Bio-Inspired Network for Diagnosing Liver Steatosis in Ultrasound Images
Abstract
1. Introduction
- A Bio-inspired network (BiNet) for liver ultrasound image classification is presented by simulating the selective mechanism and feedback regulation mechanism of the ventral pathway visual cortex using a self-attention mechanism, and realized the extraction of important features in ultrasound images. In addition, a receptive field feature extraction module is designed based on the inhibition characteristics of the V1 neuron nCRF response to the CRF response, which further improves the accuracy of liver ultrasound image classification;
- A new parallel attention module is proposed. Unlike the previous attention methods that process input features sequentially, the parallel attention block has the same input. The input features are processed by two different attention paths at the same time, after which the outputs of both are fused and passed to the next stage as the input. By integrating more characteristic information, the module makes different information fully integrated and improves the overall performance of the model;
- A new dataset for fatty liver ultrasound image classification is constructed to train, validate, and test the proposed method. A total of 250 liver ultrasound images are collected in the new dataset, including 100 normal liver ultrasound images and 150 abnormal liver ultrasound images.
2. Materials and Methods
2.1. Datasets
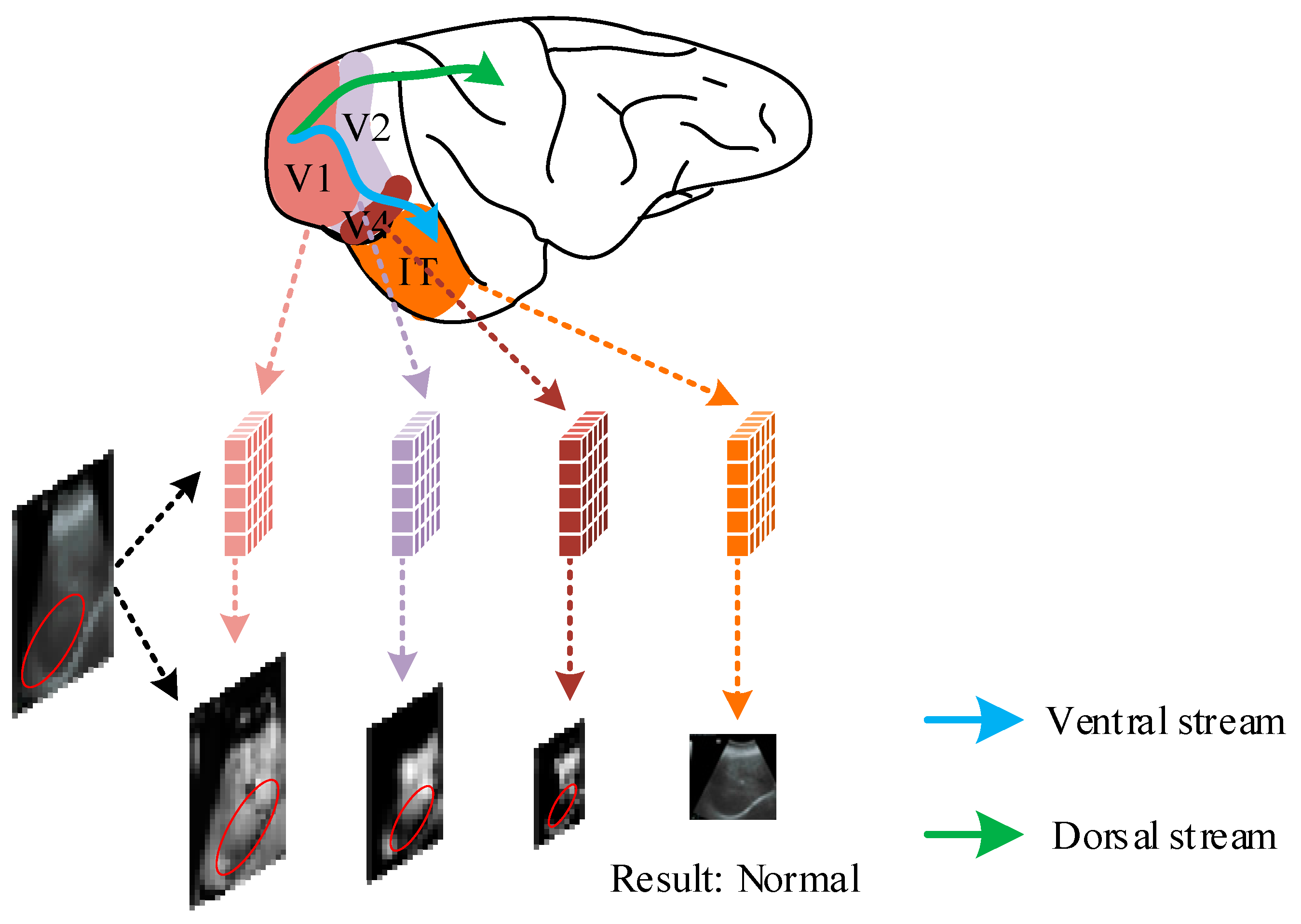
2.2. Selective Mechanisms of the Visual Cortex in the Biological Visual System
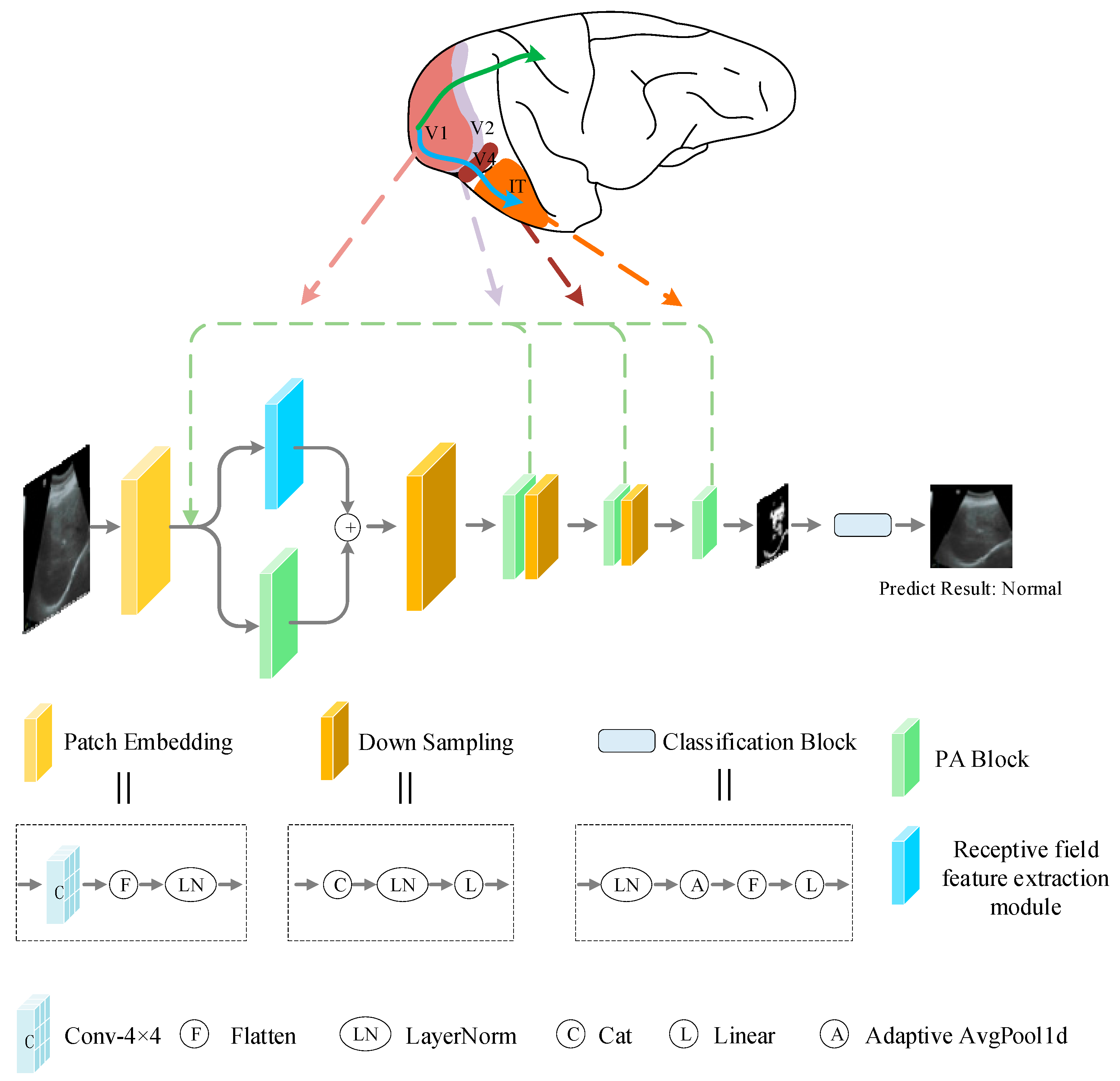
2.3. Overall Network Structure
2.4. Receptive Field Feature Extraction Module
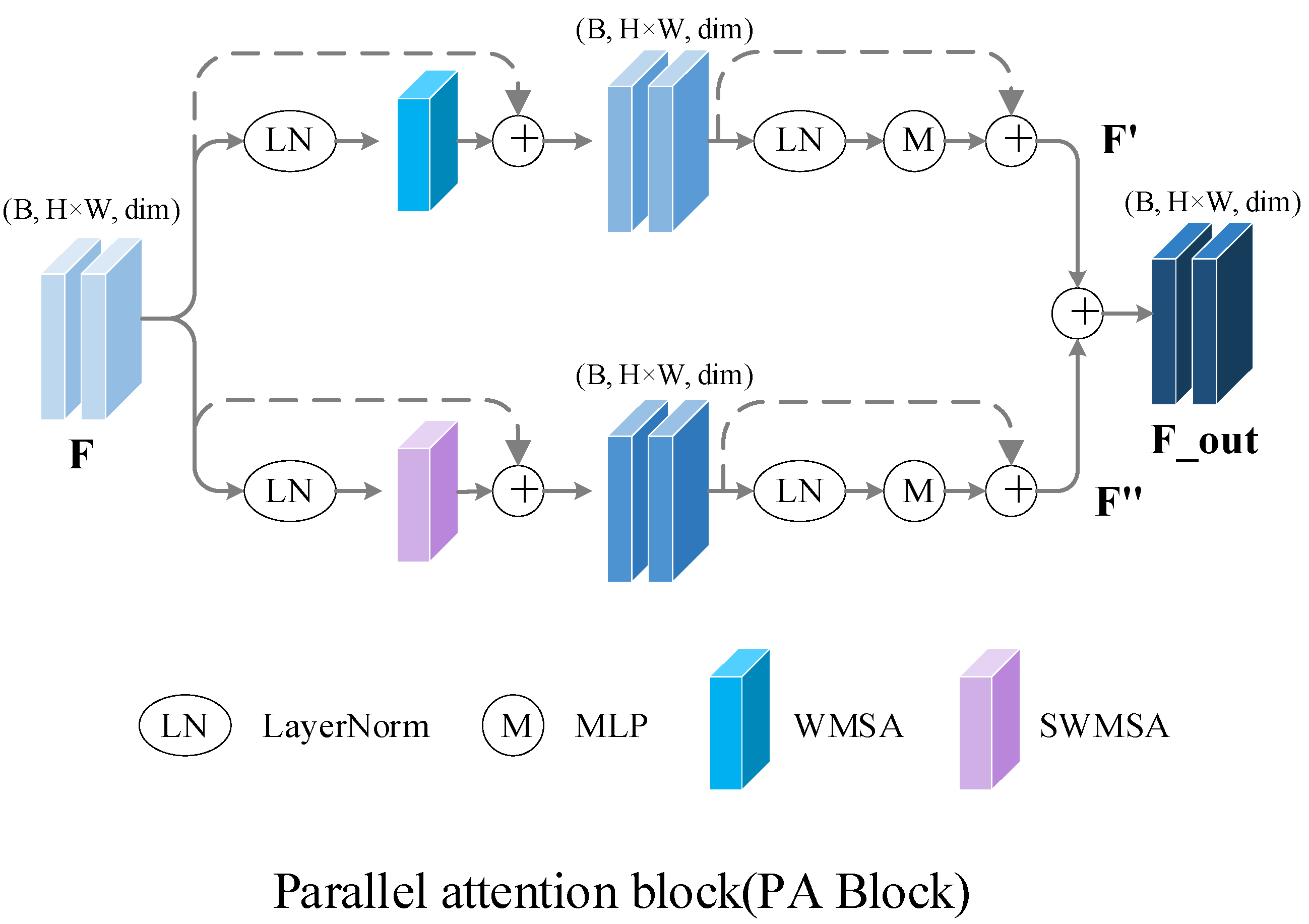
2.5. Parallel Attention Block
2.6. Implementation Details and Evaluation Metrics Methods
3. Results
3.1. Comparison of Results under Different Parameters
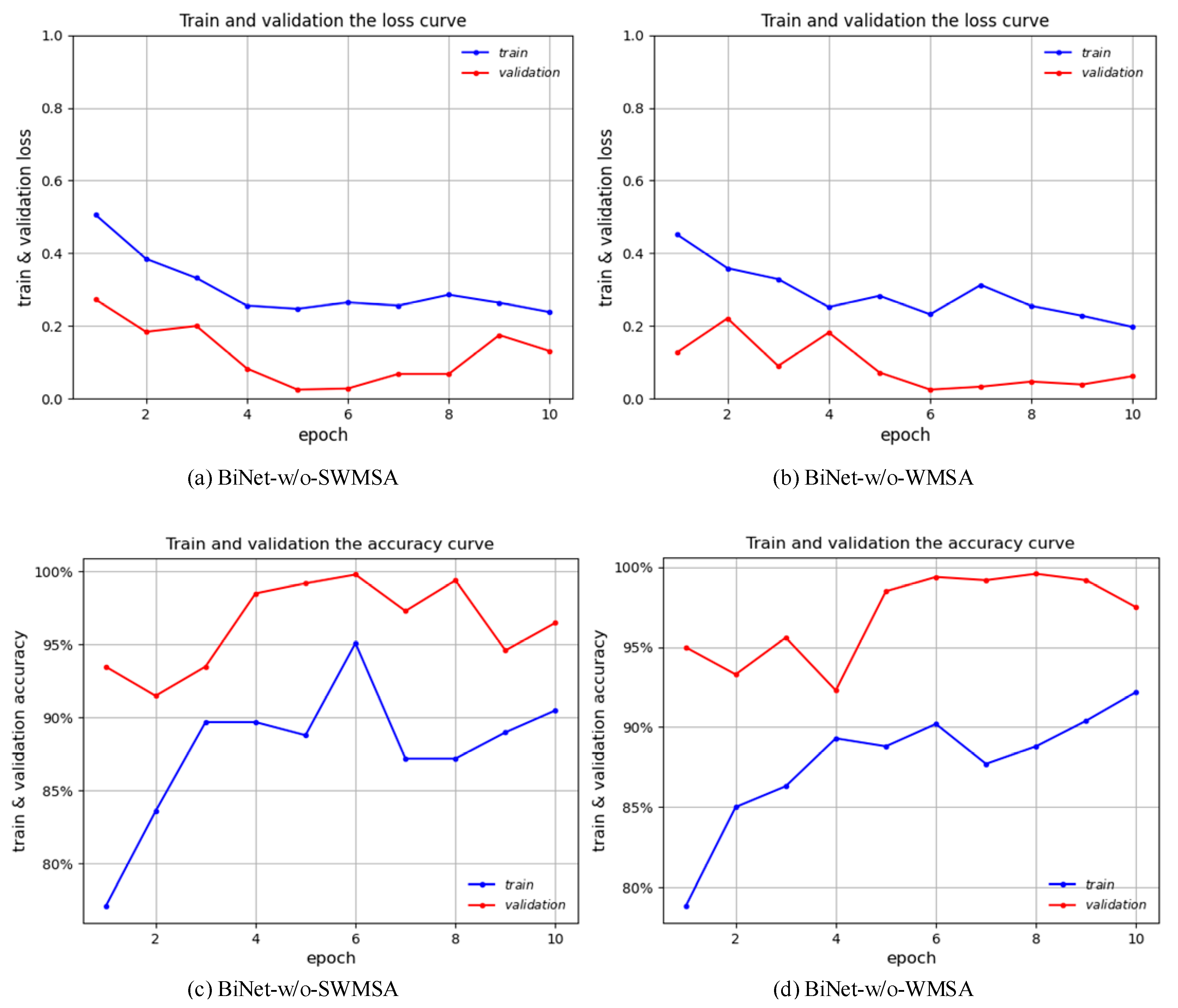
3.2. Result Verification of Parallel Attention Blocks
3.3. Comparison with Other Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Che, H.; Brown, L.G.; Foran, D.J.; Nosher, J.L.; Hacihaliloglu, I. Liver disease classification from ultrasound using multi-scale CNN. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Rhyou, S.-Y.; Yoo, J.-C. Cascaded Deep Learning Neural Network for Automated Liver Steatosis Diagnosis Using Ultrasound Images. Sensors 2021, 21, 5304. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Raghavendra, U.; Fujita, H.; Hagiwara, Y.; Koh, J.E.; Hong, T.J.; Sudarshan, V.K.; Vijayananthan, A.; Yeong, C.H.; Gudigar, A. Automated characterization of fatty liver disease and cirrhosis using curvelet transform and entropy features extracted from ultrasound images. Comput. Biol. Med. 2016, 79, 250–258. [Google Scholar] [CrossRef]
- Strauss, S.; Gavish, E.; Gottlieb, P.; Katsnelson, L. Interobserver and intraobserver variability in the sonographic assessment of fatty liver. Am. J. Roentgenol. 2007, 189, W320–W323. [Google Scholar] [CrossRef] [PubMed]
- Sudha, S.; Suresh, G.; Sukanesh, R. Speckle noise reduction in ultrasound images by wavelet thresholding based on weighted variance. Int. J. Comput. Theory Eng. 2009, 1, 7. [Google Scholar] [CrossRef]
- Yang, J.; Fan, J.; Ai, D.; Wang, X.; Zheng, Y.; Tang, S.; Wang, Y. Local statistics and non-local mean filter for speckle noise reduction in medical ultrasound image. Neurocomputing 2016, 195, 88–95. [Google Scholar] [CrossRef]
- Khov, N.; Sharma, A.; Riley, T.R. Bedside ultrasound in the diagnosis of nonalcoholic fatty liver disease. World J. Gastroenterol. WJG 2014, 20, 6821. [Google Scholar] [CrossRef]
- Sabih, D.; Hussain, M. Automated classification of liver disorders using ultrasound images. J. Med. Syst. 2012, 36, 3163–3172. [Google Scholar]
- Ho, M.-C.; Chen, A.; Tsui, P.-H.; Jeng, Y.-M.; Chen, C.-N. Clinical validation of ultrasound backscatter statistics for the assessment of liver fibrosis. Ultrasound Med. Biol. 2019, 45, S94. [Google Scholar] [CrossRef]
- Zhu, H.; Liu, Y.; Gao, X.; Zhang, L. Combined CNN and Pixel Feature Image for Fatty Liver Ultrasound Image Classification. Comput. Math. Methods Med. 2022, 2022, 9385734. [Google Scholar] [CrossRef]
- Pushpa, B.; Baskaran, B.; Vivekanandan, S.; Gokul, P. Liver fat analysis using optimized support vector machine with support vector regression. Technol. Health Care 2023, 31, 867–886. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.H.; Eissa, M.; Bluth, E.I.; Gulotta, P.M.; Davis, N.K. Hepatorenal index as an accurate, simple, and effective tool in screening for steatosis. Am. J. Roentgenol. 2012, 199, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Andrade, A.; Silva, J.S.; Santos, J.; Belo-Soares, P. Classifier approaches for liver steatosis using ultrasound images. Procedia Technol. 2012, 5, 763–770. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, H.; Yang, T. Deep Neural Networks for fatty liver ultrasound images classification. In Proceedings of the 2019 Chinese Control And Decision Conference (CCDC), IEEE, Nanchang, China, 3–5 June 2019; pp. 4641–4646. [Google Scholar]
- Reddy, D.S.; Bharath, R.; Rajalakshmi, P. Classification of nonalcoholic fatty liver texture using convolution neural networks. In Proceedings of the 2018 IEEE 20th International Conference on e-Health Networking, Applications and Services (Healthcom), IEEE, Ostrava, Czech Republic, 17–20 September 2018; pp. 1–5. [Google Scholar]
- Biswas, M.; Kuppili, V.; Edla, D.R.; Suri, H.S.; Saba, L.; Marinhoe, R.T.; Sanches, J.M.; Suri, J.S. Symtosis: A liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput. Methods Programs Biomed. 2018, 155, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Byra, M.; Styczynski, G.; Szmigielski, C.; Kalinowski, P.; Michałowski, Ł.; Paluszkiewicz, R.; Ziarkiewicz-Wróblewska, B.; Zieniewicz, K.; Sobieraj, P.; Nowicki, A. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1895–1903. [Google Scholar] [CrossRef]
- Kuppili, V.; Biswas, M.; Sreekumar, A.; Suri, H.S.; Saba, L.; Edla, D.R.; Marinhoe, R.T.; Sanches, J.M.; Suri, J.S. Extreme learning machine framework for risk stratification of fatty liver disease using ultrasound tissue characterization. J. Med. Syst. 2017, 41, 152. [Google Scholar] [CrossRef]
- Meng, D.; Zhang, L.; Cao, G.; Cao, W.; Zhang, G.; Hu, B. Liver fibrosis classification based on transfer learning and FCNet for ultrasound images. IEEE Access 2017, 5, 5804–5810. [Google Scholar] [CrossRef]
- Tang, Q.; Sang, N.; Liu, H. Learning nonclassical receptive field modulation for contour detection. IEEE Trans. Image Process. 2019, 29, 1192–1203. [Google Scholar] [CrossRef]
- Lin, C.; Zhang, Z.; Hu, Y. Bio-inspired feature enhancement network for edge detection. Appl. Intell. 2022, 52, 11027–11042. [Google Scholar] [CrossRef]
- Fan, X.; Jiang, M.; Shahid, A.R.; Yan, H. Hierarchical scale convolutional neural network for facial expression recognition. Cogn. Neurodyn. 2022, 16, 847–858. [Google Scholar] [CrossRef]
- Grigorescu, C.; Petkov, N.; Westenberg, M.A. Contour detection based on nonclassical receptive field inhibition. IEEE Trans. Image Process. 2003, 12, 729–739. [Google Scholar] [CrossRef]
- Yang, K.-F.; Gao, S.-B.; Guo, C.-F.; Li, C.-Y.; Li, Y.-J. Boundary detection using double-opponency and spatial sparseness constraint. IEEE Trans. Image Process. 2015, 24, 2565–2578. [Google Scholar] [CrossRef]
- Wang, W.; Xie, E.; Li, X.; Fan, D.-P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 568–578. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Chen, Y.; Lin, C.; Qiao, Y. DPED: Bio-inspired dual-pathway network for edge detection. Front. Bioeng. Biotechnol. 2022, 10, 1008140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lin, C.; Qiao, Y.; Pan, Y. Edge detection networks inspired by neural mechanisms of selective attention in biological visual cortex. Front. Neurosci. 2022, 16, 1073484. [Google Scholar] [CrossRef] [PubMed]
- Bear, M.; Connors, B.; Paradiso, M.A. Neuroscience: Exploring the Brain, Enhanced Edition: Exploring the Brain; Jones & Bartlett Learning: Burlington, MA, USA, 2020. [Google Scholar]
- Mishkin, M.; Ungerleider, L.G.; Macko, K.A. Object vision and spatial vision: Two cortical pathways. Trends Neurosci. 1983, 6, 414–417. [Google Scholar] [CrossRef]
- Ungerleider, L.G.; Haxby, J.V. ‘What’and ‘where’in the human brain. Curr. Opin. Neurobiol. 1994, 4, 157–165. [Google Scholar] [CrossRef]
- Yoshioka, T.; Dow, B.M.; Vautin, R.G. Neuronal mechanisms of color categorization in areas V1, V2 and V4 of macaque monkey visual cortex. Behav. Brain Res. 1996, 76, 51–70. [Google Scholar] [CrossRef]
- Luck, S.J.; Chelazzi, L.; Hillyard, S.A.; Desimone, R. Neural mechanisms of spatial selective attention in areas V1, V2, and V4 of macaque visual cortex. J. Neurophysiol. 1997, 77, 24–42. [Google Scholar] [CrossRef]
- Marcus, D.S.; Van Essen, D.C. Scene segmentation and attention in primate cortical areas V1 and V2. J. Neurophysiol. 2002, 88, 2648–2658. [Google Scholar] [CrossRef]
- Allen, H.A.; Humphreys, G.W.; Colin, J.; Neumann, H. Ventral extra-striate cortical areas are required for human visual texture segmentation. J. Vis. 2009, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Gross, C.G.; Rocha-Miranda, C.d.; Bender, D. Visual properties of neurons in inferotemporal cortex of the Macaque. J. Neurophysiol. 1972, 35, 96–111. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K. Inferotemporal cortex and object vision. Annu. Rev. Neurosci. 1996, 19, 109–139. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Ba, J.L.; Kiros, J.R.; Hinton, G.E. Layer normalization. arXiv 2016, arXiv:1607.06450. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, IEEE, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Gaber, A.; Youness, H.A.; Hamdy, A.; Abdelaal, H.M.; Hassan, A.M. Automatic classification of fatty liver disease based on supervised learning and genetic algorithm. Appl. Sci. 2022, 12, 521. [Google Scholar] [CrossRef]
- Wu, C.-H.; Hung, C.-L.; Lee, T.-Y.; Wu, C.-Y.; Chu, W.C.-C. Fatty Liver Diagnosis Using Deep Learning in Ultrasound Image. In Proceedings of the 2022 IEEE International Conference on Digital Health (ICDH), IEEE, Barcelona, Spain, 10–16 July 2022; pp. 185–192. [Google Scholar]
- Acharya, U.R.; Sree, S.V.; Ribeiro, R.; Krishnamurthi, G.; Marinho, R.T.; Sanches, J.; Suri, J.S. Data mining framework for fatty liver disease classification in ultrasound: A hybrid feature extraction paradigm. Med. Phys. 2012, 39 Pt 1, 4255–4264. [Google Scholar] [CrossRef]
- Sharma, V.; Juglan, K. Automated classification of fatty and normal liver ultrasound images based on mutual information feature selection. IRBM 2018, 39, 313–323. [Google Scholar] [CrossRef]
- Rivas, E.C.; Moreno, F.; Benitez, A.; Morocho, V.; Vanegas, P.; Medina, R. Hepatic Steatosis detection using the co-occurrence matrix in tomography and ultrasound images. In Proceedings of the 2015 20th Symposium on Signal Processing, Images and Computer Vision (STSIVA), IEEE, Bogota, Colombia, 2–4 September 2015; pp. 1–7. [Google Scholar]




| Method | Epoch | Accuracy (Validation) | Accuracy (Test) | Sensitivity | Specificity | F1-Score |
|---|---|---|---|---|---|---|
| BiNet | 5 | 94.0% | 96.0% | 100.0% | 92.0% | 0.96 |
| BiNet | 8 | 98.5% | 96.0% | 100.0% | 92.0% | 0.96 |
| BiNet | 10 | 99.8% | 98.0% | 100.0% | 96.0% | 0.98 |
| Method | Lr | Accuracy (Validation) | Accuracy (Test) | Sensitivity | Specificity | F1-Score |
|---|---|---|---|---|---|---|
| BiNet | 0.001 | 81.5% | 82.0% | 64.0% | 100.0% | 0.78 |
| BiNet | 0.00001 | 98.3% | 90.0% | 80.0% | 100.0% | 0.89 |
| BiNet | 0.0001 | 99.8% | 98.0% | 100.0% | 96.0% | 0.98 |
| Method | Accuracy (Validation) | Accuracy (Test) | Sensitivity | Specificity | F1-score |
|---|---|---|---|---|---|
| Swin_original | 99.4% | 96.0% | 92.0% | 100.0% | 0.96 |
| BiNet | 99.8% | 98.0% | 100.0% | 96.0% | 0.98 |
| BiNet-w/o-SWMSA | 99.8% | 96.0% | 92.0% | 100.0% | 0.96 |
| BiNet-w/o-WMSA | 99.6% | 98.0% | 96.0% | 100.0% | 0.98 |
| Authors | Dataset | Accuracy | Sensitivity | Specificity | F1-Score |
|---|---|---|---|---|---|
| Acharya et al. [44] | Private | 93.3% † | - | - | - |
| Sharma et al. [45] | Delta Diagnostic Centre Patiala, India, Private | 95.55% † | - | - | - |
| Andrea et al. [46] | Coimbra University Hospital, Private | kNN:74.05% † ANN:76.92% † SVM: 79.77% † | - | - | - |
| Gaber et al. [42] | Private | 95.71% † | 97.05% † | 94.44% † | 0.956 |
| Zhang et al. [14] | Private | 90.0% † | 81.0% † | 92.0% † | - |
| Byra et al. [17] | Medical University of Warsaw, Poland, Publicly available | 96.3% † | 100.0% † | 88.20% † | - |
| BiNet (ours) | Medical University of Warsaw, Poland, Publicly available | 99.1% | 100.0% | 98.7% | 0.986 |
| BiNet (ours) | Private | 98.0% | 100.0% | 96.0% | 0.980 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.; Zhang, Z.; Peng, B.; Tang, J. Bio-Inspired Network for Diagnosing Liver Steatosis in Ultrasound Images. Bioengineering 2023, 10, 768. https://doi.org/10.3390/bioengineering10070768
Yao Y, Zhang Z, Peng B, Tang J. Bio-Inspired Network for Diagnosing Liver Steatosis in Ultrasound Images. Bioengineering. 2023; 10(7):768. https://doi.org/10.3390/bioengineering10070768
Chicago/Turabian StyleYao, Yuan, Zhenguang Zhang, Bo Peng, and Jin Tang. 2023. "Bio-Inspired Network for Diagnosing Liver Steatosis in Ultrasound Images" Bioengineering 10, no. 7: 768. https://doi.org/10.3390/bioengineering10070768
APA StyleYao, Y., Zhang, Z., Peng, B., & Tang, J. (2023). Bio-Inspired Network for Diagnosing Liver Steatosis in Ultrasound Images. Bioengineering, 10(7), 768. https://doi.org/10.3390/bioengineering10070768






