ECG Recordings as Predictors of Very Early Autism Likelihood: A Machine Learning Approach
Abstract
1. Introduction
2. Methods
2.1. Acquisition Protocol
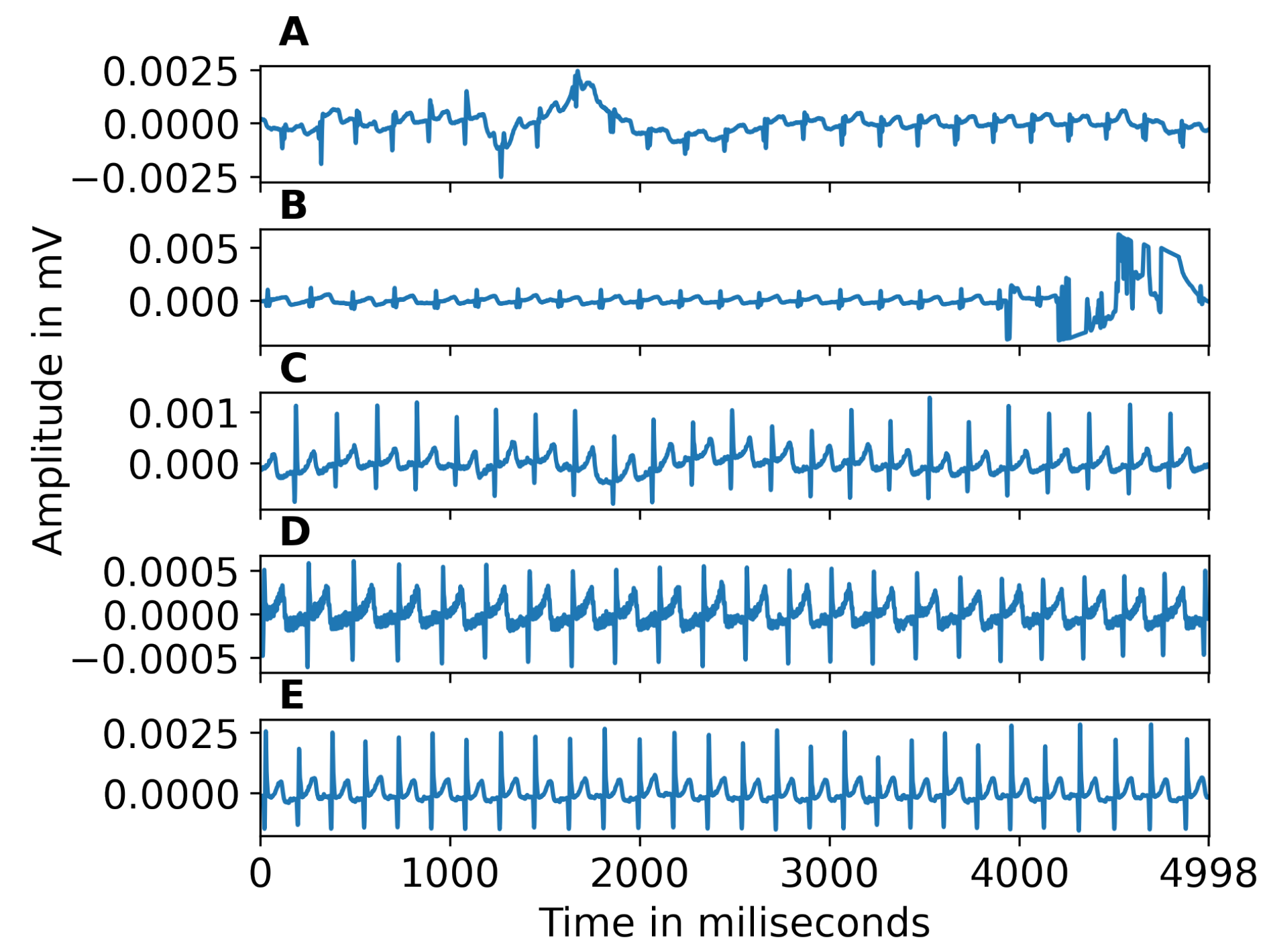
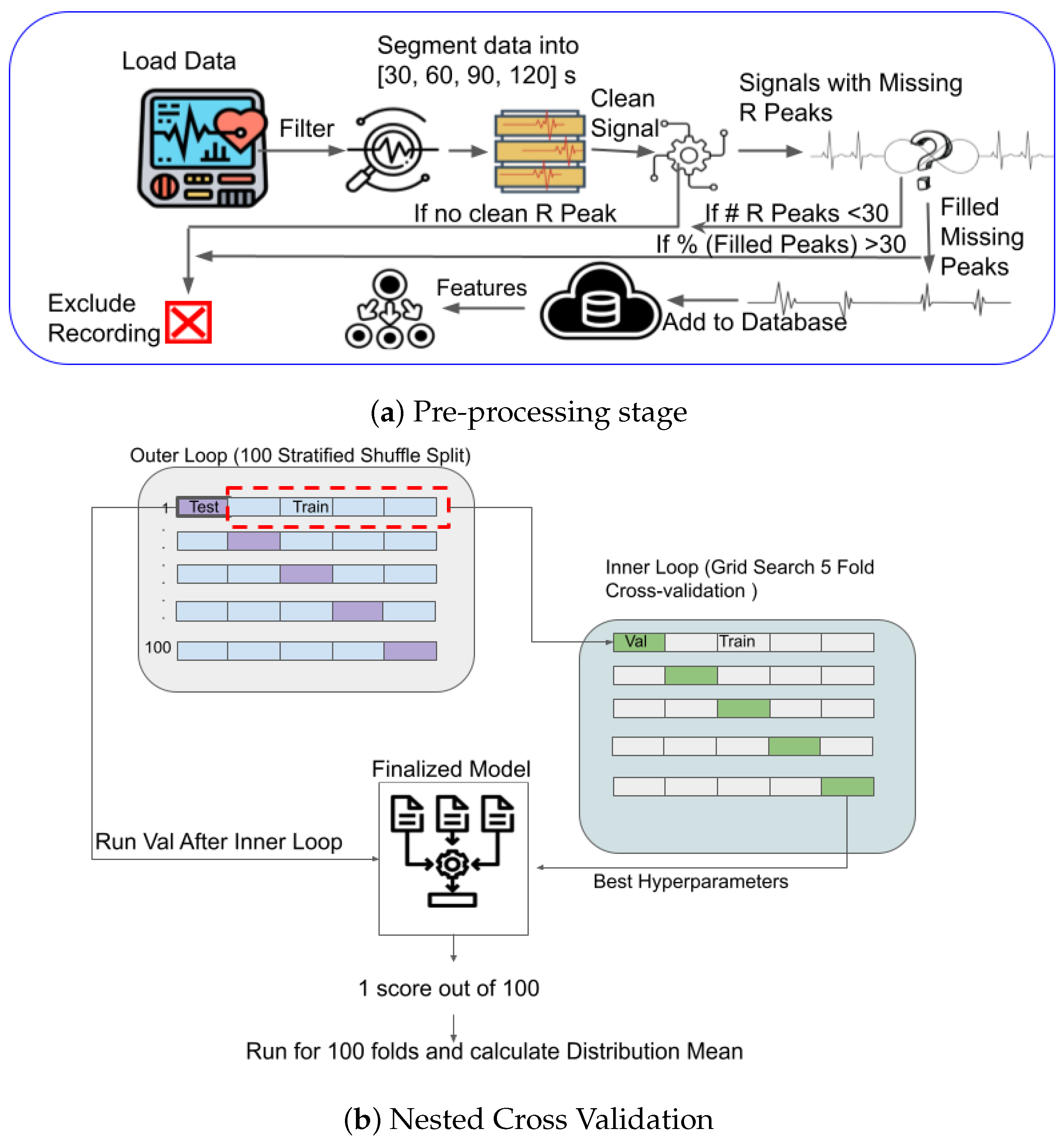
2.2. ECG Pre-Processing
2.3. Feature Extraction and Preliminary Analysis
2.4. EL vs. TL Classification Using Machine Learning
2.5. Cross Validation
2.6. Analysis Pipeline
3. Results
4. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADI | Autism Diagnostic Interview |
| ADOS | Autism Diagnostic Observation Schedule |
| ANS | Autonomic Nervous System |
| ASD | Autism Spectrum Disorder |
| CVI | Cardiovagal Index |
| CSI | Cardiac Sympathetic Index |
| ECG | Electrocardiogram |
| EDA | Electrodermal activity |
| EEG | Electroencephalogram |
| EL | Elevated Likelihood |
| fMRI | functional Magnetic Resonance Imaging |
| HRV | Heart Rate Variability |
| IQR | Interquartile Range |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| OIX | Object Only Intreactions |
| PIX | Caregiver Only Interactions |
| TL | Typical Likelihood |
References
- Carpenter, B. DSM-5; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar] [CrossRef]
- Guthrie, W.; Wetherby, A.M.; Woods, J.; Schatschneider, C.; Holland, R.D.; Morgan, L.; Lord, C.E. The earlier the better: An RCT of treatment timing effects for toddlers on the autism spectrum. Autism 2023, 13623613231159153. [Google Scholar] [CrossRef] [PubMed]
- Speaks, A. Autism Statistics and Facts. 2023. Available online: https://www.autismspeaks.org/autism-statistics-asd (accessed on 4 April 2023).
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Larsson, H.; Hultman, C.M.; Reichenberg, A. The Familial Risk of Autism. JAMA 2014, 311, 1770–1777. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.; Schendel, D.; Francis, R.; Windham, G.; Bresnahan, M.; Levine, S.; Reichenberg, A.; Gissler, M.; Kodesh, A.; Bai, D.; et al. Recurrence Risk of Autism in Siblings and Cousins: A Multi-National, Population-Based Study. J. Am. Acad. Child Adolesc. Psychiatry 2019, 58, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.; Del Rosario, M.M.; Nosco, E.; Li, Y.; Senturk, D.; Mcdonald, N.; Wilson, R.; Dapretto, M.; Jeste, S. Early concerns in parents of infants at risk for autism. Dev. Med. Child Neurol. 2021, 63, 1410–1416. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.; Steiner, A.; Gengoux, G.; Koegel, L. Feasibility and Effectiveness of Very Early Intervention for Infants At-Risk for Autism Spectrum Disorder: A Systematic Review. J. Autism Dev. Disord. 2014, 45, 778–794. [Google Scholar] [CrossRef] [PubMed]
- Frigaux, A.; Evrard, R.; Lighezzolo-Alnot, J. ADI-R and ADOS and the differential diagnosis of autism spectrum disorders: Interests, limits and openings. L’Encéphale 2019, 45, 441–448. [Google Scholar] [CrossRef]
- Hurwitz, S.; Yirmiya, N. Autism Diagnostic Observation Schedule (ADOS) and Its Uses in Research and Practice. In Comprehensive Guide to Autism; Springer: New York, NY, USA, 2014. [Google Scholar] [CrossRef]
- Bildt, A.; Sytema, S.; Zander, E.; Bölte, S.; Sturm, H.; Yirmiya, N.; Yaari, M.; Charman, T.; Salomone, E.; LeCouteur, A.; et al. Autism Diagnostic Interview-Revised (ADI-R) Algorithms for Toddlers and Young Preschoolers: Application in a Non-US Sample of 1,104 Children. J. Autism Dev. Disord. 2015, 45, 2076–2091. [Google Scholar] [CrossRef]
- Bosl, W.; Tager-Flusberg, H.; Nelson, C. “EEG” Analytics for Early Detection of Autism Spectrum Disorder: A data-driven approach. Sci. Rep. 2018, 8, 6828. [Google Scholar] [CrossRef]
- Estelle, H.; Mento, G.; Desnous, B.; François, C. Challenges and new perspectives of developmental cognitive EEG studies. NeuroImage 2022, 260, 119508. [Google Scholar] [CrossRef]
- Pereira, A.; Campos, B.; Coan, A.; Pegoraro, L.; Rezende, T.; Obeso, I.; Dalgalarrondo, P.; Dacosta, J.; Dreher, J.C.; Cendes, F. Differences in Cortical Structure and Functional MRI Connectivity in High Functioning Autism. Front. Neurol. 2018, 9, 539. [Google Scholar] [CrossRef]
- Bauman, M.; Kemper, T. Neuroanatomic observations of the brain in autism: A review and future directions. Int. J. Dev. Neurosci. 2005, 23, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Ecker, C. The neuroanatomy of autism spectrum disorder: An overview of structural neuroimaging findings and their translatability to the clinical setting. Autism 2016, 21, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Fenning, R.; Erath, S.; Baker, J.; Messinger, D.; Moffitt, J.; Baucom, B.; Kaeppler, A. Sympathetic-Parasympathetic Interaction and Externalizing Problems in Children with Autism Spectrum Disorder. Autism Res. 2019, 12, 1805–1816. [Google Scholar] [CrossRef] [PubMed]
- Panju, S.; Brian, J.; Dupuis, A.; Anagnostou, E.; Kushki, A. Atypical sympathetic arousal in children with autism spectrum disorder and its association with anxiety symptomatology. Mol. Autism 2015, 6, 64. [Google Scholar] [CrossRef]
- Prince, E.; Kim, E.; Wall, C.; Gisin, E.; Goodwin, M.; Schoen Simmons, E.; Chawarska, K.; Shic, F. The relationship between autism symptoms and arousal level in toddlers with autism spectrum disorder, as measured by electrodermal activity. Autism 2016, 21, 504–508. [Google Scholar] [CrossRef]
- MacNeil, B.; Lopes, V.; Minnes, P. Anxiety in children and adolescents with Autism Spectrum Disorders. Res. Autism Spectr. Disord. 2009, 3, 1–21. [Google Scholar] [CrossRef]
- Alvares, G.; Quintana, D.; Kemp, A.; Zwieten, A.; Balleine, B.; Hickie, I.; Guastella, A. Reduced Heart Rate Variability in Social Anxiety Disorder: Associations with Gender and Symptom Severity. PLoS ONE 2013, 8, e70468. [Google Scholar] [CrossRef]
- Benevides, T.; Lane, S. A Review of Cardiac Autonomic Measures: Considerations for Examination of Physiological Response in Children with Autism Spectrum Disorder. J. Autism Dev. Disord. 2013, 45, 560–575. [Google Scholar] [CrossRef]
- Eilam-Stock, T.; Xu, P.; Cao, M.; Gu, X.; Van Dam, N.; Anagnostou, E.; Kolevzon, A.; Soorya, L.; Park, Y.; Siller, M.; et al. Abnormal autonomic and associated brain activities during rest in autism spectrum disorder. Brain 2014, 137, 153–171. [Google Scholar] [CrossRef]
- Herry, C.L.; Burns, P.; Desrochers, A.; Fecteau, G.; Durosier, L.D.; Cao, M.; Seely, A.J.; Frasch, M.G. Vagal contributions to fetal heart rate variability: An omics approach. Physiol. Meas. 2019, 40, 065004. [Google Scholar] [CrossRef]
- Sassi, R.; Cerutti, S.; Lombardi, F.; Malik, M.; Huikuri, H.V.; Peng, C.K.; Schmidt, G.; Yamamoto, Y.; Reviewers:, D.; Gorenek, B.; et al. Advances in heart rate variability signal analysis: Joint position statement by the e-Cardiology ESC Working Group and the European Heart Rhythm Association co-endorsed by the Asia Pacific Heart Rhythm Society. Europace 2015, 17, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, F.; Ginsberg, J.P. An overview of heart rate variability metrics and norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef] [PubMed]
- Billeci, L.; Tonacci, A.; Narzisi, A.; Manigrasso, Z.; Varanini, M.; Fulceri, F.; Lattarulo, C.; Calderoni, S.; Muratori, F. Heart rate variability during a joint attention task in toddlers with autism spectrum disorders. Front. Physiol. 2018, 9, 467. [Google Scholar] [CrossRef] [PubMed]
- Bricout, V.A.; Pace, M.; Dumortier, L.; Favre-Juvin, A.; Guinot, M. Autonomic responses to head-up tilt test in children with autism spectrum disorders. J. Abnorm. Child Psychol. 2018, 46, 1121–1128. [Google Scholar] [CrossRef]
- Corbett, B.A.; Muscatello, R.A.; Baldinger, C. Comparing stress and arousal systems in response to different social contexts in children with ASD. Biol. Psychol. 2019, 140, 119–130. [Google Scholar] [CrossRef]
- Pagnozzi, A.; Conti, E.; Calderoni, S.; Fripp, J.; Rose, S. A systematic review of structural MRI biomarkers in Autism Spectrum Disorder: A Machine Learning perspective. Int. J. Dev. Neurosci. 2018, 71, 68–82. [Google Scholar] [CrossRef]
- Yassin, W.; Nakatani, H.; Zhu, Y.; Kojima, M.; Owada, K.; Kuwabara, H.; Gonoi, W.; Aoki, Y.; Takao, H.; Natsubori, T.; et al. Machine-learning classification using neuroimaging data in schizophrenia, autism, ultra-high risk and first-episode psychosis. Transl. Psychiatry 2020, 10, 278. [Google Scholar] [CrossRef]
- Eslami, T.; Almuqhim, F.; Raiker, J.S.; Saeed, F. Machine Learning Methods for Diagnosing Autism Spectrum Disorder and Attention- Deficit/Hyperactivity Disorder Using Functional and Structural MRI: A Survey. Front. Neuroinform. 2021, 14, 62. [Google Scholar] [CrossRef]
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.; Saeed, F. ASD-DiagNet: A hybrid learning approach for detection of Autism Spectrum Disorder using f“MRI” data. Front. Neuroinform. 2019, 13, 70. [Google Scholar] [CrossRef]
- Han, Y.; Rizzo, D.; Hanley, J.; Coderre, E.; Prelock, P. Identifying neuroanatomical and behavioral features for autism spectrum disorder diagnosis in children using machine learning. PLoS ONE 2022, 17, e0269773. [Google Scholar] [CrossRef]
- Just, M.; Cherkassky, V.; Buchweitz, A.; Keller, T.; Tom, M. Identifying Autism from Neural Representations of Social Interactions: Neurocognitive Markers of Autism. PLoS ONE 2014, 9, e113879. [Google Scholar] [CrossRef] [PubMed]
- Nair, B.; Ashok, G.; Sreekumar, N. Classification of autism based on feature extraction from segmented brain MRI. Int. J. Recent Technol. Eng. 2019, 7, 85–89. [Google Scholar]
- Song, J.W.; Yoon, N.R.; Jang, S.M.; Lee, G.Y.; Kim, B. Neuroimaging-Based Deep Learning in Autism Spectrum Disorder and Attention-Deficit/Hyperactivity Disorder. J. Korean Acad. Child Adolesc. Psychiatry 2020, 31, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Frazier, T.; Strauss, M.; Klingemier, E.; Zetzer, E.; Hardan, A.; Eng, C.; Youngstrom, E. A Meta-Analysis of Gaze Differences to Social and Nonsocial Information Between Individuals With and Without Autism. J. Am. Acad. Child Adolesc. Psychiatry 2017, 56, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Kaliukhovich, D.; Manyakov, N.; Bangerter, A.; Ness, S.; Skalkin, A.; Goodwin, M.; Dawson, G.; Hendren, R.; Leventhal, B.; Hudac, C.; et al. Social attention to activities in children and adults with autism spectrum disorder: Effects of context and age. Mol. Autism 2020, 11, 79. [Google Scholar] [CrossRef]
- Brihadiswaran, G.; Haputhanthri, D.; Gunathilaka, S.; Meedeniya, D.; Jayarathna, S. “EEG”-based processing and classification methodologies for Autism Spectrum Disorder: A Review. J. Comput. Sci. 2019, 15, 1161–1183. [Google Scholar] [CrossRef]
- Hyde, K.; Novack, M.; LaHaye, N.; Parlett-Pelleriti, C.; Anden, R.; Dixon, D.; Linstead, E. Applications of Supervised Machine Learning in Autism Spectrum Disorder Research: A Review. Rev. J. Autism Dev. Disord. 2019, 6, 128–146. [Google Scholar] [CrossRef]
- Tonnsen, B.; Richards, J.; Roberts, J. Heart rate-defined sustained attention in infants at risk for autism. J. Neurodev. Disord. 2018, 10, 7. [Google Scholar] [CrossRef]
- van Gent, P.; Farah, H.; Nes, N.; Arem, B. HeartPy: A novel heart rate algorithm for the analysis of noisy signals. Transp. Res. Part F Traffic Psychol. Behav. 2019, 66, 368–378. [Google Scholar] [CrossRef]
- Simjanoska, M.; Papa, G.; Seljak, B.K.; Eftimov, T. Comparing Different Settings of Parameters Needed for Pre-processing of ECG Signals used for Blood Pressure Classification. In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), Prague, Czech Republic, 22–24 February 2019; pp. 62–72. [Google Scholar] [CrossRef]
- Makowski, D.; Pham, T.; Lau, Z.J.; Brammer, J.C.; Lespinasse, F.; Pham, H.; Schölzel, C.; Chen, S.H.A. NeuroKit2: A Python toolbox for neurophysiological signal processing. Behav. Res. Methods 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- Hnatkova, K.; Copie, X.; Staunton, A.; Malik, M. Numeric processing of Lorenz plots of R-R intervals from long-term ECGs: Comparison with time-domain measures of heart rate variability for risk stratification after myocardial infarction. J. Electrocardiol. 1995, 28, 74–80. [Google Scholar] [CrossRef] [PubMed]
- kumar, C.; Mullagiri, M.; Maruthy, K.; Siva Kumar, A.V.; Kuppusamy, M. Association of Heart rate variability measured by RR interval from ECG and pulse to pulse interval from Photoplethysmography. Clin. Epidemiol. Glob. Health 2021, 10, 100698. [Google Scholar] [CrossRef]
- Anandhi, B.; Selvaraj, J.; Anusuya, I.; Das, H. Time Domain Analysis of Heart Rate Variability Signals in Valence Recognition for Children with Autism Spectrum Disorder (ASD). IRBM 2021, 43, 380–390. [Google Scholar] [CrossRef]
- Dodo, N.; Hashimoto, R. Autonomic Nervous System Activity During a Speech Task. Front. Neurosci. 2019, 13, 406. [Google Scholar] [CrossRef] [PubMed]
- Low, P.; Tomalia, V.; Park, K.J. Autonomic Function Tests: Some Clinical Applications. J. Clin. Neurol. 2013, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2012, 12, 2825–2830. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Tohka, J.; Gils, M. Evaluation of machine learning algorithms for Health and Wellness applications: A tutorial. Comput. Biol. Med. 2021, 132, 104324. [Google Scholar] [CrossRef]
- Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef]
- He, H.; Ma, Y. Imbalanced Learning: Foundations, Algorithms, and Applications, 1st ed.; Wiley-IEEE Press: Hoboken, NJ, USA, 2013; p. 27. [Google Scholar] [CrossRef]
- Frasch, M.; Shen, C.; Wu, H.T.; Müller, A.; Neuhaus, E.; Bernier, R.; Kamara, D.; Beauchaine, T. Can a composite heart rate variability biomarker shed new insights about autism spectrum disorder in school-age children? J. Autism Dev. Disord. 2020, 51, 346–356. [Google Scholar] [CrossRef]
- Ebrahimi, Z.; Loni, M.; Daneshtalab, M.; Gharehbaghi, A. A Review on Deep Learning Methods for ECG Arrhythmia Classification. Expert Syst. Appl. X 2020, 7, 100033. [Google Scholar] [CrossRef]
- Doussard-Roosevelt, J.A.; Porges, S.W.; Scanlon, J.W.; Alemi, B.; Scanlon, K.B. Vagal regulation of heart rate in the prediction of developmental outcome for very low birth weight preterm infants. Child Dev. 1997, 68, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Patriquin, M.; Lorenzi, J.; Scarpa, A.; Bell, M.A. Developmental trajectories of respiratory sinus arrhythmia: Associations with social responsiveness. Dev. Psychobiol. 2014, 56, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Taylor, Z.; Eisenberg, N.; Spinrad, T. Respiratory Sinus Arrhythmia, Effortful Control, and Parenting as Predictors of Children’s Sympathy Across Early Childhood. Dev. Psychol. 2014, 51, 17–25. [Google Scholar] [CrossRef]
- Bal, E.; Harden, E.; Lamb, D.; Van Hecke, A.; Denver, J.; Porges, S. Emotion Recognition in Children with Autism Spectrum Disorders: Relations to Eye Gaze and Autonomic State. J. Autism Dev. Disord. 2010, 40, 358–370. [Google Scholar] [CrossRef]
- Bell, M.A.; Cuevas, K. Using EEG to Study Cognitive Development: Issues and Practices. J. Cogn. Dev. 2012, 13, 281–294. [Google Scholar] [CrossRef]

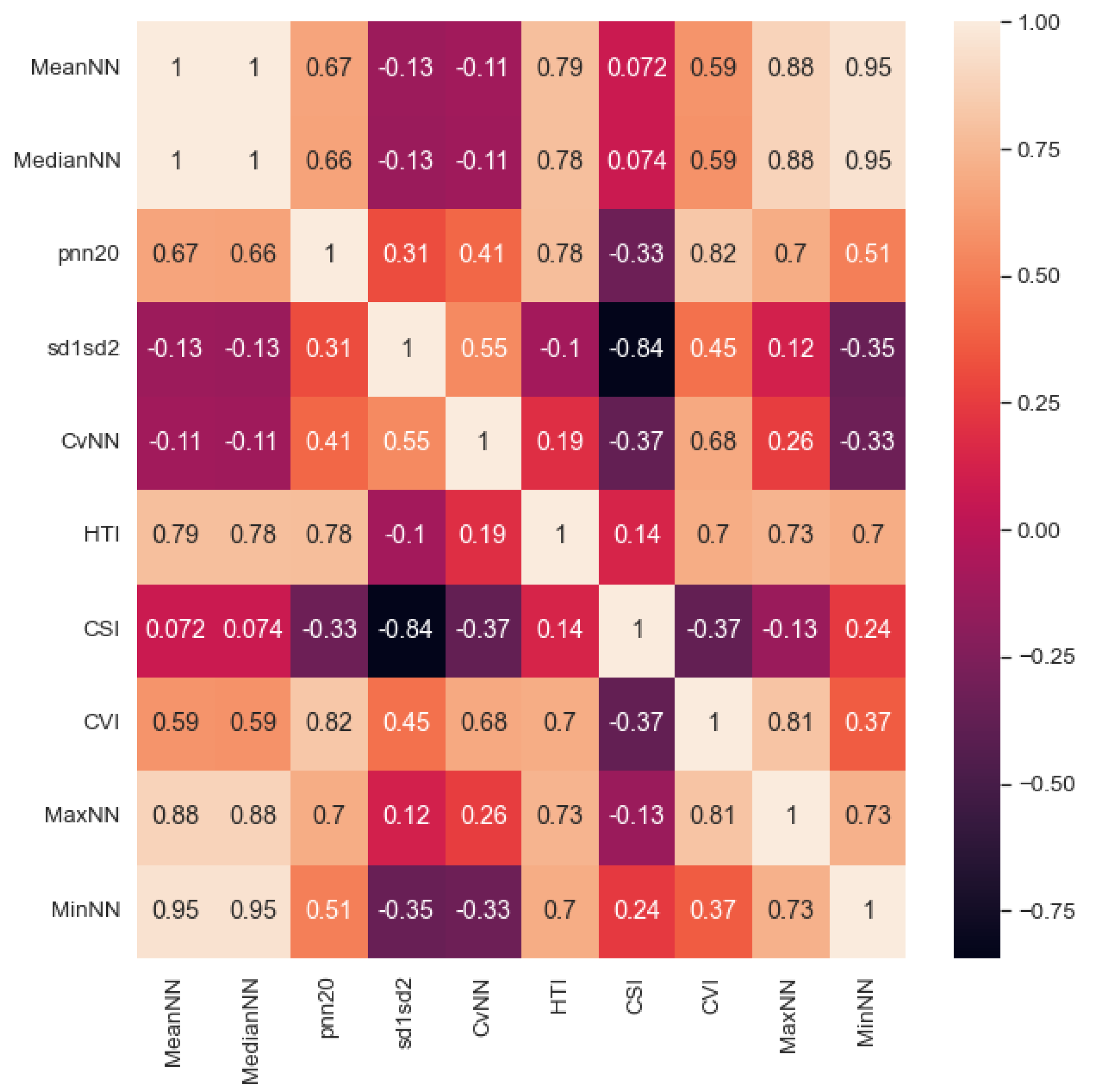


| Features | Feature Description |
|---|---|
| MeanNN | Mean of the NN [46,47]. |
| MedianNN | Median of the NN [46,47]. |
| pNN20 | Proportion of successive NN with a difference larger than 20 ms. This measures the relative frequency of changes in heart rhythm [22]. |
| sd1sd2 | In a Poincaré plot, sd1 is standard deviation perpendicular to the line of identity and sd2 is standard deviation along the line of identity. sd1sd2 is the ratio of (sd1/sd2), an indicator of the unpredictability of the NN to measure autonomic balance when there is sympathetic activation [22]. |
| CVNN | Coefficient of variation of NN, calculated as standard deviation of NN divided by mean of NN [26]. |
| HTI | HRV Triangular Index is the integral of the NN density histrogram divided by its height [22]. |
| CSI | The CSI quantifies the sympathetic nervous system activity indicating increased arousal [48]. |
| CVI | The CVI quantifies the parasympathetic nervous system activity [49]. |
| MaxNN | Max of the NN [46,47]. |
| MinNN | Min of NN [46,47]. |
| ADB | |||||||
|---|---|---|---|---|---|---|---|
| Segment Length | Accuracy p-val | Accuracy (Mean ± std) | ROC p-val | ROC (Mean ± std) | Sensitivity (Mean ± std) | Precision (Mean ± std) | F1 (Mean ± std) |
| 30 | 0.07 | 0.665 ± 0.111 | 0.12 | 0.65 ± 0.124 | 0.665 ± 0.111 | 0.68 ± 0.128 | 0.657 ± 0.116 |
| 60 | 0.06 | 0.693 ± 0.136 | 0.07 | 0.706 ± 0.139 | 0.693 ± 0.136 | 0.739 ± 0.135 | 0.691 ± 0.141 |
| 90 | 0.06 | 0.68 ± 0.128 | 0.1 | 0.677 ± 0.135 | 0.68 ± 0.128 | 0.697 ± 0.14 | 0.674 ± 0.132 |
| 120 | 0.1 | 0.648 ± 0.129 | 0.14 | 0.649 ± 0.133 | 0.648 ± 0.129 | 0.67 ± 0.139 | 0.641 ± 0.13 |
| Full Length | 0.04 | 0.705 ± 0.116 | 0.07 | 0.697 ± 0.129 | 0.705 ± 0.116 | 0.727 ± 0.127 | 0.702 ± 0.119 |
| KNN | |||||||
| Segment Length | Accuracy p-val | Accuracy (Mean ± std) | ROC p-val | ROC (Mean ± std) | Sensitivity (Mean ± std) | Precision (Mean ± std) | F1 (Mean ± std) |
| 30 | 0.19 | 0.60 ± 0.108 | 0.27 | 0.561 ± 0.108 | 0.600 ± 0.108 | 0.597 ± 0.132 | 0.579 ± 0.111 |
| 60 | 0.13 | 0.637 ± 0.114 | 0.19 | 0.625 ± 0.125 | 0.637 ± 0.114 | 0.659 ± 0.129 | 0.632 ± 0.117 |
| 90 | 0.02 | 0.70 ± 0.117 | 0.06 | 0.686 ± 0.122 | 0.70 ± 0.117 | 0.717 ± 0.128 | 0.689 ± 0.124 |
| 120 | 0.08 | 0.659 ± 0.13 | 0.09 | 0.655 ± 0.13 | 0.659 ± 0.13 | 0.675 ± 0.133 | 0.653 ± 0.133 |
| Full Length | 0.17 | 0.617 ± 0.126 | 0.24 | 0.594 ± 0.136 | 0.617 ± 0.126 | 0.675 ± 0.133 | 0.653 ± 0.133 |
| DT | |||||||
| Segment Length | Accuracy p-val | Accuracy (Mean ± std) | ROC p-val | ROC (Mean ± std) | Sensitivity (Mean ± std) | Precision (Mean ± std) | F1 (Mean ± std) |
| 30 | 0.11 | 0.641 ± 0.116 | 0.16 | 0.621 ± 0.132 | 0.641 ± 0.116 | 0.644 ± 0.145 | 0.627 ± 0.125 |
| 60 | 0.06 | 0.683 ± 0.133 | 0.1 | 0.691 ± 0.144 | 0.683 ± 0.133 | 0.719 ± 0.158 | 0.676 ± 0.144 |
| 90 | 0.04 | 0.670 ± 0.121 | 0.09 | 0.677 ± 0.125 | 0.678 ± 0.121 | 0.698 ± 0.136 | 0.672 ± 0.126 |
| 120 | 0.05 | 0.666 ± 0.127 | 0.13 | 0.666 ± 0.131 | 0.666 ± 0.127 | 0.685 ± 0.139 | 0.659 ± 0.133 |
| Full Length | 0.11 | 0.696 ± 0.137 | 0.15 | 0.687 ± 0.154 | 0.696 ± 0.137 | 0.713 ± 0.157 | 0.692 ± 0.143 |
| Methodology | Precision | Sensitivity | F1-Score | ROC | Accuracy |
|---|---|---|---|---|---|
| Ensemble [47] | - | - | - | - | 0.75 |
| (avg. accuracy) | |||||
| XGB (their highest performing model) [55] | 0.57 ± 0.22 | 0.59 ± 0.14 | - | 0.88 ± 0.12 | 0.59 ± 0.22 |
| Feature | MeanNN | HTI |
|---|---|---|
| TL | 571.33 ± 214.77 | 5.64 ± 2.38 |
| EL | 419.47 ± 175.86 | 5.01 ± 2.02 |
| Feature | MedianNN | CSI |
| TL | 569.13 ± 213.75 | 3.38 ± 1.23 |
| EL | 417.09 ± 176.22 | 3.56 ± 1.61 |
| Feature | pnn20 | CVI |
| TL | 11.7 ± 11.2 | 3.67 ± 0.52 |
| EL | 8.75 ± 11.86 | 3.57 ± 0.58 |
| Feature | sd1sd2 | MaxNN |
| TL | 0.35 ± 0.15 | 704.71 ± 313.01 |
| EL | 0.39 ± 0.27 | 520.11 ± 225.89 |
| Feature | CvNN | MinNN |
| TL | 0.04 ± 0.02 | 501.48 ± 198.36 |
| EL | 0.06 ± 0.03 | 347.18 ± 149.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tilwani, D.; Bradshaw, J.; Sheth, A.; O’Reilly, C. ECG Recordings as Predictors of Very Early Autism Likelihood: A Machine Learning Approach. Bioengineering 2023, 10, 827. https://doi.org/10.3390/bioengineering10070827
Tilwani D, Bradshaw J, Sheth A, O’Reilly C. ECG Recordings as Predictors of Very Early Autism Likelihood: A Machine Learning Approach. Bioengineering. 2023; 10(7):827. https://doi.org/10.3390/bioengineering10070827
Chicago/Turabian StyleTilwani, Deepa, Jessica Bradshaw, Amit Sheth, and Christian O’Reilly. 2023. "ECG Recordings as Predictors of Very Early Autism Likelihood: A Machine Learning Approach" Bioengineering 10, no. 7: 827. https://doi.org/10.3390/bioengineering10070827
APA StyleTilwani, D., Bradshaw, J., Sheth, A., & O’Reilly, C. (2023). ECG Recordings as Predictors of Very Early Autism Likelihood: A Machine Learning Approach. Bioengineering, 10(7), 827. https://doi.org/10.3390/bioengineering10070827







