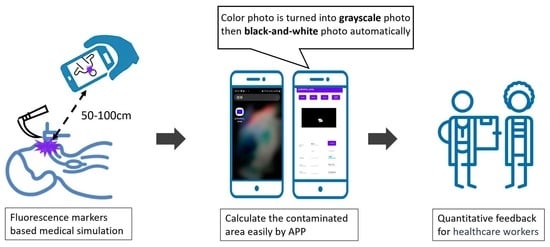
Prediction of Contaminated Areas Using Ultraviolet Fluorescence Markers for Medical Simulation: A Mobile Phone Application Approach
Abstract
1. Introduction
2. Materials and Methods
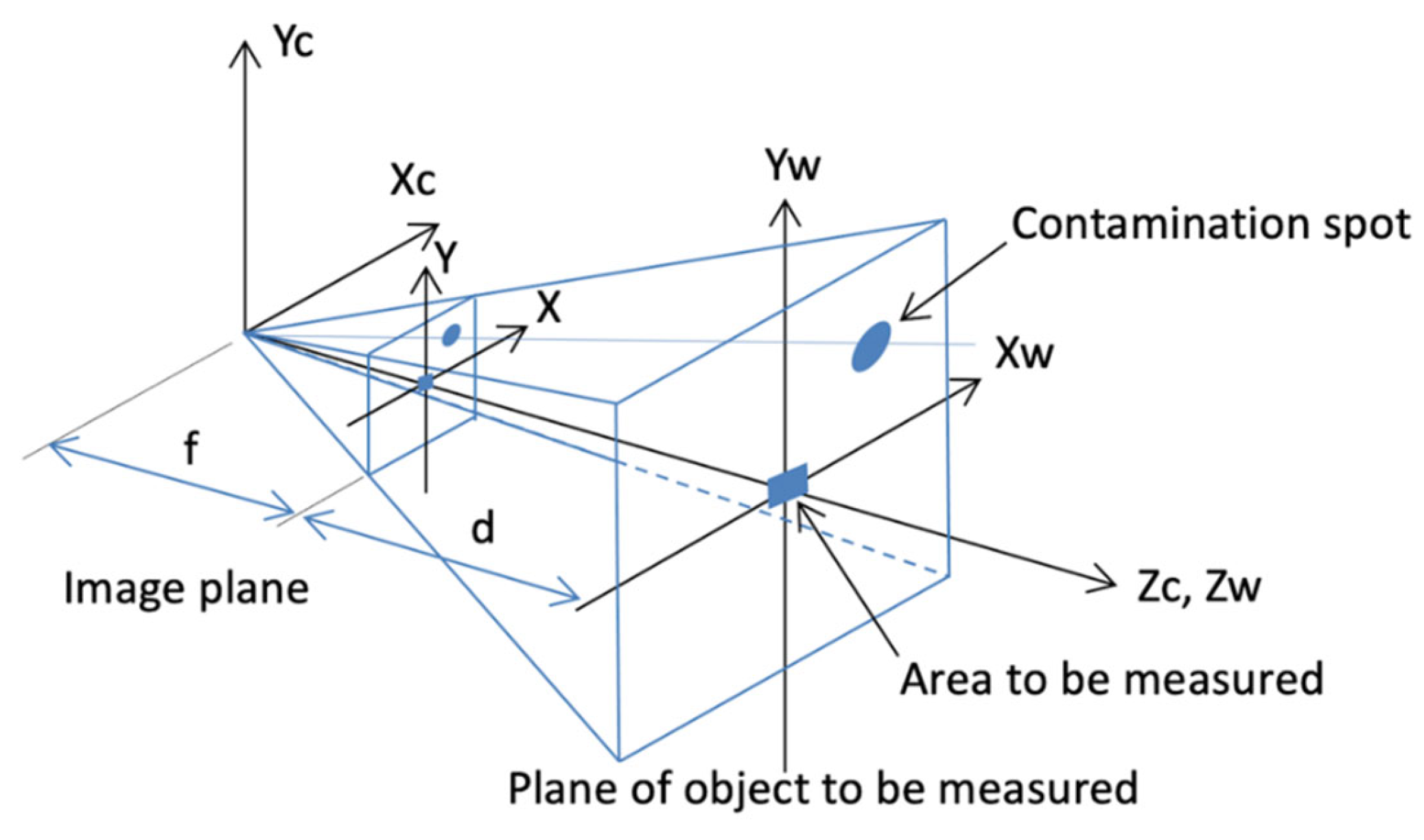
2.1. Mathematical Theory of Contamination Area in Photographs
2.2. Image Processing Theory and Method
2.2.1. Grayscale Processing
2.2.2. Image Binarization
2.2.3. Calculation of Binarized Area
2.3. Estimation of Pollution Area
2.3.1. Preliminary Analysis Results
2.3.2. Contaminated Area Based on Least Squares Regression with Image Linearity
2.3.3. Pixel Linear Interpolation Based on Least Squares Regression
3. Results
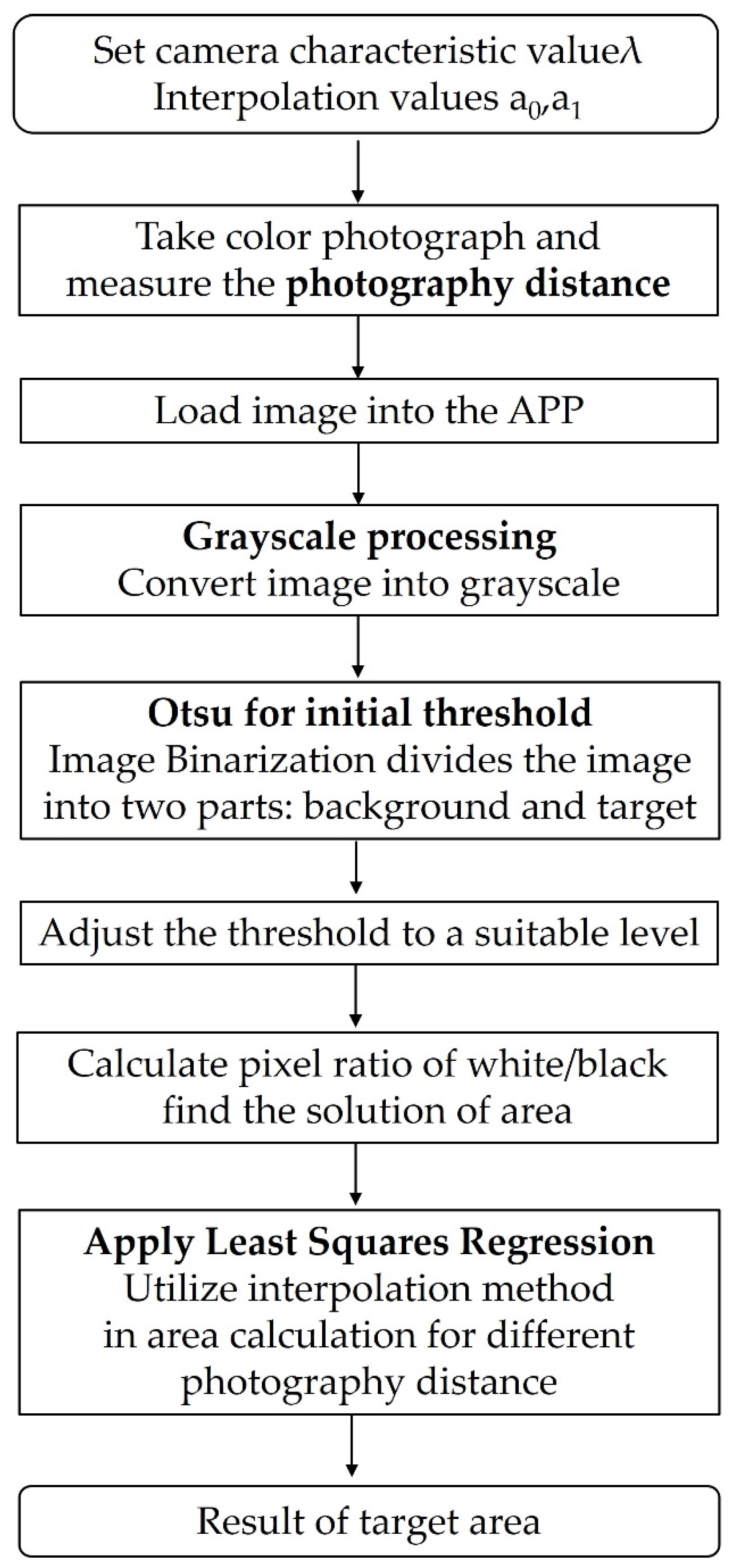
3.1. Binarization of Photo to Calculate Target Area
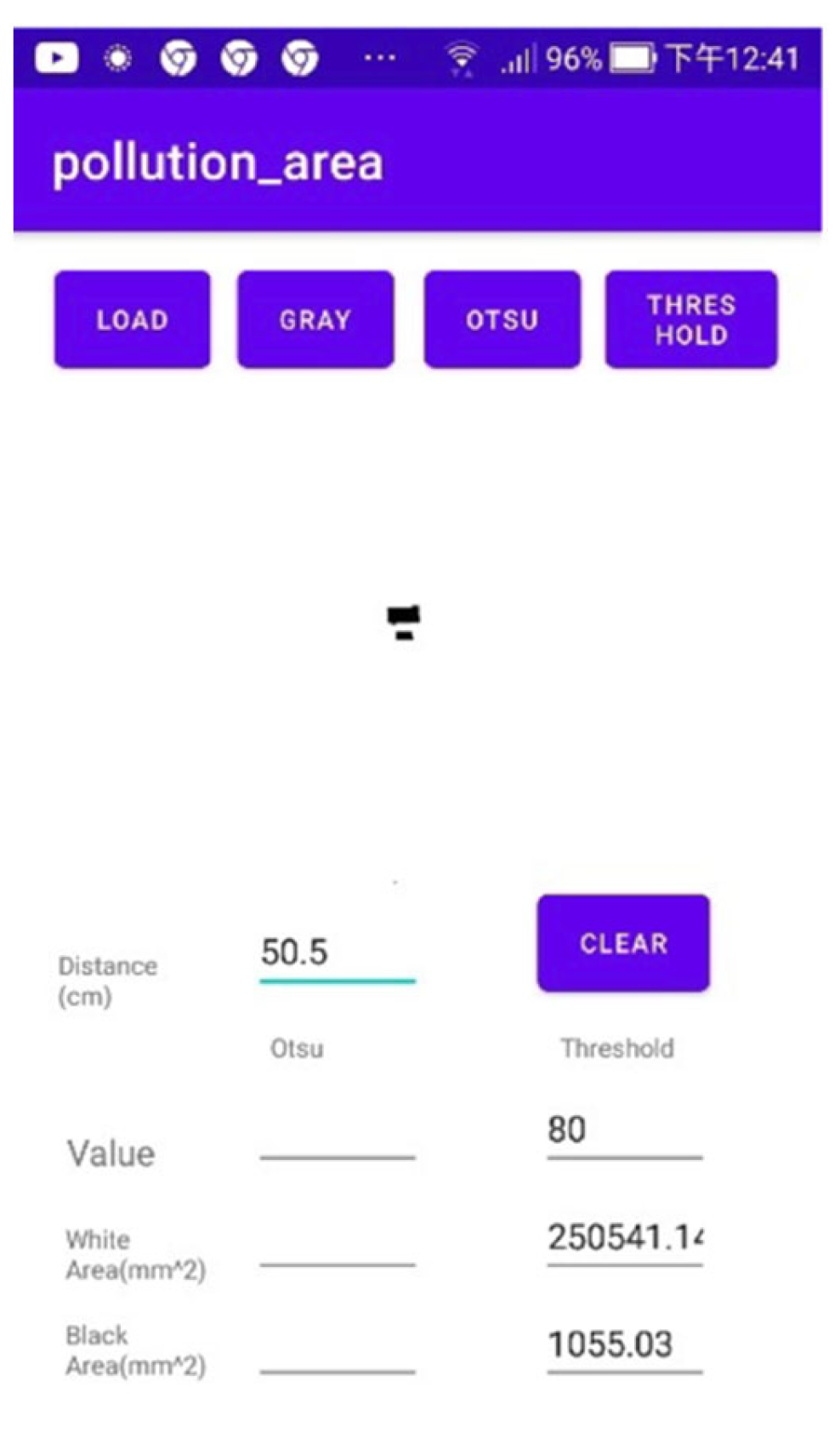
- The LOAD button was pressed to input the sample image and the GRAY button was pressed to convert the image into grayscale, as shown in Figure 7b.
- The OTSU button was pressed to obtain the initial threshold value based on the OTSU method. The threshold given was 139, which is evidently too high for effective processing (see Figure 7c).
- A threshold lower than 139 was input, such as 110, and the THRESHOLD button was pressed to obtain the shade in the lower half.
- The threshold value was adjusted further until the shade in the lower half of the resulting image disappeared. Eventually, a threshold value of 80 was reached. The THRESHOLD button was pressed to obtain Figure 7d. The target value obtained had an area of 10,022.14 mm2.
3.2. Error Analysis
3.3. Application in a Medical Simulation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Porteous, G.H.; Bean, H.A.; Woodward, C.M.; Beecher, R.P.; Bernstein, J.R.; Wilkerson, S.; Porteous, I.; Hsiung, R.L. A simulation study to evaluate improvements in anesthesia work environment contamination after implementation of an infection prevention bundle. Anesth. Analg. 2018, 127, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Andonian, J.; Kazi, S.; Therkorn, J.; Benishek, L.; Billman, C.; Schiffhauer, M.; Nowakowski, E.; Osei, P.; Gurses, A.P.; Hsu, Y.J.; et al. Effect of an intervention package and teamwork training to prevent healthcare personnel self-contamination during personal protective equipment doffing. Clin. Infect. Dis. 2019, 69 (Suppl. S3), S248–S255. [Google Scholar] [CrossRef]
- Roff, M.W. Accuracy and reproducibility of calibrations on the skin using the fives fluorescence monitor. Ann. Occup. Hyg. 1997, 41, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Veal, D.A.; Deere, D.; Ferrari, B.; Piper, J.; Attfield, P.V. Attfield a Fluorescence staining and flow cytometry for monitoring microbial cells. J. Immunol. Methods 2000, 243, 191–210. [Google Scholar] [CrossRef] [PubMed]
- Canelli, R.; Connor, C.W.; Gonzalez, M.; Nozari, A.; Ortega, R. Barrier enclosure during endotracheal intubation. N. Engl. J. Med. 2020, 382, 1957–1958. [Google Scholar] [CrossRef]
- Thomas, L.S.V.; Schaefer, F.; Gehrig, J. Fiji plugins for qualitative image annotations: Routine analysis and application to image classification. F1000Research 2020, 9, 1248. [Google Scholar] [CrossRef]
- Bray, M.A.; Carpenter, A.E. CellProfiler Tracer: Exploring and validating high-throughput, time-lapse microscopy image data. BMC Bioinform. 2015, 16, 368. [Google Scholar] [CrossRef]
- Meijering, E.; Dzyubachyk, O.; Smal, I. Methods for cell and particle tracking. Methods Enzymol. 2012, 504, 183–200. [Google Scholar] [CrossRef]
- de Chaumont, F.; Dallongeville, S.; Chenouard, N.; Hervé, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; Lecomte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef]
- Available online: https://icy.bioimageanalysis.org/ (accessed on 21 October 2022).
- Hartig, S.M. Basic image analysis and manipulation in ImageJ. Curr. Protoc. Mol. Biol. 2013, 102, 14.15.1–14.15.12. [Google Scholar] [CrossRef]
- Schroeder, A.B.; Dobson, E.T.A.; Rueden, C.T.; Tomancak, P.; Jug, F.; Eliceiri, K.W. The ImageJ ecosystem: Open-source software for image visualization, processing, and analysis. Protein Sci. 2021, 30, 234–249. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; Dezonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Weng, C.H.; Chiu, P.W.; Kao, C.L.; Lin, Y.Y.; Lin, C.H. Combating COVID-19 during airway management: Validation of a protection tent for containing aerosols and droplets. Appl. Sci. 2021, 11, 7245. [Google Scholar] [CrossRef]
- Polo Freitag, G.; Freitag de Lima, L.G.; Ernandes Kozicki, L.; Simioni Felicio, L.C.; Romualdo Weiss, R. Use of a smartphone camera attached to a light microscope to determine equine sperm concentration in ImageJ Software. Arch. Vet. Sci. 2020, 25, 33–45. [Google Scholar]
- Rabiolo, A.; Bignami, F.; Rama, P.; Ferrari, G. VesselJ: A new tool for semiautomatic measurement of corneal neovascularization. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8199–8206. [Google Scholar] [CrossRef]
- Available online: https://imagej.net/imagej-wiki-static/Android (accessed on 21 October 2022).
- Cardona, A.; Tomancak, P. Current challenges in open-source bioimage informatics. Nat. Methods 2012, 9, 661–665. [Google Scholar] [CrossRef]
- Tsai, R.Y. A versatile camera calibration technique for high-accuracy 3D machine vision metrology using off-the-shelf TV cameras and lenses. IEEE J. Robot. Automat. 1987, 3, 323–344. [Google Scholar] [CrossRef]
- Zhang, Z. A flexible new technique for camera calibration. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1330–1334. [Google Scholar] [CrossRef]
- Cui, C.; Ngan, K.N. Plane-based external camera calibration with accuracy measured by relative deflection angle. Signal Process. Image Commun. 2010, 25, 224–234. [Google Scholar] [CrossRef]
- Kwak, K.; Huber, D.F.; Badino, H.; Kanade, T. Extrinsic calibration of a single line scanning lidar and a camera. In Proceedings of the 2011 IEEE/RSJ International Conference on Intelligent Robots and Systems, San Francisco, CA, USA, 25–30 September 2011; IEEE Publications: San Francisco, CA, USA, 2011; pp. 3283–3289. [Google Scholar]
- Buades, A.; Coll, B.; Morel, J.M. A review of image denoising algorithms, with a new one. Multiscale Model. Simul. 2005, 4, 490–530. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E. Digital Image Processing, 4th ed.; Pearson: London, UK, 2018. [Google Scholar]
- Barney Smith, E.H.; Likforman-Sulem, L.; Darbon, J. Effect of pre-processing on binarization. SPIE Proc. 2010, 7534, 154–161. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Hall, S.; Poller, B.; Bailey, C.; Gregory, S.; Clark, R.; Roberts, P.; Tunbridge, A.; Poran, V.; Evans, C.; Crook, B. Use of ultraviolet-fluorescence-based simulation in evaluation of personal protective equipment worn for first assessment and care of a patient with suspected high-consequence infectious disease. J. Hosp. Infect. 2018, 99, 218–228. [Google Scholar] [CrossRef]
- Blue, J.; O’Neill, C.; Speziale, P.; Revill, J.; Ramage, L.; Ballantyne, L. Use of a fluorescent chemical as a quality indicator for a hospital cleaning program. Can. J. Infect. Control 2008, 23, 216–219. [Google Scholar]
- Dewangan, A.; Gaikwad, U. Comparative evaluation of a novel fluorescent marker and environmental surface cultures to assess the efficacy of environmental cleaning practices at a tertiary care hospital. J. Hosp. Infect. 2020, 104, 261–268. [Google Scholar] [CrossRef]





| Area (mm2) | 50 cm | 75 cm | 100 cm |
|---|---|---|---|
| 50 | 2127 | 736 | 356 |
| 100 | 3846 | 1334 | 793 |
| 200 | 10,547 | 2672 | 1979 |
| 400 | 14,263 | 5220 | 2944 |
| 450 | 15,480 | 5579 | 3807 |
| 800 | 28,019 | 10,309 | 5721 |
| 900 | 31,204 | 11,978 | 6499 |
| 1600 | 57,437 | 22,185 | 12,013 |
| Distance (cm) | 48.5 | 49.5 | 50.5 | 51.5 | 52.5 |
| Area (mm2) | 923.48 | 989.25 | 1055.03 | 1120.80 | 1186.57 |
| Sites | Face | Hands | Chest Wall |
|---|---|---|---|
| Area (mm2) | 3351.3 | 5040.6 | 89.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, P.-W.; Hsu, C.-T.; Huang, S.-P.; Chiou, W.-Y.; Lin, C.-H. Prediction of Contaminated Areas Using Ultraviolet Fluorescence Markers for Medical Simulation: A Mobile Phone Application Approach. Bioengineering 2023, 10, 530. https://doi.org/10.3390/bioengineering10050530
Chiu P-W, Hsu C-T, Huang S-P, Chiou W-Y, Lin C-H. Prediction of Contaminated Areas Using Ultraviolet Fluorescence Markers for Medical Simulation: A Mobile Phone Application Approach. Bioengineering. 2023; 10(5):530. https://doi.org/10.3390/bioengineering10050530
Chicago/Turabian StyleChiu, Po-Wei, Chien-Te Hsu, Shao-Peng Huang, Wu-Yao Chiou, and Chih-Hao Lin. 2023. "Prediction of Contaminated Areas Using Ultraviolet Fluorescence Markers for Medical Simulation: A Mobile Phone Application Approach" Bioengineering 10, no. 5: 530. https://doi.org/10.3390/bioengineering10050530
APA StyleChiu, P.-W., Hsu, C.-T., Huang, S.-P., Chiou, W.-Y., & Lin, C.-H. (2023). Prediction of Contaminated Areas Using Ultraviolet Fluorescence Markers for Medical Simulation: A Mobile Phone Application Approach. Bioengineering, 10(5), 530. https://doi.org/10.3390/bioengineering10050530