Retinal Vascular Image Segmentation Using Improved UNet Based on Residual Module
Abstract
1. Introduction
2. Related Works
2.1. U-Net, Residual Module and Inception Block
2.2. Medical Image Segmentation Methods Based on Deep Learning
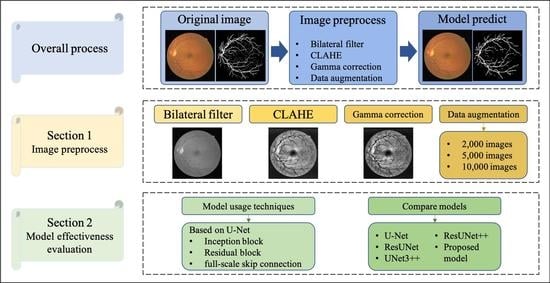
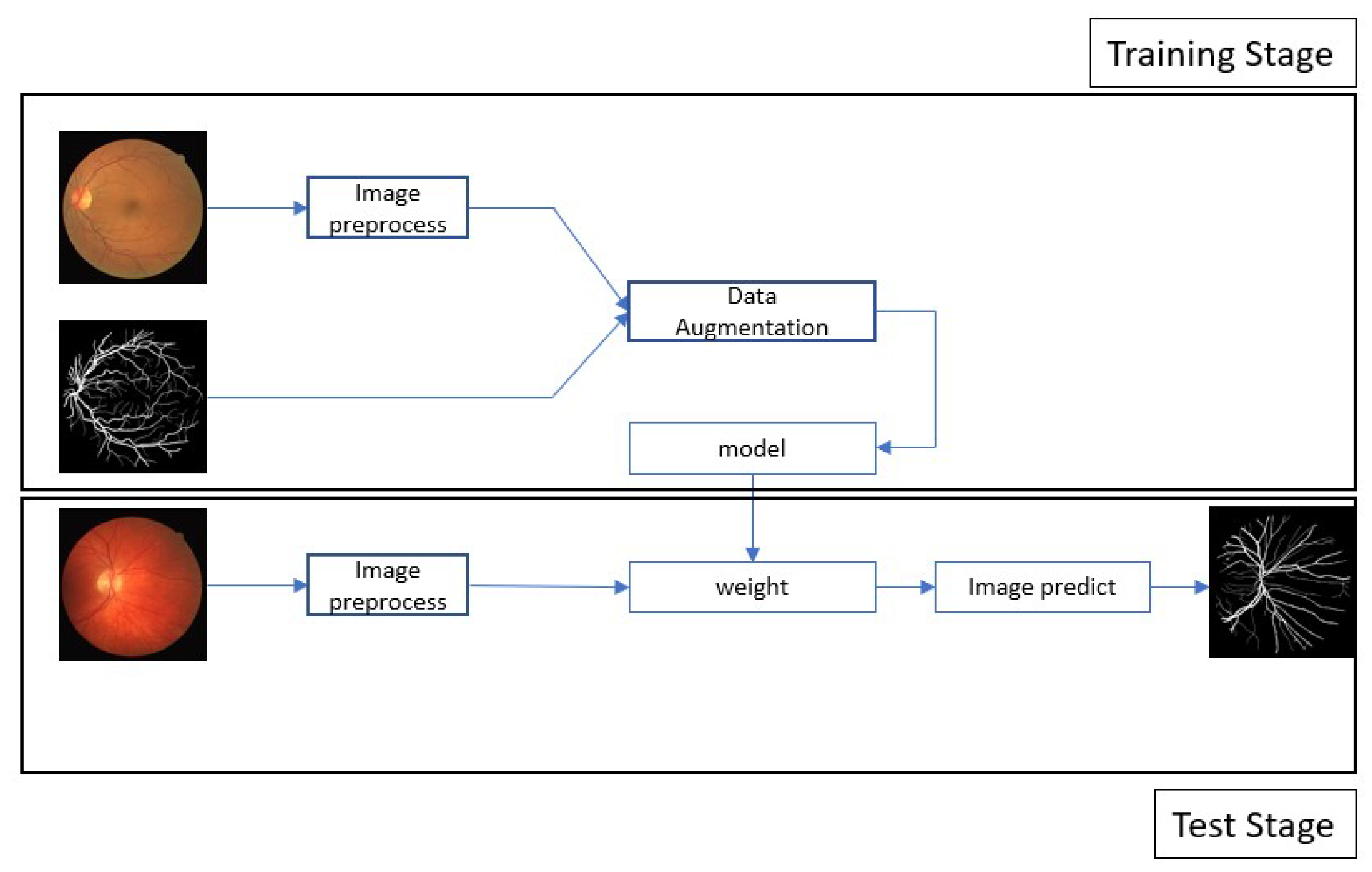
3. Proposed Method
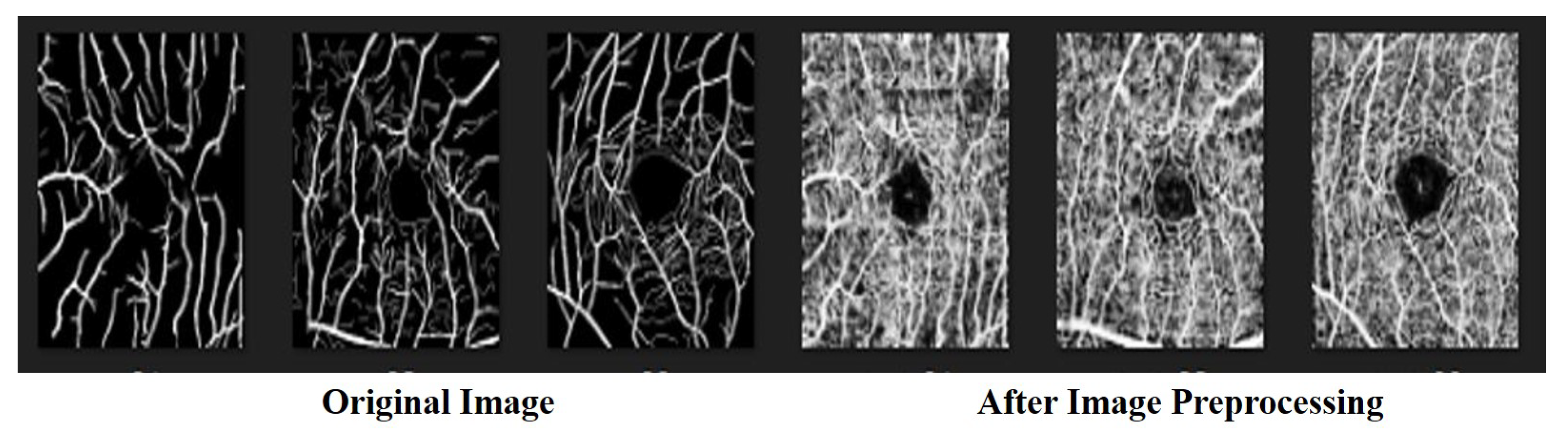
3.1. Data Preprocessing
3.2. Model Architecture
3.3. Residual Block
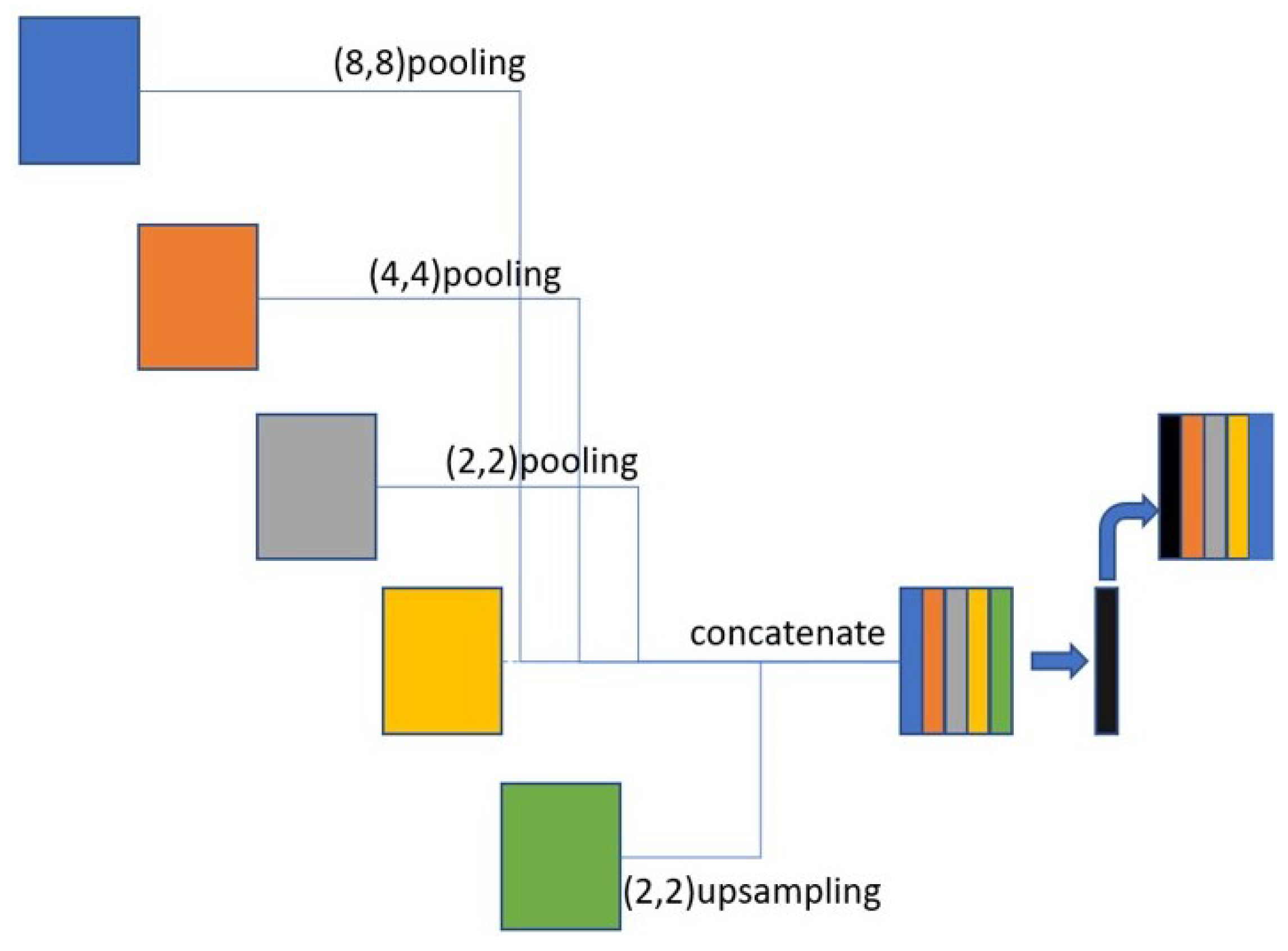
3.4. Skip Connection
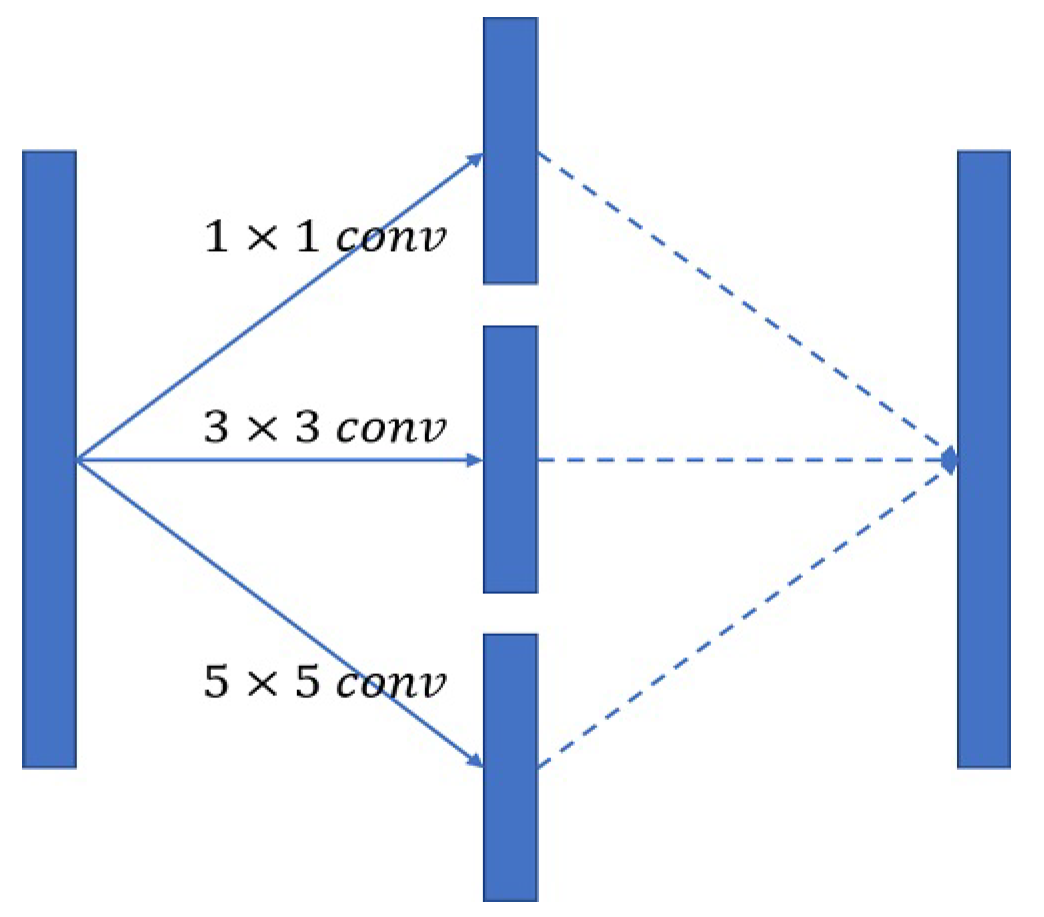
3.5. Inception Block
4. Experiment
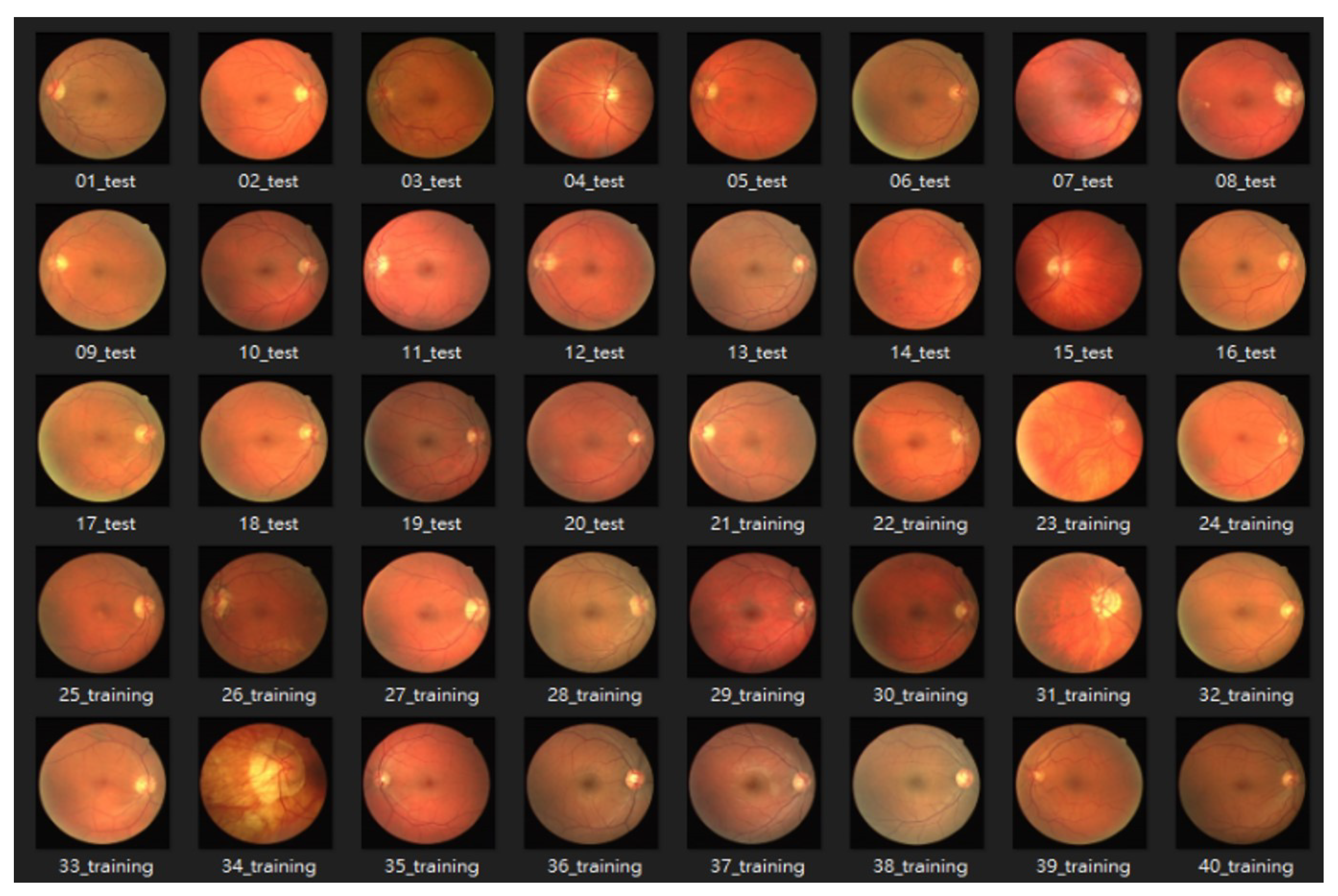
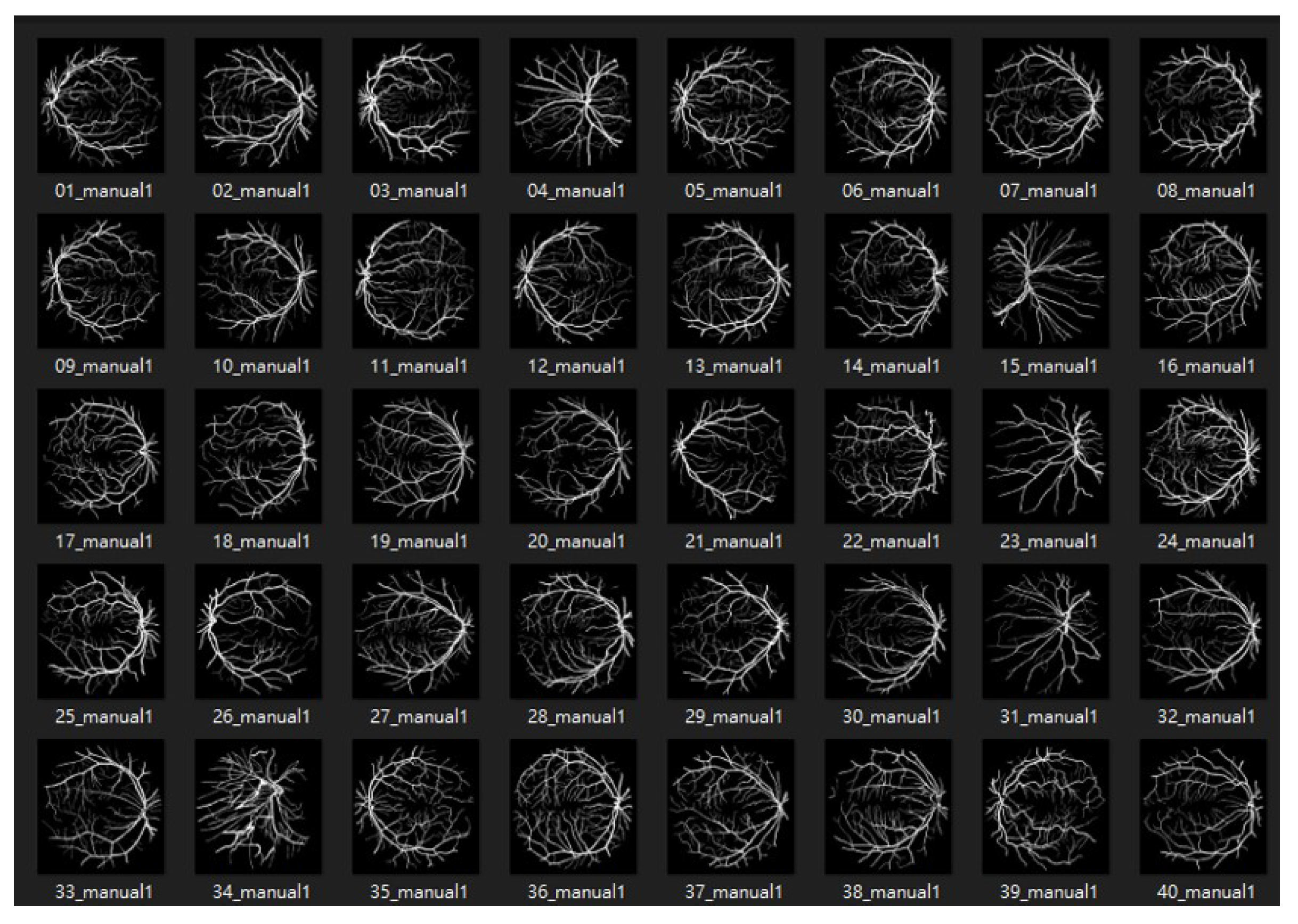
4.1. Dataset
4.1.1. DRIVE: Digital Retinal Images for Vessel Extraction
4.1.2. ROSE: A Retinal OCT-Angiography Vessel Segmentation Dataset
4.2. Data Augmentation
4.3. Evaluation Indexes
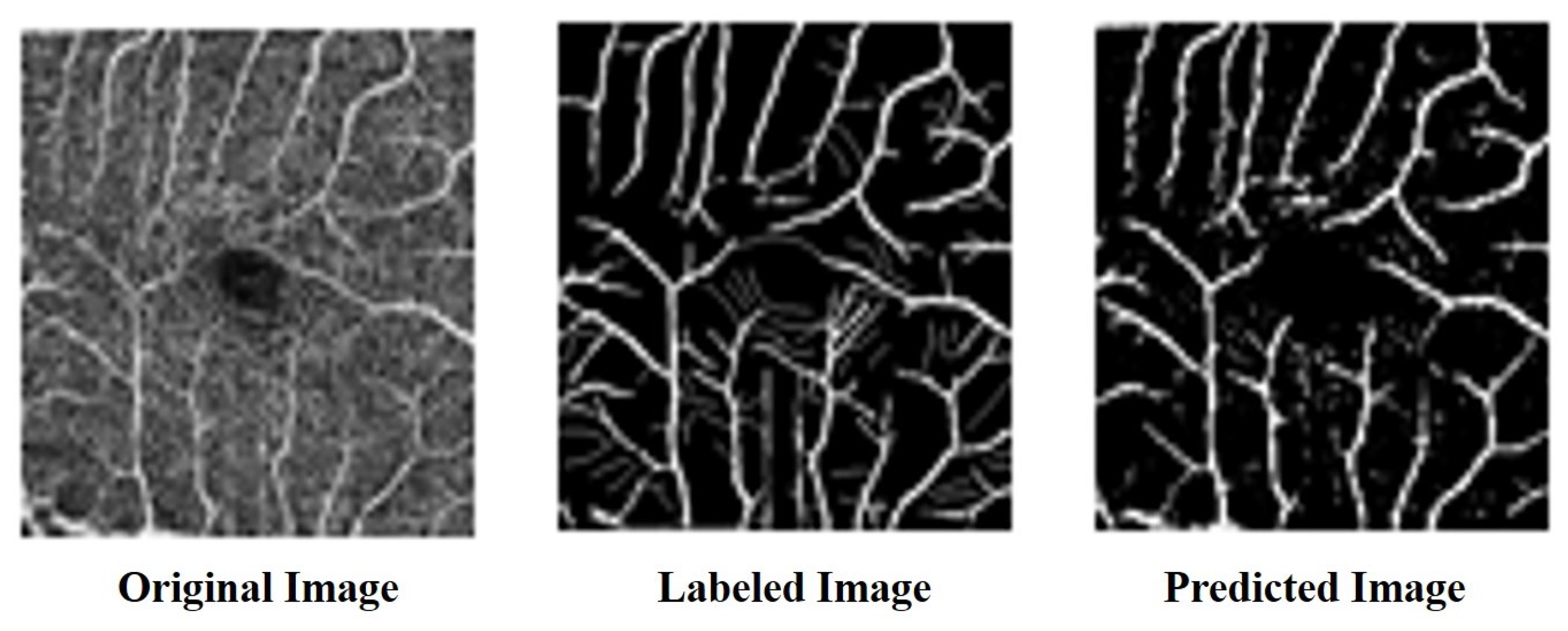
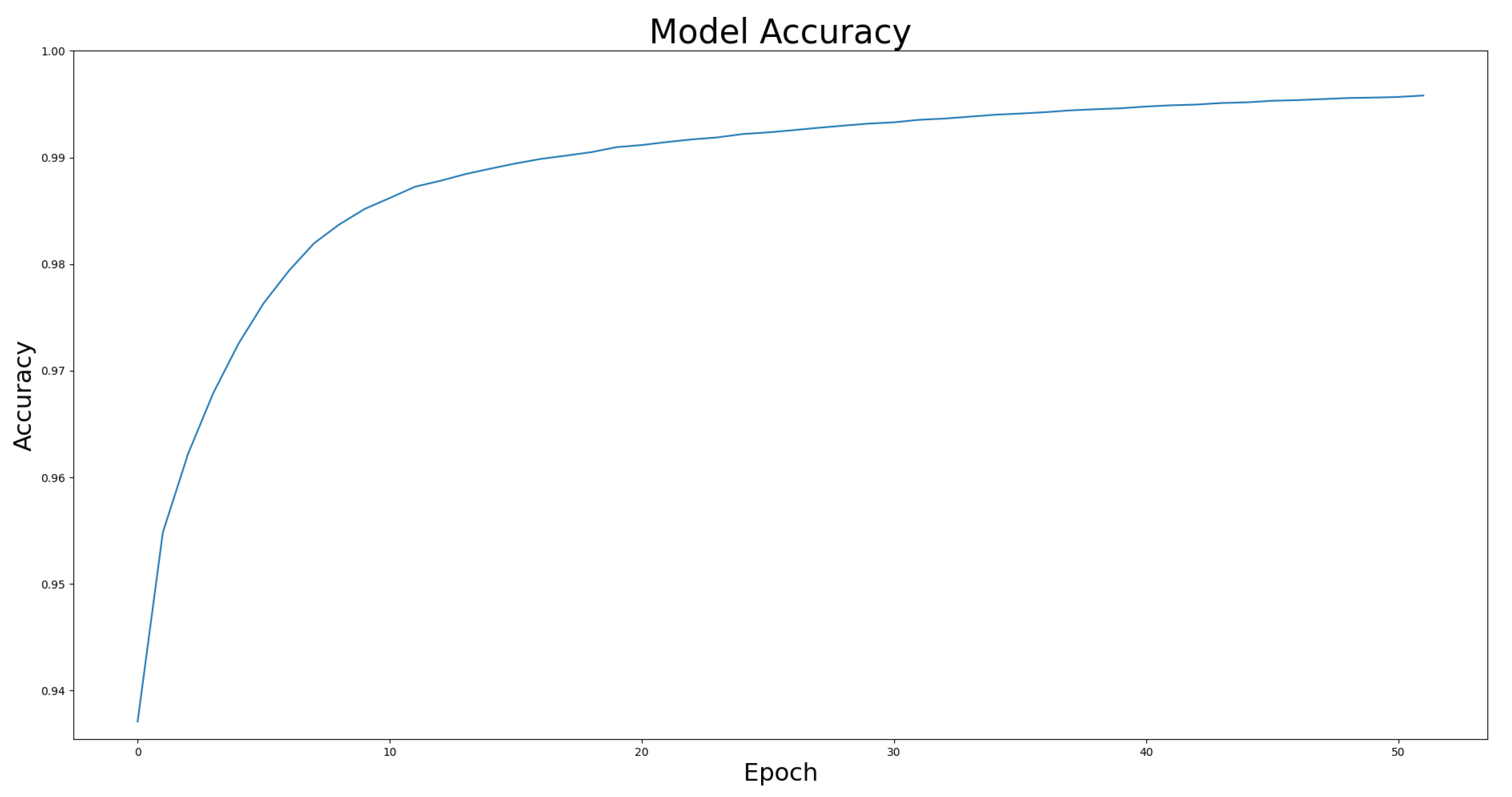
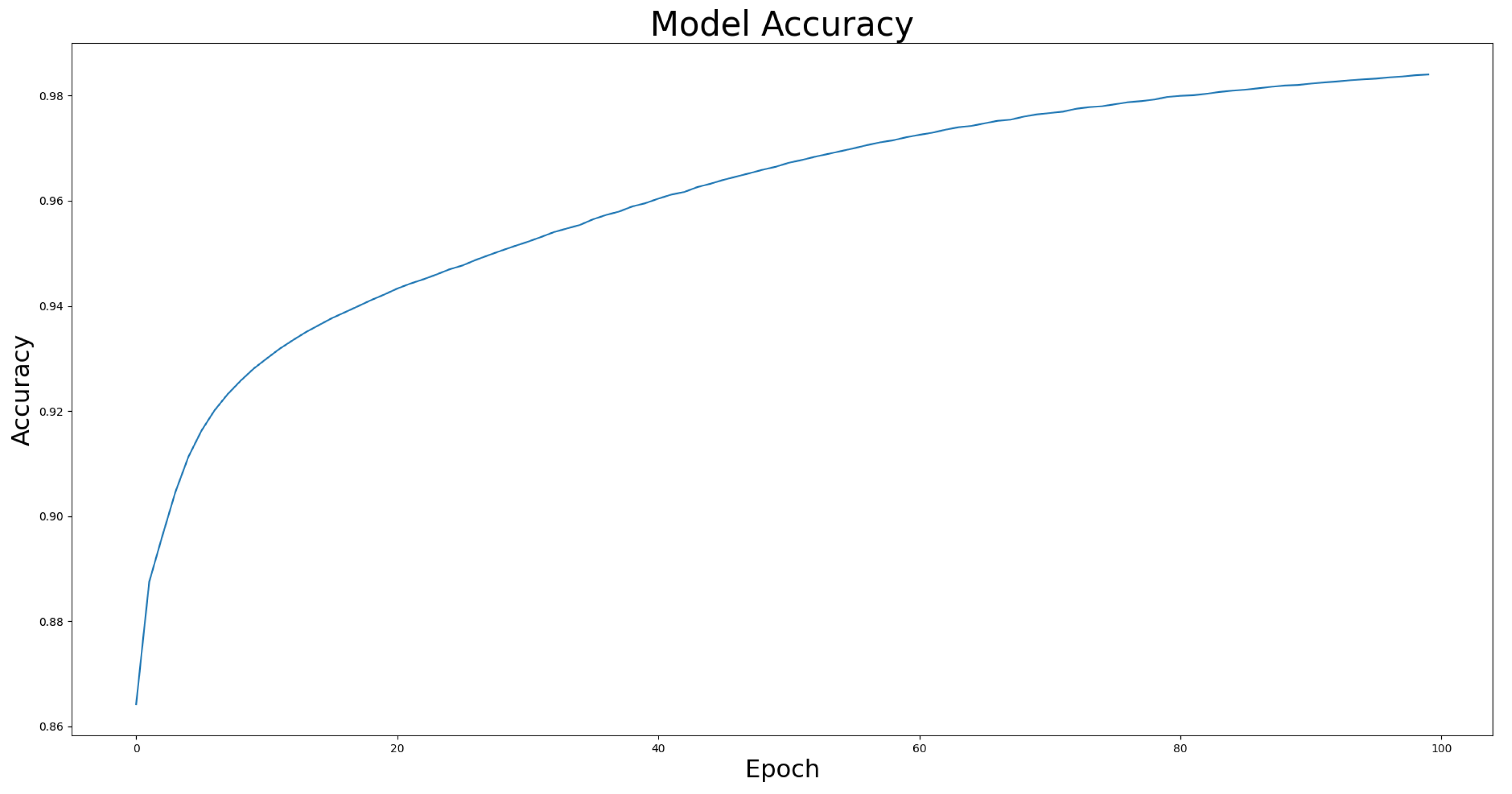
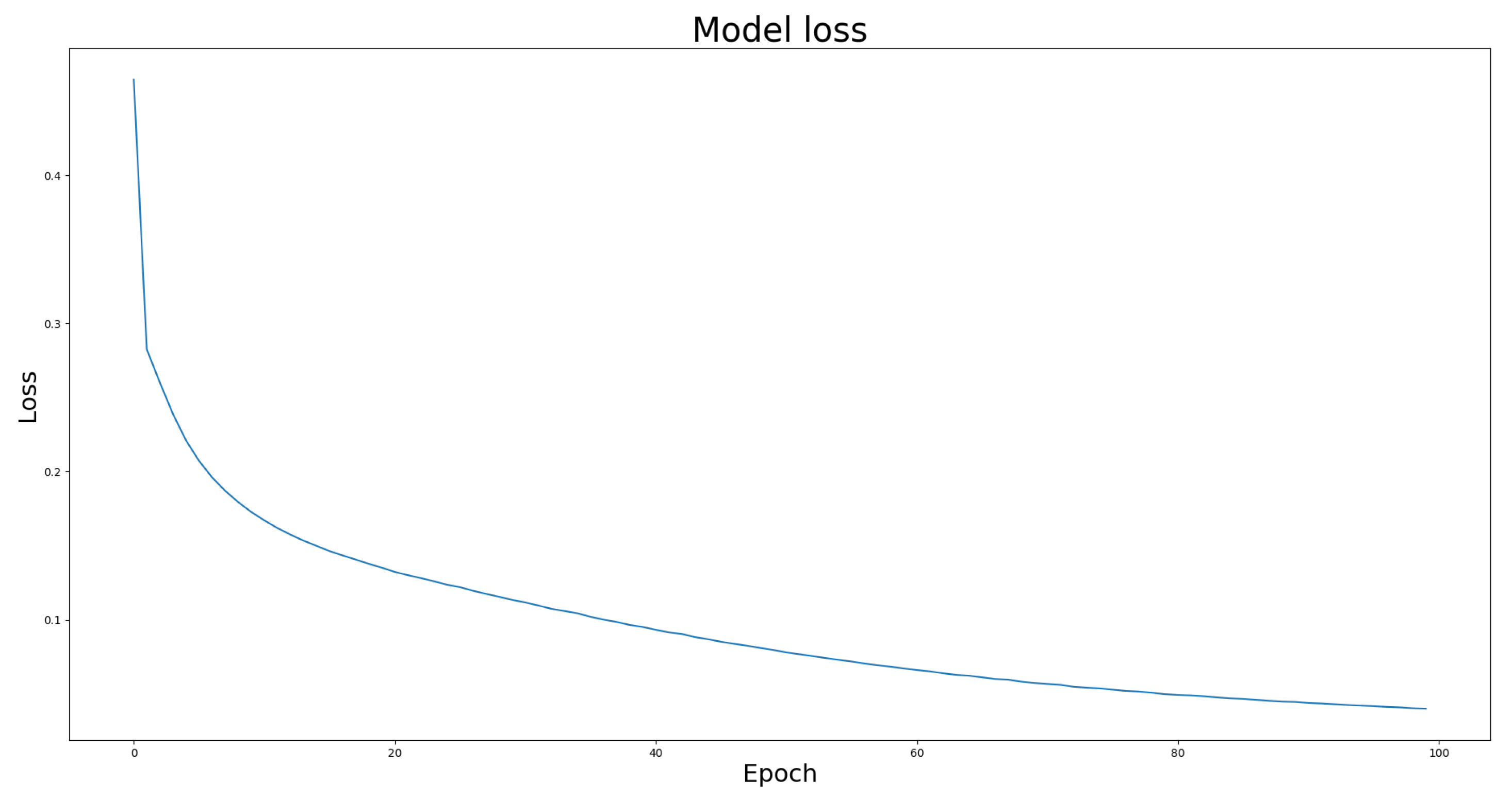
4.4. Model Effectiveness Evaluation
4.5. Ablation Experiment-Image Preprocessing
4.6. Ablation Experiment-Data Augmentation
4.7. Compare with State-of-the-Art Model
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 3523–3542. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Bin, X.W.; Yong, K.W. Information Identification Technology; Mechanical Industry Press: New York, NY, USA, 2006. [Google Scholar]
- Shin, E.S.; Sorenson, C.M.; Sheibani, N. Diabetes and retinal vascular dysfunction. J. Ophthalmic Vis. Res. 2014, 9, 362–373. [Google Scholar]
- Nemeth, S.; Joshi, V.; Agurto, C.; Soliz, P.; Barriga, S. Detection of hypertensive retinopathy using vessel measurements and textural features. In Proceedings of the 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014. [Google Scholar]
- Soares, J.V.B.; Leandro, J.J.G.; Cesar, R.M.; Jelinek, H.F.; Cree, M.J. Retinal vessel segmentation using the 2-D Gabor wavelet and supervised classification. IEEE Trans. Med. Imag. 2006, 25, 1214–1222. [Google Scholar] [CrossRef]
- Marín, D.; Aquino, A.; Gegúndez-Arias, M.E.; Bravo, J.M. A new supervised method for blood vessel segmentation in retinal images by using gray-level and moment invariants-based features. IEEE Trans. Med. Imag. 2011, 30, 146–158. [Google Scholar] [CrossRef]
- Soomro, T.A.; Khan, T.M.; Khan, M.A.U.; Gao, J.; Paul, M.; Zheng, L. Impact of ICA-based image enhancement technique on retinal blood vessels segmentation. IEEE Access 2018, 6, 3524–3538. [Google Scholar] [CrossRef]
- Tong, H.; Fang, Z.; Wei, Z.; Cai, Q.; Gao, Y. SAT-Net: A side attention network for retinal image segmentation. Appl. Intell. 2021, 51, 5146–5156. [Google Scholar] [CrossRef]
- Wu, H.; Wang, W.; Zhong, J.; Lei, B.; Wen, Z.; Qin, J. Scs-net: A scale and context sensitive network for retinal vessel segmentation. Med. Image Anal. 2021, 70, 102025. [Google Scholar] [CrossRef]
- Chala, M.; Nsiri, B.; El yousfi Alaoui, M.H.; Soulaymani, A.; Mokhtari, A.; Benaji, B. An automatic retinal vessel segmentation approach based on Convolutional Neural Networks. Expert Syst. Appl. 2021, 184, 115459. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Cui, W.; Lei, B.; Kuang, X.; Zhang, T. Dual encoder-based dynamic-channel graph convolutional network with edge enhancement for retinal vessel segmentation. IEEE Trans. Med. Imaging 2022, 41, 1975–1989. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, S.; Agrawal, S.; Mishro, P.K.; Pachori, R.B. A novel framework for retinal vessel segmentation using optimal improved frangi filter and adaptive weighted spatial FCM. Comput. Biol. Med. 2022, 147, 105770. [Google Scholar] [CrossRef]
- Zhai, Z.; Feng, S.; Yao, L.; Li, P. Retinal vessel image segmentation algorithm based on encoder-decoder structure. Multimed. Tools Appl. 2022, 81, 33361–33373. [Google Scholar] [CrossRef]
- Ni, J.; Sun, H.; Xu, J.; Liu, J.; Chen, Z. A feature aggregation and feature fusion network for retinal vessel segmentation. Biomed. Signal Process. Control 2023, 85, 104829. [Google Scholar] [CrossRef]
- Du, L.; Liu, H.; Zhang, L.; Lu, Y.; Li, M.; Hu, Y.; Zhang, Y. Deep ensemble learning for accurate retinal vessel segmentation. Comput. Biol. Med. 2023, 158, 106829. [Google Scholar] [CrossRef]
- Kumar, K.S.; Singh, N.P. Retinal disease prediction through blood vessel segmentation and classification using ensemble-based deep learning approaches. Neural Comput. Appl. 2023, 35, 12495–12511. [Google Scholar] [CrossRef]
- Girish, G.N.; Thakur, B.; Chowdhury, S.R.; Kothari, A.R.; Rajan, J. Segmentation of Intra-Retinal Cysts from Optical Coherence Tomography Images Using a Fully Convolutional Neural Network Model. IEEE J. Biomed. Health Inform. 2019, 23, 296–304. [Google Scholar] [CrossRef]
- Park, K.-B.; Choi, S.H.; Lee, J.Y. M-GAN: Retinal Blood Vessel Segmentation by Balancing Losses through Stacked Deep Fully Convolutional Networks. IEEE Access 2020, 8, 146308–146322. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.C.; Bengio, Y. Generative adversarial nets. In Proceedings of the Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 2672–2680. [Google Scholar]
- Mao, Y.; Ye, J.; Liu, L.; Zhang, S.; Shen, L.; Sun, M. Automatic Diagnosis of Familial Exudative Vitreoretinopathy Using a Fusion Neural Network for Wide-Angle Retinal Images. IEEE Access 2020, 8, 162–173. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Beeche, C.; Singh, J.P.; Leader, J.K.; Gezer, N.S.; Oruwari, A.P.; Dansingani, K.K.; Pu, J. Super U-Net: A modularized generalizable architecture. Pattern Recognit. 2022, 128, 108669. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hong, D.; Chanussot, J. UIU-Net: U-Net in U-Net for infrared small object detection. IEEE Trans. Image Process. 2022, 32, 364–376. [Google Scholar] [CrossRef]
- Lin, A.; Chen, B.; Xu, J.; Zhang, Z.; Lu, G.; Zhang, D. Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Trans. Instrum. Meas. 2022, 71, 1–15. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Cheng, Y. Towards an extended EfficientNet-based U-Net framework for joint optic disc and cup segmentation in the fundus image. Biomed. Signal Process. Control 2023, 85, 104906. [Google Scholar] [CrossRef]
- Allah, A.M.G.; Sarhan, A.M.; Elshennawy, N.M. Edge U-Net: Brain tumor segmentation using MRI based on deep U-Net model with boundary information. Expert Syst. Appl. 2023, 213, 118833. [Google Scholar] [CrossRef]
- Mu, N.; Lyu, Z.; Rezaeitaleshmahalleh, M.; Tang, J.; Jiang, J. An attention residual U-Net with differential preprocessing and geometric postprocessing: Learning how to segment vasculature including intracranial aneurysms. Med. Image Anal. 2023, 84, 102697. [Google Scholar] [CrossRef]
- Islam, M.T.; Al-Absi, H.R.H.; Ruagh, E.A.; Alam, T. DiaNet: A Deep Learning Based Architecture to Diagnose Diabetes Using Retinal Images Only. IEEE Access 2021, 9, 15686–15695. [Google Scholar] [CrossRef]
- He, J.; Jiang, D. Fully Automatic Model Based on SE-ResNet for Bone Age Assessment. IEEE Access 2021, 9, 62460–62466. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Chang, J.; Zhang, X.; Ye, M.; Huang, D.; Wang, P.; Yao, C. Brain Tumor Segmentation Based on 3D Unet with Multi-Class Focal Loss. In Proceedings of the 2018 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Beijing, China, 13–15 October 2018; pp. 1–5. [Google Scholar]
- Richter, T.; Kim, K.J. A MS-SSIM Optimal JPEG 2000 Encoder. In Proceedings of the 2009 Data Compression Conference, Snowbird, UT, USA, 16–18 March 2009; pp. 401–410. [Google Scholar]
- Zhai, H.; Cheng, J.; Wang, M. Rethink the IoU-based loss functions for bounding box regression. In Proceedings of the 2020 IEEE 9th Joint International Information Technology and Artificial Intelligence Conference (ITAIC), Chongqing, China, 11–13 December 2020; pp. 1522–1528. [Google Scholar]
- Xiuqin, P.; Zhang, Q.; Zhang, H.; Li, S. A Fundus Retinal Vessels Segmentation Scheme Based on the Improved Deep Learning U-Net Model. IEEE Access 2019, 7, 122634–122643. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems, Proceedings of the 31st Conference on Neural Information Processing Systems NIPS 2017, Long Beach, CA, USA, 4–9 December 2017; Curran Associates, Inc.: Red Hook, NY, USA, 2017; pp. 5998–6008. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets atrous convolution and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Malhotra, P.; Gupta, S.; Koundal, D.; Zaguia, A.; Enbeyle, W. Deep neural networks for medical image segmentation. J. Healthc. Eng. 2022, 2022, 9580991. [Google Scholar] [CrossRef]
- Wang, K.; Zhan, B.; Zu, C.; Wu, X.; Zhou, J.; Zhou, L.; Wang, Y. Semi-supervised medical image segmentation via a tripled-uncertainty guided mean teacher model with contrastive learning. Med. Image Anal. 2022, 79, 102447. [Google Scholar] [CrossRef]
- Xun, S.; Li, D.; Zhu, H.; Chen, M.; Wang, J.; Li, J.; Huang, P. Generative adversarial networks in medical image segmentation: A review. Comput. Biol. Med. 2022, 140, 105063. [Google Scholar] [CrossRef]
- Wu, Y.; Liao, K.; Chen, J.; Wang, J.; Chen, D.Z.; Gao, H.; Wu, J. D-former: A u-shaped dilated transformer for 3d medical image segmentation. Neural Comput. Appl. 2023, 35, 1931–1944. [Google Scholar] [CrossRef]
- Yuan, F.; Zhang, Z.; Fang, Z. An effective CNN and Transformer complementary network for medical image segmentation. Pattern Recognit. 2023, 136, 109228. [Google Scholar] [CrossRef]
- Zhou, T.; Li, L.; Bredell, G.; Li, J.; Unkelbach, J.; Konukoglu, E. Volumetric memory network for interactive medical image segmentation. Med. Image Anal. 2023, 83, 102599. [Google Scholar] [CrossRef]
- Tomasi, C.; Manduchi, R. Bilateral filtering for gray and color images. In Proceedings of the Sixth International Conference on Computer Vision, Bombay, India, 7 January 1998; pp. 839–846. [Google Scholar]
- Reza, A.M. Realization of the contrast limited adaptive histogram equalization (CLAHE) for real-time image enhancement. J. VLSI Signal Process. Syst. Signal Image Video Technol. 2014, 38, 35–44. [Google Scholar] [CrossRef]
- Rahman, S.; Rahman, M.M.; Abdullah-Al-Wadud, M.; Al-Quaderi, G.D.; Shoyaib, M. An adaptive gamma correction for image enhancement. EURASIP J. Image Video Process. 2016, 1, 1–13. [Google Scholar] [CrossRef]
- Staal, J.; Abramoff, M.D.; Niemeijer, M. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imag. 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Ma, Y.; Hao, H.; Xie, J.; Fu, H.; Zhang, J.; Yang, J.; Zhao, Y. ROSE: A retinal OCT-angiography vessel segmentation dataset and new model. IEEE Trans. Med. Imaging 2020, 40, 928–939. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Y.; Ji, Z.; Xie, K.; Yuan, S.; Liu, Q.; Chen, Q. Ipn-v2 and octa-500: Methodology and dataset for retinal image segmentation. arXiv 2020, arXiv:2012.07261. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Lou, A.; Guan, S.; Ko, H.; Loew, M.H. CaraNet: Context axial reverse attention network for segmentation of small medical objects. Med Imaging 2022 Image Process. 2022, 12032, 81–92. [Google Scholar]
- Gao, S.H.; Cheng, M.M.; Zhao, K.; Zhang, X.Y.; Yang, M.H.; Torr, P. Res2net: A new multi-scale backbone architecture. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Lou, A.; Loew, M. Cfpnet: Channel-wise feature pyramid for real-time semantic segmentation. In Proceedings of the 2021 IEEE International Conference on Image Processing (ICIP), Anchorage, AK, USA, 19–22 September 2021; pp. 1894–1898. [Google Scholar]







| Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | IOU (%) | |
|---|---|---|---|---|---|
| U-NET | 97.5 | 69.3 | 86.7 | 76.9 | 58.5 |
| ResUNet | 97.3 | 68.3 | 85.6 | 75.3 | 57.9 |
| UNET3+ | 97.3 | 70.6 | 89.7 | 76 | 58.1 |
| ResUNet++ | 97.6 | 73.1 | 85.8 | 76.8 | 59.8 |
| proposed model | 97.5 | 73.1 | 85.4 | 77.8 | 60.8 |
| Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | IOU (%) | |
|---|---|---|---|---|---|
| U-NET | 94.2 | 66.1 | 94.1 | 73 | 58.8 |
| ResUNet | 94.2 | 63.1 | 88.4 | 72.7 | 57.2 |
| UNET3+ | 94.1 | 66.3 | 88.4 | 73.2 | 58.5 |
| ResUNet++ | 94.5 | 67.2 | 77.8 | 74.8 | 58.8 |
| proposed model | 95 | 72.3 | 80.3 | 74.4 | 59.3 |
| Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | IOU (%) | ||
|---|---|---|---|---|---|---|
| U-NET | w/o | 96.4 | 52.8 | 81.8 | 64.1 | 47.2 |
| w/ | 97.5 | 69.3 | 86.4 | 76.9 | 58.5 | |
| ResUNet | w/o | 96.1 | 51.6 | 77.1 | 51.6 | 44 |
| w/ | 97.3 | 68.3 | 83.9 | 75.3 | 57.9 | |
| UNET3+ | w/o | 96 | 50.2 | 77.1 | 60.8 | 43.7 |
| w/ | 97.3 | 70.6 | 83.4 | 75.8 | 58.1 | |
| ResUNet++ | w/o | 96.5 | 58.2 | 88.1 | 69.5 | 51.5 |
| w/ | 97.6 | 71.9 | 85.8 | 77.5 | 59.8 | |
| proposed model | w/o | 96.7 | 60.7 | 88.9 | 72.2 | 56.5 |
| w/ | 97.5 | 73.1 | 85.4 | 77.8 | 60.8 |
| Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | IOU (%) | ||
|---|---|---|---|---|---|---|
| U-NET | 2000 | 97.3 | 69.2 | 84.1 | 75.9 | 58.2 |
| 5000 | 97.4 | 68.3 | 86.7 | 76.4 | 58.4 | |
| 10,000 | 97.5 | 69.3 | 86.4 | 76.9 | 58.5 | |
| ResUNet | 2000 | 97.1 | 64.8 | 83.8 | 73.7 | 57.3 |
| 5000 | 97.3 | 66.3 | 85.6 | 74.7 | 57.4 | |
| 10,000 | 97.3 | 68.3 | 83.9 | 75.3 | 57.9 | |
| UNET3+ | 2000 | 96.7 | 60.5 | 89.7 | 72.2 | 56.8 |
| 5000 | 97.3 | 70.5 | 82.6 | 76 | 57.8 | |
| 10,000 | 97.3 | 70.6 | 83.4 | 75.8 | 58.1 | |
| ResUNet++ | 2000 | 96.8 | 68.5 | 83.5 | 75.8 | 59.1 |
| 5000 | 97.3 | 69.6 | 84.9 | 76.8 | 59.5 | |
| 10,000 | 97.6 | 71.9 | 85.8 | 77.5 | 59.8 | |
| proposed model | 2000 | 97.3 | 69.1 | 84.4 | 76 | 60.5 |
| 5000 | 97.5 | 71.5 | 85.4 | 77.8 | 60.8 | |
| 10,000 | 97.4 | 73.1 | 85.8 | 77.7 | 60.8 |
| Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | IOU (%) | ||
|---|---|---|---|---|---|---|
| U-NET | 2000 | 93.5 | 58.3 | 89.9 | 70.7 | 57.2 |
| 5000 | 94.1 | 64.5 | 94.1 | 72.9 | 57.4 | |
| 10,000 | 94.2 | 66.1 | 82.4 | 73 | 58.8 | |
| ResUNet | 2000 | 94.1 | 60.2 | 88.4 | 71.6 | 55.8 |
| 5000 | 94.1 | 62.3 | 85.8 | 72.2 | 56.5 | |
| 10,000 | 94.2 | 63.1 | 85.8 | 72.7 | 57.2 | |
| UNET3+ | 2000 | 94.1 | 63.1 | 88.4 | 72.6 | 58.5 |
| 5000 | 94 | 63.9 | 83.1 | 72.3 | 58.4 | |
| 10,000 | 94 | 66.3 | 81.7 | 73.2 | 58 | |
| ResUNet++ | 2000 | 94.2 | 65.3 | 76.5 | 72.5 | 57.3 |
| 5000 | 94.2 | 65.8 | 77.3 | 73.1 | 58 | |
| 10,000 | 94.5 | 67.2 | 77.8 | 74.8 | 58.8 | |
| proposed model | 2000 | 94.7 | 70 | 77.7 | 73.7 | 59.3 |
| 5000 | 94.7 | 72.3 | 76.2 | 74.2 | 57.6 | |
| 10,000 | 95 | 69.4 | 80.3 | 74.4 | 59.3 |
| Dataset (%) | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | IOU (%) | |
|---|---|---|---|---|---|---|
| Proposed model | Drive | 97.5 | 73.1 | 85.4 | 64.1 | 60.8 |
| Rose | 95 | 72.3 | 80.3 | 76.9 | 59.3 | |
| CaraNet | Drive | 70.3 | 40.3 | 96.8 | 56.9 | 28.4 |
| Rose | 59.5 | 41.7 | 99 | 58.6 | 29.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, K.-W.; Yang, Y.-R.; Huang, Z.-H.; Liu, Y.-Y.; Lee, S.-H. Retinal Vascular Image Segmentation Using Improved UNet Based on Residual Module. Bioengineering 2023, 10, 722. https://doi.org/10.3390/bioengineering10060722
Huang K-W, Yang Y-R, Huang Z-H, Liu Y-Y, Lee S-H. Retinal Vascular Image Segmentation Using Improved UNet Based on Residual Module. Bioengineering. 2023; 10(6):722. https://doi.org/10.3390/bioengineering10060722
Chicago/Turabian StyleHuang, Ko-Wei, Yao-Ren Yang, Zih-Hao Huang, Yi-Yang Liu, and Shih-Hsiung Lee. 2023. "Retinal Vascular Image Segmentation Using Improved UNet Based on Residual Module" Bioengineering 10, no. 6: 722. https://doi.org/10.3390/bioengineering10060722
APA StyleHuang, K.-W., Yang, Y.-R., Huang, Z.-H., Liu, Y.-Y., & Lee, S.-H. (2023). Retinal Vascular Image Segmentation Using Improved UNet Based on Residual Module. Bioengineering, 10(6), 722. https://doi.org/10.3390/bioengineering10060722