanifeh, M.H.; Bokharaei-Salim, F.; Mirzaei, H.; et al. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: A review. Int. J. Cancer 2020, 146, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, X.; Zhou, X.; Jia, J.; Chen, H.; Zhao, W. miR-106a Regulates Cell Proliferation and Autophagy by Targeting LKB1 in HPV-16-Associated Cervical Cancer. Mol. Cancer Res. 2020, 18, 1129–1141. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Q.; Tao, L.; Yu, C. MicroRNA-106a promotes cell migration and invasion by targeting tissue inhibitor of matrix metalloproteinase 2 in cervical cancer. Oncol. Rep. 2017, 38, 1774–1782. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.H.; Jin, M.; Wang, L.Q.; Xu, G.J.; Lin, Z.Y.; Yu, D.D.; Yang, S.L.; Ran, R.Z.; Wu, G.; Zhang, T. Long noncoding RNA TCL6 binds to miR-106a-5p to regulate hepatocellular carcinoma cells through PI3K/AKT signaling pathway. J. Cell. Physiol. 2020, 235, 6154–6166. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Zhi, Q.; Zhao, H.; Han, Y.; Gao, L.; Wang, B.; Kou, Z.; Guo, Z.; He, S.; Xue, X.; et al. Upregulated expression of miR-106a by DNA hypomethylation plays an oncogenic role in hepatocellular carcinoma. Tumour Biol. 2015, 36, 3093–3100. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Y.; Dong, K.; Ma, Y.; Jin, Q.; Yin, S.; Zhu, X.; Wang, S. Effects of FER1L4-miR-106a-5p/miR-372-5p-E2F1 regulatory axis on drug resistance in liver cancer chemotherapy. Mol. Ther. Nucleic Acids 2021, 24, 449–461. [Google Scholar] [CrossRef]
- Xue, X.; Zhao, Y.; Wang, X.; Qin, L.; Hu, R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J. Cell. Biochem. 2019, 120, 135–142. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Z.; Qiao, G.; Tang, B.; Li, P. Visualization of endoplasmic reticulum viscosity in the liver of mice with nonalcoholic fatty liver disease by a near-infrared fluorescence probe. Chin. Chem. Lett. 2021, 32, 3641–3645. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Hou, Y.; Yang, Q.; Chen, S.; Wang, X.; Wang, Z.; Yang, Y.; Chen, C.; Wang, Z.; et al. The PDGF-D/miR-106a/Twist1 pathway orchestrates epithelial-mesenchymal transition in gemcitabine resistance hepatoma cells. Oncotarget 2015, 6, 7000–7010. [Google Scholar] [CrossRef]
- Deng, P.; Wu, Y. Knockdown of miR-106a suppresses migration and invasion and enhances radiosensitivity of hepatocellular carcinoma cells by upregulating FBXW7. Int. J. Clin. Exp. Pathol. 2019, 12, 1184–1193. [Google Scholar]
- Liang, Z.; Li, X.; Duan, F.; Song, L.; Wang, Z.; Li, X.; Yang, P.; Li, L. Protein tyrosine phosphatase non-receptor type 12 (PTPN12), negatively regulated by miR-106a-5p, suppresses the progression of hepatocellular carcinoma. Hum. Cell 2022, 35, 299–309. [Google Scholar] [CrossRef]
- Wang, J.L.; Li, H.; Zhang, J.B.; Zhang, C.H.; Hou, X.Q. Suppression of connexin 43 expression by miR-106a promotes melanoma cell proliferation. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Lai, Y.C.; Husna, A.A.; Chen, H.W.; Tanaka, Y.; Kawaguchi, H.; Miyoshi, N.; Nakagawa, T.; Fukushima, R.; Miura, N. Micro RNA Transcriptome Profile in Canine Oral Melanoma. Int. J. Mol. Sci. 2019, 20, 4832. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.; Raymond, G.; Tzou, K.S.; Baxi, S.; Mani, R.R.; Kumar Govind, S.; Chandramoorthy, H.C.; Sivanandy, P.; Rajagopal, M.; Samiappan, S.; et al. Molecular Investigation of miRNA Biomarkers as Chemoresistance Regulators in Melanoma: A Protocol for Systematic Review and Meta-Analysis. Genes 2022, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Wroblewska, J.P.; Lach, M.S.; Ustaszewski, A.; Kulcenty, K.; Ibbs, M.; Jagiello, I.; Suchorska, W.M.; Marszalek, A. The Potential Role of Selected miRNA in Uveal Melanoma Primary Tumors as Early Biomarkers of Disease Progression. Genes 2020, 11, 271. [Google Scholar] [CrossRef]
- Ming, J.; Zhou, Y.; Du, J.; Fan, S.; Pan, B.; Wang, Y.; Fan, L.; Jiang, J. miR-381 suppresses C/EBPalpha-dependent Cx43 expression in breast cancer cells. Biosci. Rep. 2015, 35, e00266. [Google Scholar] [CrossRef]
- Luan, W.; Zhou, Z.; Ni, X.; Xia, Y.; Wang, J.; Yan, Y.; Xu, B. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J. Cancer Res. Clin. Oncol. 2018, 144, 531–542. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, T.; Yang, X.; Liu, C.; Li, P.; Wang, Z.; Zhi, S. MicroRNA106a regulates the proliferation and invasion of human osteosarcoma cells by targeting VNN2. Oncol. Rep. 2018, 40, 2251–2259. [Google Scholar] [CrossRef]
- Guo, J.; Dou, D.; Zhang, T.; Wang, B. HOTAIR Promotes Cisplatin Resistance of Osteosarcoma Cells by Regulating Cell Proliferation, Invasion, and Apoptosis via miR-106a-5p/STAT3 Axis. Cell Transplant. 2020, 29, 1–12. [Google Scholar] [CrossRef]
- Nakka, M.; Allen-Rhoades, W.; Li, Y.; Kelly, A.J.; Shen, J.; Taylor, A.M.; Barkauskas, D.A.; Yustein, J.T.; Andrulis, I.L.; Wunder, J.S.; et al. Biomarker significance of plasma and tumor miR-21, miR-221, and miR-106a in osteosarcoma. Oncotarget 2017, 8, 96738–96752. [Google Scholar] [CrossRef]
- Huang, J.; Takeda, Y.; Watanabe, T.; Sendo, F. A sandwich ELISA for detection of soluble GPI-80, a glycosylphosphatidyl-inositol (GPI)-anchored protein on human leukocytes involved in regulation of neutrophil adherence and migration—Its release from activated neutrophils and presence in synovial fluid of rheumatoid arthritis patients. Microbiol. Immunol. 2001, 45, 467–471. [Google Scholar] [CrossRef]
- Zhao, H.; Yan, P.; Wang, J.; Zhang, Y.; Zhang, M.; Wang, Z.; Fu, Q.; Liang, W. Clinical significance of tumor miR-21, miR-221, miR-143, and miR-106a as biomarkers in patients with osteosarcoma. Int. J. Biol. Markers 2019, 34, 184–193. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Zhao, T.; Xu, Z.; Chi, W.; Chai, X.; Ai, J.; Zhang, J.; Huo, F.; Strongin, R.M.; Yin, C. Enlarging the Stokes Shift by Weakening the pi-Conjugation of Cyanines for High Signal-to-Noise Ratiometric Imaging. Adv. Sci. 2023, 10, e2205080. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Yang, W.; Feng, W.; Cao, L.; Wang, X.; Niu, L.; Li, Y.; Zhou, W.; Zhang, Y.; Liu, J.; et al. Molecular mechanisms and clinical implications of miRNAs in drug resistance of colorectal cancer. Ther. Adv. Med. Oncol. 2020, 12, 1–28. [Google Scholar] [CrossRef]
- Ren, X.; Liao, L.; Yang, Z.; Li, H.; Li, X.; Wang, Y.; Ye, Y.; Song, X. Rational design of a bifunctional fluorescent probe for distinguishing Hcy/Cys from GSH with ideal properties. Chin. Chem. Lett. 2021, 32, 1061–1065. [Google Scholar] [CrossRef]
- Ghasabi, M.; Mansoori, B.; Mohammadi, A.; Duijf, P.H.; Shomali, N.; Shirafkan, N.; Mokhtarzadeh, A.; Baradaran, B. MicroRNAs in cancer drug resistance: Basic evidence and clinical applications. J. Cell. Physiol. 2019, 234, 2152–2168. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Liu, Y.; Shang, L.; Zhou, F.; Yang, S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun. 2021, 41, 199–217. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zheng, Y.; Ma, L.; Tian, L.; Sun, Q. Clinically-Relevant ABC Transporter for Anti-Cancer Drug Resistance. Front. Pharmacol. 2021, 12, 648407. [Google Scholar] [CrossRef]
- Lok, A.S.; McMahon, B.J. Chronic hepatitis B: Update 2009. Hepatology 2009, 50, 661–662. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, D.; Tout, I.; Narguet, S.; Benazzouz, S.M.; Mansouri, A.; Asselah, T. miRNAs as Potential Biomarkers for Viral Hepatitis B and C. Viruses 2020, 12, 1440. [Google Scholar] [CrossRef]
- Morgan, M.; Keeffe, E.B. Diagnosis and treatment of chronic hepatitis B: 2009 update. Minerva Gastroenterol. Dietol. 2009, 55, 5–22. [Google Scholar] [PubMed]
- Boltjes, A.; Groothuismink, Z.M.; van Oord, G.W.; Janssen, H.L.; Woltman, A.M.; Boonstra, A. Monocytes from chronic HBV patients react in vitro to HBsAg and TLR by producing cytokines irrespective of stage of disease. PLoS ONE 2014, 9, e97006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Gao, Y.; Du, C.; Markowitz, G.J.; Fu, J.; Zhang, Z.; Liu, C.; Qin, W.; Wang, H.; Wang, F.; et al. Hepatitis B-Induced IL8 Promotes Hepatocellular Carcinoma Venous Metastasis and Intrahepatic Treg Accumulation. Cancer Res. 2021, 81, 2386–2398. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Hong, H.; Liu, J.; Zheng, X.; Huang, M.; Li, C.; Xia, J. miR-106a Is Downregulated in Peripheral Blood Mononuclear Cells of Chronic Hepatitis B and Associated with Enhanced Levels of Interleukin-8. Mediat. Inflamm. 2015, 2015, 629862. [Google Scholar] [CrossRef]
- Basak, J.; Majsterek, I. miRNA-Dependent CD4(+) T Cell Differentiation in the Pathogenesis of Multiple Sclerosis. Mult. Scler. Int. 2021, 2021, 8825588. [Google Scholar] [CrossRef]
- Rahimirad, S.; Navaderi, M.; Alaei, S.; Sanati, M.H. Identification of hsa-miR-106a-5p as an impact agent on promotion of multiple sclerosis using multi-step data analysis. Neurol. Sci. 2021, 42, 3791–3799. [Google Scholar] [CrossRef]
- Sanctuary, M.R.; Huang, R.H.; Jones, A.A.; Luck, M.E.; Aherne, C.M.; Jedlicka, P.; de Zoeten, E.F.; Collins, C.B. miR-106a deficiency attenuates inflammation in murine IBD models. Mucosal Immunol. 2019, 12, 200–211. [Google Scholar] [CrossRef]
- Zailaie, S.A.; Siddiqui, J.J.; Al Saadi, R.M.; Anbari, D.M.; Alomari, A.S.; Cupler, E.J. Serum Based miRNA as a Diagnostic Biomarker for Multiple Sclerosis: A Systematic Review and Meta-Analysis. Immunol. Investig. 2022, 51, 947–962. [Google Scholar] [CrossRef]
- Zhu, D.; Pan, C.; Li, L.; Bian, Z.; Lv, Z.; Shi, L.; Zhang, J.; Li, D.; Gu, H.; Zhang, C.Y.; et al. MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein alpha. J. Allergy Clin. Immunol. 2013, 132, 426–436 e8. [Google Scholar] [CrossRef]
- Bo, C.; Wang, J.; Zhang, H.; Cao, Y.; Lu, X.; Wang, T.; Wang, Y.; Li, S.; Kong, X.; Sun, X.; et al. Global pathway view analysis of microRNA clusters in myasthenia gravis. Mol. Med. Rep. 2019, 19, 2350–2360. [Google Scholar] [CrossRef]
- Saghazadeh, A.; Rezaei, N. MicroRNA expression profiles of peripheral blood and mononuclear cells in myasthenia gravis: A systematic review. Int. Immunopharmacol. 2022, 112, 109205. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Bao, Z.; Liang, D.; Li, M.; Wei, M.; Ge, X.; Liu, J.; Li, J. Plasma exosomal miR-106a-5p expression in myasthenia gravis. Muscle Nerve 2020, 61, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Holvoet, P. Non-Coding RNAs in Cardiomyopathy and Heart Failure. In Non-Coding RNAs at the Cross-Road of Cardiometabolic Diseases and Cancer; Springer International Publishing: Cham, Switzerland, 2021; pp. 119–147. [Google Scholar]
- Guan, X.; Wang, L.; Liu, Z.; Guo, X.; Jiang, Y.; Lu, Y.; Peng, Y.; Liu, T.; Yang, B.; Shan, H.; et al. miR-106a promotes cardiac hypertrophy by targeting mitofusin 2. J. Mol. Cell. Cardiol. 2016, 99, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Li, C.; Liu, J.; Wang, Z.; Liu, Y.; Luo, C.; Chen, Y.; Wen, S. Expression Profile of microRNAs in Hypertrophic Cardiomyopathy and Effects of microRNA-20 in Inducing Cardiomyocyte Hypertrophy Through Regulating Gene MFN2. DNA Cell Biol. 2019, 38, 796–807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.B.; Qi, Y.F.; Xiao, Z.X.; Chen, H.; Liu, S.H.; Li, Z.Z.; Zeng, Z.F.; Wu, H.F. CircHIPK3 Regulates Vascular Smooth Muscle Cell Calcification Via the miR-106a-5p/MFN2 Axis. J. Cardiovasc. Transl. Res. 2022, 15, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Wu, W.; Qu, J.; Wang, X.; Lei, S.; Yuan, L.; Liu, X. Inhibition of Mitofusin-2 Promotes Cardiac Fibroblast Activation via the PERK/ATF4 Pathway and Reactive Oxygen Species. Oxidative Med. Cell Longev. 2019, 2019, 3649808. [Google Scholar] [CrossRef]
- Jung, J.H.; Ikeda, G.; Tada, Y.; von Bornstadt, D.; Santoso, M.R.; Wahlquist, C.; Rhee, S.; Jeon, Y.J.; Yu, A.C.; O’Brien, C.G.; et al. miR-106a-363 cluster in extracellular vesicles promotes endogenous myocardial repair via Notch3 pathway in ischemic heart injury. Basic. Res. Cardiol. 2021, 116, 19. [Google Scholar] [CrossRef]
- Sethi, S.; Mehta, P.; Pandey, A.; Gupta, G.; Rajender, S. miRNA Profiling of Major Testicular Germ Cells Identifies Stage-Specific Regulators of Spermatogenesis. Reprod. Sci. 2022, 29, 3477–3493. [Google Scholar] [CrossRef]
- Sharma, P.; Ghanghas, P.; Kaushal, N.; Kaur, J.; Kaur, P. Epigenetics and oxidative stress: A twin-edged sword in spermatogenesis. Andrologia 2019, 51, e13432. [Google Scholar] [CrossRef]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef]
- He, Z.; Jiang, J.; Kokkinaki, M.; Tang, L.; Zeng, W.; Gallicano, I.; Dobrinski, I.; Dym, M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells 2013, 31, 2205–2217. [Google Scholar] [CrossRef] [PubMed]
- Hackl, M.; Brunner, S.; Fortschegger, K.; Schreiner, C.; Micutkova, L.; Muck, C.; Laschober, G.T.; Lepperdinger, G.; Sampson, N.; Berger, P.; et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 2010, 9, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Sandiford, O.A.; Moore, C.A.; Du, J.; Boulad, M.; Gergues, M.; Eltouky, H.; Rameshwar, P. Human Aging and Cancer: Role of miRNA in Tumor Microenvironment. Adv. Exp. Med. Biol. 2018, 1056, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Grillari, J.; Grillari-Voglauer, R. Novel modulators of senescence, aging, and longevity: Small non-coding RNAs enter the stage. Exp. Gerontol. 2010, 45, 302–311. [Google Scholar] [CrossRef]
- Ren, S.; Lin, P.; Wang, J.; Yu, H.; Lv, T.; Sun, L.; Du, G. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as an miRNA Sponge. Mol. Ther. Methods Clin. Dev. 2020, 18, 215–229. [Google Scholar] [CrossRef]
- Tai, L.; Huang, C.J.; Choo, K.B.; Cheong, S.K.; Kamarul, T. Oxidative Stress Down-Regulates MiR-20b-5p, MiR-106a-5p and E2F1 Expression to Suppress the G1/S Transition of the Cell Cycle in Multipotent Stromal Cells. Int. J. Med. Sci. 2020, 17, 457–470. [Google Scholar] [CrossRef]
- ElSharawy, A.; Keller, A.; Flachsbart, F.; Wendschlag, A.; Jacobs, G.; Kefer, N.; Brefort, T.; Leidinger, P.; Backes, C.; Meese, E.; et al. Genome-wide miRNA signatures of human longevity. Aging Cell 2012, 11, 607–616. [Google Scholar] [CrossRef]
- Tiberio, P.; Callari, M.; Angeloni, V.; Daidone, M.G.; Appierto, V. Challenges in using circulating miRNAs as cancer biomarkers. Biomed. Res. Int. 2015, 2015, 731479. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Singh, R.; Ramasubramanian, B.; Kanji, S.; Chakraborty, A.R.; Haque, S.J.; Chakravarti, A. Circulating microRNAs in cancer: Hope or hype? Cancer Lett. 2016, 381, 113–121. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Ghaffari, S.H. Why have microRNA biomarkers not been translated from bench to clinic? Future Oncol. 2019, 15, 801–803. [Google Scholar] [CrossRef] [PubMed]
- Kotowska-Zimmer, A.; Pewinska, M.; Olejniczak, M. Artificial miRNAs as therapeutic tools: Challenges and opportunities. Wiley Interdiscip. Rev. RNA 2021, 12, e1640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cheng, Z.; Wang, Y.; Han, T. The Risks of miRNA Therapeutics: In a Drug Target Perspective. Drug Des. Dev. Ther. 2021, 15, 721–733. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).