Deep Learning-Assisted Measurements of Photoreceptor Ellipsoid Zone Area and Outer Segment Volume as Biomarkers for Retinitis Pigmentosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Dataset—OCT Volume Scans and Manual Segmentation
2.2. Deep Learning Model and Training Datasets
2.3. Deep Learning Model Training and Validation
2.4. Segmentation of Test Volume Scans by DLM-MC and DLM Only
2.5. Photoreceptor Outer Segment (OS) Metrics Measurements
2.6. Data Analysis
3. Results
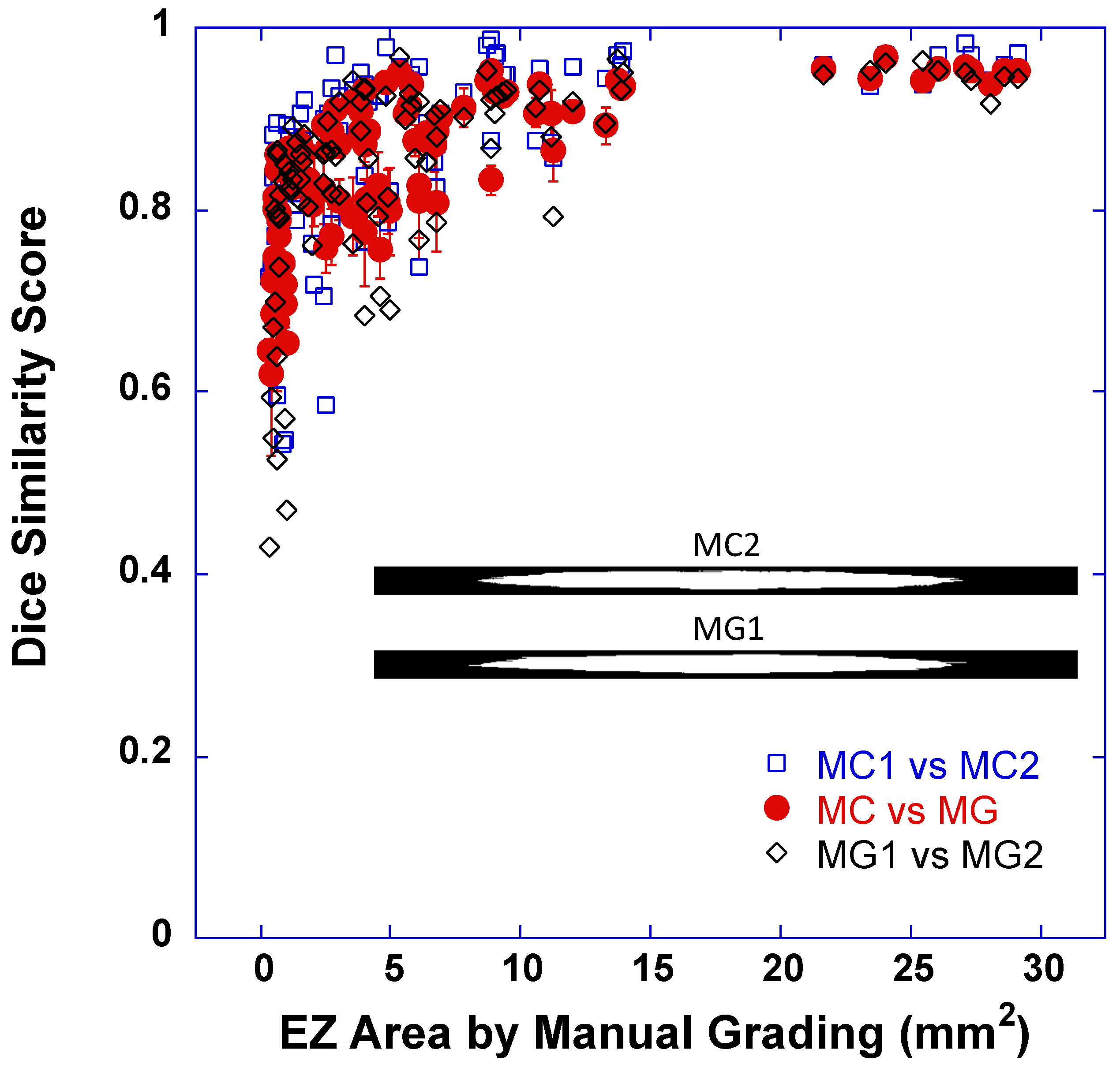
3.1. Dice Scores between EZ Band Segmentations by DLM-MC and by MG
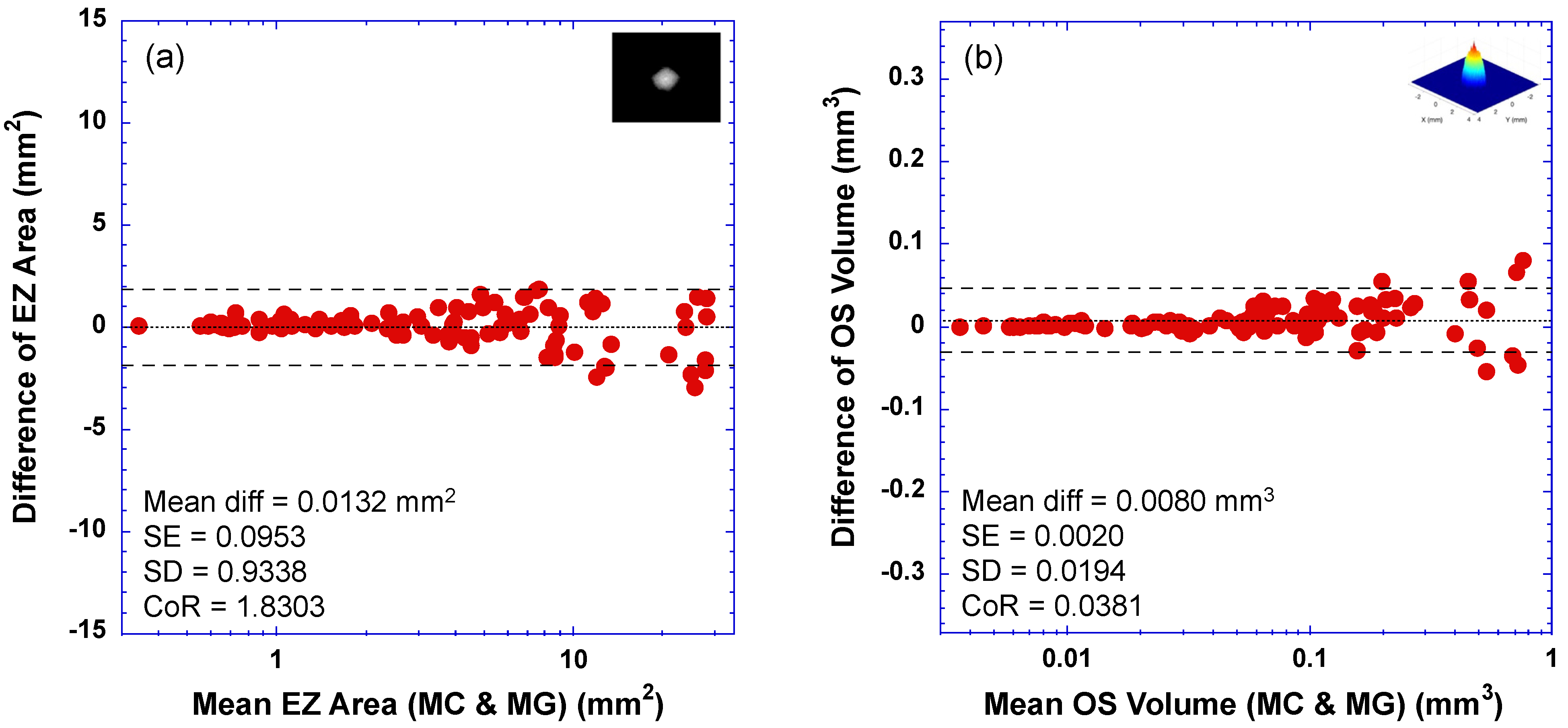
3.2. Bland–Altman Plots—Limit of Agreement between DLM-MC and MG
3.3. Correlation and Linear Regression between DLM-MC and MG
3.4. DLM Only vs. MG
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2D | two-dimensional |
| 3D | three-dimensional |
| ART | automatic real-time tracking |
| BM | Bruch’s membrane |
| CI | confidence interval |
| CNN | convolutional neural network |
| CoR | coefficient of repeatability |
| dINL | distal inner nuclear layer |
| DLM | deep learning model |
| DLM-MC | deep learning model segmentation with manual correction |
| EZ | photoreceptor ellipsoid zone |
| HVSL | Hood Visual Science Laboratory |
| ILM | inner limiting membrane |
| MC | manual correction |
| MG | conventional manual grading |
| OCT | optical coherence tomography |
| OS | photoreceptor outer segment |
| pRPE | proximal (apical) retinal pigment epithelium |
| R2 | coefficients of determination |
| ReLU | rectified linear unit |
| RP | retinitis pigmentosa |
| RPE | retinal pigment epithelium |
| RPGR | Retinitis Pigmentosa GTPase Regulator |
| SD | standard deviation |
| SE | standard error |
| SW | sliding-window |
| XLRP | x-linked retinitis pigmentosa |
References
- Aleman, T.S.; Cideciyan, A.V.; Sumaroka, A.; Windsor, E.A.M.; Herrera, W.; White, D.A.; Kaushal, S.; Naidu, A.; Roman, A.J.; Schwartz, S.B.; et al. Retinal Laminar Architecture in Human Retinitis Pigmentosa Caused by Rhodopsin Gene Mutations. Investig. Opthalmol. Vis. Sci. 2008, 49, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, S.G.; Aleman, T.S.; Sumaroka, A.; Cideciyan, A.V.; Roman, A.J.; Windsor, E.A.M.; Schwartz, S.B.; Rehm, H.L.; Kimberling, W.J. Disease Boundaries in the Retina of Patients with Usher Syndrome Caused by MYO7A Gene Mutations. Investig. Ophthalmol. Vis. Sci. 2009, 50, 1886–1894. [Google Scholar] [CrossRef] [PubMed]
- Witkin, A.J.; Ko, T.H.; Fujimoto, J.G.; Chan, A.; Drexler, W.; Schuman, J.S.; Reichel, E.; Duker, J.S. Ultra-high Resolution Optical Coherence Tomography Assessment of Photoreceptors in Retinitis Pigmentosa and Related Diseases. Arch. Ophthalmol. 2006, 142, 945–952.e1. [Google Scholar] [CrossRef]
- Birch, D.G.; Locke, K.G.; Wen, Y.; Locke, K.I.; Hoffman, D.R.; Hood, D.C. Spectral-Domain Optical Coherence Tomography Measures of Outer Segment Layer Progression in Patients with X-Linked Retinitis Pigmentosa. JAMA Ophthalmol. 2013, 131, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.B.; Parker, M.A.; Steinkamp, P.N.; Romo, A.; Erker, L.R.; Lujan, B.J.; Smith, N. Reliability of Spectral-Domain OCT Ellipsoid Zone Area and Shape Measurements in Retinitis Pigmentosa. Transl. Vis. Sci. Technol. 2019, 8, 37. [Google Scholar] [CrossRef]
- Menghini, M.; Jolly, J.K.; Nanda, A.; Wood, L.; Cehajic-Kapetanovic, J.; MacLaren, R.E. Early Cone Photoreceptor Outer Segment Length Shortening in RPGR X-Linked Retinitis Pigmentosa. Ophthalmology 2020, 244, 281–290. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Birch, D.G. Performance of Deep Learning Models in Automatic Measurement of Ellipsoid Zone Area on Baseline Optical Coherence Tomography (OCT) Images From the Rate of Progression of USH2A-Related Retinal Degeneration (RUSH2A) Study. Front. Med. 2022, 9, 932498. [Google Scholar] [CrossRef]
- Varela, M.D.; Sen, S.; De Guimaraes, T.A.C.; Kabiri, N.; Pontikos, N.; Balaskas, K.; Michaelides, M. Artificial intelligence in retinal disease: Clinical application, challenges, and future directions. Graefe’s Arch. Clin. Exp. Ophthalmol. 2023, 261, 3283–3297. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2019, 103, 167–175. [Google Scholar] [CrossRef]
- Gargeya, R.; Leng, T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration. Ophthalmol. Retina 2017, 1, 322–327. [Google Scholar] [CrossRef] [PubMed]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Christopher, M.; Bowd, C.; Belghith, A.; Goldbaum, M.H.; Weinreb, R.N.; Fazio, M.A.; Girkin, C.A.; Liebmann, J.M.; Zangwill, L.M. Deep Learning Approaches Predict Glaucomatous Visual Field Damage from OCT Optic Nerve Head En Face Images and Retinal Nerve Fiber Layer Thickness Maps. Ophthalmology 2020, 127, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Kawczynski, M.G.; Bengtsson, T.; Dai, J.; Hopkins, J.J.; Gao, S.S.; Willis, J.R. Development of Deep Learning Models to Predict Best-Corrected Visual Acuity from Optical Coherence Tomography. Transl. Vis. Sci. Technol. 2020, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Cunefare, D.; Wang, C.; Guymer, R.H.; Li, S.; Farsiu, S. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed. Opt. Express 2017, 8, 2732–2744. [Google Scholar] [CrossRef]
- Kugelman, J.; Alonso-Caneiro, D.; Read, S.A.; Hamwood, J.; Vincent, S.J.; Chen, F.K.; Collins, M.J. Automatic choroidal segmentation in OCT images using supervised deep learning methods. Sci. Rep. 2019, 9, 13298. [Google Scholar] [CrossRef]
- Lee, C.S.; Tyring, A.J.; Deruyter, N.P.; Wu, Y.; Rokem, A.; Lee, A.Y. Deep-learning based, automated segmentation of macular edema in optical coherence tomography. Biomed. Opt. Express 2017, 8, 3440–3448. [Google Scholar] [CrossRef]
- Schlegl, T.; Waldstein, S.M.; Bogunovic, H.; Endstrasser, F.; Sadeghipour, A.; Philip, A.M.; Podkowinski, D.; Gerendas, B.S.; Langs, G.; Schmidt-Erfurth, U. Fully Automated Detection and Quantification of Macular Fluid in OCT Using Deep Learning. Ophthalmology 2018, 125, 549–558. [Google Scholar] [CrossRef]
- Masood, S.; Fang, R.; Li, P.; Li, H.; Sheng, B.; Mathavan, A.; Wang, X.; Yang, P.; Wu, Q.; Qin, J.; et al. Automatic Choroid Layer Segmentation from Optical Coherence Tomography Images Using Deep Learning. Sci. Rep. 2019, 9, 3058. [Google Scholar] [CrossRef]
- Ngo, L.; Cha, J.; Han, J.H. Deep Neural Network Regression for Automated Retinal Layer Segmentation in Optical Coherence Tomography Images. IEEE Trans. Image Process. 2020, 29, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Pekala, M.; Joshi, N.; Liu, T.Y.A.; Bressler, N.M.; DeBuc, D.C.; Burlina, P. Deep learning based retinal OCT segmentation. Comput. Biol. Med. 2019, 114, 103445. [Google Scholar] [CrossRef]
- Shah, A.; Zhou, L.; Abramoff, M.D.; Wu, X. Multiple surface segmentation using convolution neural nets: Application to retinal layer segmentation in OCT images. Biomed. Opt. Express 2018, 9, 4509–4526. [Google Scholar] [CrossRef] [PubMed]
- Viedma, I.A.; Alonso-Caneiro, D.; Read, S.A.; Collins, M.J. Deep learning in retinal optical coherence tomography (OCT): A comprehensive survey. Neurocomputing 2022, 507, 247–264. [Google Scholar] [CrossRef]
- Camino, A.; Wang, Z.; Wang, J.; Pennesi, M.E.; Yang, P.; Huang, D.; Li, D.; Jia, Y. Deep learning for the segmentation of preserved photoreceptors on en face optional coherence tomogrpahy in two inherited retinal diseases. Biomed. Opt. Express 2018, 9, 3092–3105. [Google Scholar] [CrossRef] [PubMed]
- Loo, J.M.; Jaffe, G.J.; Duncan, J.L.; Birch, D.G.; Farsiu, S. Validation of a Deep Learning-Based Algorithm for Segmentation of The Ellipsoid Zone on Optical Coherence Tomography Images of an Ush2a-Related Retinal Degeneration Clinical Trial. Retina 2022, 42, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Z.; Wu, W.; Birch, D.G. A Hybrid Model Composed of Two Convolutional Neural Networks (CNNs) for Automatic Retinal Layer Segmentation of OCT Images in Retinitis Pigmentosa (RP). Transl. Vis. Sci. Technol. 2021, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Camino, A.; Hagag, A.M.; Wang, J.; Weleber, R.G.; Yang, P.; Pennesi, M.E.; Huang, D.; Li, D.; Jia, Y. Automated detection of preserved photoreceptor on optical coherence tomography in choroideremia based on machine learning. J. Biophotonics 2018, 11, e201700313. [Google Scholar] [CrossRef]
- Loo, J.; Fang, L.; Cunefare, D.; Jaffe, G.J.; Farsiu, S. Deep longitudinal transfer learning-based automatic segmentation of photoreceptor ellipsoid zone defects on optical coherence tomography images of macular telangiectasia type 2. Biomed. Opt. Express 2018, 9, 2681–2698. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Galles, D.; Klein, M.; Locke, K.G.; Birch, D.G. Application of a Deep Machine Learning Model for Automatic Measurement of EZ Width in SD-OCT Images of RP. Transl. Vis. Sci. Technol. 2020, 9, 15. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Juroch, K.; Chen, Y.; Ying, G.S.; Birch, D.G. Deep learning facilitated study of the rate of change in photoreceptor outer segment (OS) metrics in x-linked retinitis pigmentosa (xlRP). Investig. Ophthalmol. Vis. Sci. 2023, 64, 31. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, D.R.; Hughbanks-Wheaton, D.K.; Pearson, N.S.; Fish, G.E.; Spencer, R.; Takacs, A.; Klein, M.; Locke, K.G.; Birch, D.G. Four-Year Placebo-Controlled Trial of Docosahexaenoic Acid in X-Linked Retinitis Pigmentosa (DHAX Trial). JAMA Ophthalmol. 2014, 132, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Ciresan, D.C.; Gambardella, L.M.; Giusti, A.; Schmidhuber, J. Deep neural networks segment neuronal membranes in electron microscopy images. In NIPS’12: Proceedings of the NIPS’12: 25th International Conference on Neural Information Processing System, Lake Tahoe, NV, USA, 3–6 December 2012; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates Inc.: Reed Hook, NY, USA, 2012; Volume 2, pp. 2843–2851. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning, Proceedings of Machine Learning Research, Lille, France, 7–9 July 2015. [Google Scholar]
- Yang, Q.; Reisman, C.A.; Chan, K.; Ramachandran, R.; Raza, A.; Hood, D.C. Automated segmentation of outer retinal layers in macular OCT images of patients with retinitis pigmentosa. Biomed. Opt. Express 2011, 2, 2493–2503. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, J.; He, H.; Huang, X. A fast deep learning system using GPU. In Proceedings of the 2014 IEEE International Symposium on Circuits and Systems (ISCAS), Melbourne, VIC, Australia, 1–5 June 2014; pp. 1552–1555. [Google Scholar] [CrossRef]
- Pandey, M.; Fernandez, M.; Gentile, F.; Isayev, O.; Tropsha, A.; Stern, A.C.; Cherkasov, A. The transformational role of GPU computing and deep learning in drug discovery. Nat. Mach. Intell. 2022, 4, 211–221. [Google Scholar] [CrossRef]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Zhao, H.; Sun, N. Improved U-Net Model for Nerve Segmentation. In Image and Graphics; Zhao, Y., Kong, X., Taubman, D., Eds.; Springer: Cham, Switzerland, 2017; pp. 496–504. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Kugelman, J.; Allman, J.; Read, S.A.; Vincent, S.J.; Tong, J.; Kalloniatis, M.; Chen, F.K.; Collins, M.J.; Alonso-Caneiro, D. A comparison of deep learning U-Net architectures for posterior segment OCT retinal layer segmentation. Sci. Rep. 2022, 12, 14888. [Google Scholar] [CrossRef]
- Meyes, R.; Lu, M.; de Puiseau, C.W.; Meise, T. Ablation studies in artificial neural networks. arXiv 2019, arXiv:1901.08644v2. [Google Scholar]



| Training Dataset | # Patients | # Mid-Line B-Scans | Patch Horizontal Shift & # Patches for U-Net Training | # Patches for SW Model Training | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | adRP | arRP | XLRP | Isolated | Normal | Total | High-Speed (768 A-Scans) | High-Res (1536 A-Scans) | Patch Size: 256 × 32 | Patch Size: 256 × 64 | Patch Size: 256 × 128 | |||||
| Overlap (pixels) | # Patches | Overlap (pixels) | # Patches | Overlap (pixels) | # Patches | |||||||||||
| RP140 | 140 | 50 | 15 | 15 | 30 | 30 | 280 | 183 | 97 | 28 | 300,369 | 56 | 148,705 | 112 | 72,179 | 1,646,562 |
| RP240 | 240 | 50 | 30 | 20 | 120 | 20 | 480 | 305 | 175 | 28 | 527,504 | 56 | 261,658 | 112 | 127,691 | 2,878,002 |
| RP340 | 340 | 80 | 40 | 30 | 170 | 20 | 680 | 446 | 234 | 28 | 737,084 | 56 | 365,520 | 112 | 178,242 | 3,984,084 |
| RP480 | 480 | 130 | 55 | 45 | 200 | 50 | 960 | 629 | 331 | 28 | 1,037,453 | 56 | 514,225 | 112 | 250,421 | 5,630,646 |
| Dice Score (EZ Band Segmentation) | All Cases | Cases with EZ Area ≤ 1 mm2 | Cases with EZ Area > 1 mm2 | ||||
|---|---|---|---|---|---|---|---|
| Mean (SD) | Median (Q1, Q3) | Mean (SD) | Median (Q1, Q3) | Mean (SD) | Median (Q1, Q3) | ||
| MC vs. MG | 0.8524 (0.0821) | 0.8674 (0.8052, 0.9212) | 0.7478 (0.0651) | 0.7446 (0.6963, 0.7970) | 0.8799 (0.0614) | 0.8875 (0.8319, 0.9344) | |
| MG1 vs. MG2 | 0.8417 (0.1111) | 0.8642 (0.8014, 0.9201) | 0.7210 (0.1284) | 0.7905 (0.6199, 0.8054) | 0.8735 (0.0810) | 0.8928 (0.8332, 0.9306) | |
| MC1 vs. MC2 | 0.8732 (0.0969) | 0.8902 (0.8341, 0.9486) | 0.7791 (0.1083) | 0.8208 (0.7354, 0.8559) | 0.8979 (0.0771) | 0.9239 (0.8670, 0.9561) | |
| RP140 vs. MG | 256 × 32 | 0.7859 (0.1276) | 0.8124 (0.7198, 0.8896) | 0.6212 (0.0833) | 0.6249 (0.5672, 0.6803) | 0.8293 (0.0987) | 0.8513 (0.7756, 0.9092) |
| 256 × 64 | 0.7854 (0.1383) | 0.8174 (0.7185, 0.9006) | 0.6044 (0.1148) | 0.6074 (0.4722, 0.7006) | 0.8330 (0.0992) | 0.8609 (0.7721, 0.9158) | |
| 256 × 128 | 0.7847 (0.1504) | 0.8390 (0.7261, 0.9003) | 0.5614 (0.1254) | 0.5578 (0.4897, 0.6338) | 0.8435 (0.0887) | 0.8598 (0.8112, 0.9073) | |
| RP240 vs. MG | 256 × 32 | 0.8002 (0.1047) | 0.8148 (0.7382, 0.8935) | 0.6725 (0.0883) | 0.6695 (0.6055, 0.7440) | 0.8338 (0.0801) | 0.8392 (0.7917, 0.9089) |
| 256 × 64 | 0.7915 (0.1122) | 0.8051 (0.7310, 0.8847) | 0.6544 (0.0935) | 0.6711 (0.6042, 0.7210) | 0.8276 (0.0859) | 0.8328 (0.7893, 0.9064) | |
| 256 × 128 | 0.8014 (0.1105) | 0.8339 (0.7183, 0.8976) | 0.6460 (0.0816) | 0.6767 (0.5955, 0.6999) | 0.8423 (0.0752) | 0.8510 (0.8042, 0.8999) | |
| RP340 vs. MG | 256 × 32 | 0.8055 (0.1133) | 0.8293 (0.7550, 0.8949) | 0.6561 (0.0877) | 0.6600 (0.5929, 0.7064) | 0.8448 (0.0823) | 0.8518 (0.8095, 0.9150) |
| 256 × 64 | 0.8012 (0.1168) | 0.8280 (0.7339, 0.8997) | 0.6461 (0.0932) | 0.6629 (0.6221, 0.6954) | 0.8420 (0.0835) | 0.8506 (0.8106, 0.9072) | |
| 256 × 128 | 0.8096 (0.1114) | 0.8375 (0.7391, 0.9043) | 0.6608 (0.0897) | 0.6831 (0.6130, 0.7215) | 0.8487 (0.0789) | 0.8628 (0.8126, 0.9096) | |
| RP480 vs. MG | 256 × 32 | 0.8115 (0.1065) | 0.8294 (0.7557, 0.8970) | 0.6827 (0.0910) | 0.6974 (0.6245, 0.7500) | 0.8454 (0.0818) | 0.8611 (0.7945, 0.9100) |
| 256 × 64 | 0.8027 (0.1208) | 0.8336 (0.7455, 0.9005) | 0.6411 (0.1094) | 0.6568 (0.5704, 0.7371) | 0.8452 (0.0815) | 0.8559 (0.8058, 0.9101) | |
| 256 × 128 | 0.8189 (0.1004) | 0.8369 (0.7646, 0.9069) | 0.6783 (0.0791) | 0.6815 (0.6305, 0.7320) | 0.8559 (0.0672) | 0.8558 (0.8225, 0.9132) | |
| Agreement of EZ Area Measurements | CoR (mm2) | Mean Difference (SD) (mm2) | Mean Absolute Error (SD) (mm2) | Correlation Coeffiicent r (95% CI) | R2 | Linear Regression Slope (95% CI) | |
|---|---|---|---|---|---|---|---|
| MC vs. MG | 1.8303 | 0.0132 (0.9338) | 0.6561 (0.6612) | 0.9928 (0.9892–0.9952) | 0.9856 | 0.9598 (0.9399–0.9797) | |
| MG1 vs. MG2 | 2.1629 | −0.5320 (1.1035) | 0.7958 (0.9294) | 0.9913 (0.9869–0.9942) | 0.9826 | 0.9344 (0.9131–0.9556) | |
| MC1 vs. MC2 | 2.8368 | 0.0969 (1.4474) | 1.1315 (0.9003) | 0.9809 (0.9714–0.9872) | 0.9621 | 0.9736 (0.9405–1.0067) | |
| RP140 vs. MG | 256 × 32 | 4.4089 | −1.2443 (2.2494) | 1.4211 (2.1409) | 0.9662 (0.9497–0.9774) | 0.9335 | 0.7939 (0.7576–0.8302 |
| 256 × 64 | 3.9315 | −0.9377 (2.0059) | 1.3096 (1.7830) | 0.9784 (0.9678–0.9856) | 0.9573 | 0.7972 (0.7684–0.8251) | |
| 256 × 128 | 4.2348 | −1.3690 (2.1606) | 1.5214 (2.0550) | 0.9801 (0.9703–0.9867) | 0.9606 | 0.7617 (0.7353–0.7881) | |
| RP240 vs. MG | 256 × 32 | 3.4719 | −1.6201 (1.7714) | 1.6400 (1.7529) | 0.9820 (0.9730–0.9880) | 0.9643 | 0.8310 (0.8035–0.8584) |
| 256 × 64 | 3.5993 | −1.7114 (1.8364) | 1.7513 (1.7979) | 0.9799 (0.9700–0.9866) | 0.9602 | 0.8272 (0.7983–0.8560) | |
| 256 × 128 | 3.5077 | −1.5654 (1.7896) | 1.6040 (1.7548) | 0.9817 (0.9700–0.9866) | 0.9637 | 0.8283 (0.8008–0.8558) | |
| RP340 vs. MG | 256 × 32 | 3.3957 | −1.5381 (1.7325) | 1.5720 (1.7015) | 0.9821 (0.9732–0.9880) | 0.9645 | 0.8391 (0.8115–0.8667) |
| 256 × 64 | 3.5157 | −1.6574 (1.7937) | 1.6811 (1.7713) | 0.9845 (0.9768–0.9897) | 0.9692 | 0.8142 (0.7894–0.8391) | |
| 256 × 128 | 3.0787 | −1.5493 (1.5708) | 1.5714 (1.5484) | 0.9880 (0.9820–0.9920) | 0.9761 | 0.8409 (0.8184–0.8634) | |
| RP480 vs. MG | 256 × 32 | 3.1337 | −1.5201 (1.5988) | 1.5625 (1.5570) | 0.9843 (0.9765–0.9895) | 0.9688 | 0.8569 (0.8306–0.8833) |
| 256 × 64 | 3.2281 | −1.5422 (1.6470) | 1.6031 (1.5872) | 0.9826 (0.9740–0.9884) | 0.9655 | 0.8563 (0.8286–0.8840) | |
| 256 × 128 | 3.3433 | −1.5492 (1.7058) | 1.5916 (1.6659) | 0.9844 (0.9767–0.9896) | 0.9690 | 0.8324 (0.8069–0.8079) | |
| Agreement of OS Volume Measurements | CoR (mm3) | Mean Difference (SD) (mm3) | Mean Absolute Error (SD) (mm3) | Correlation Coeffiicent r (95% CI) | R2 | Linear Regression Slope (95% CI) | |
|---|---|---|---|---|---|---|---|
| MC vs. MG | 0.0381 | 0.0080 (0.0194) | 0.0137 (0.0160) | 0.9938 (0.9906–0.9958) | 0.9876 | 1.0104 (0.9909–1.0298) | |
| MG1 vs. MG2 | 0.0389 | 0.0002 (0.0198) | 0.0128 (0.0151) | 0.9933 (0.9899–0.9955) | 0.9866 | 0.9930 (0.9732–1.0129) | |
| MC1 vs. MC2 | 0.0265 | −0.0043 (0.0135) | 0.0097 (0.0103) | 0.9970 (0.9955–0.9980) | 0.9940 | 0.9874 (0.9743–1.0005) | |
| RP140 vs. MG | 256 × 32 | 0.3300 | −0.1013 (0.1683) | 0.1058 (0.1656) | 0.5822 (0.4321–0.7009) | 0.3389 | 0.0140 (0.0106–0.0173) |
| 256 × 64 | 0.3288 | −0.1027 (0.1677) | 0.1056 (0.1659) | 0.6200 (0.4791–0.7298) | 0.3845 | 0.0177 (0.0138–0.0215) | |
| 256 × 128 | 0.3290 | −0.1014 (0.1679) | 0.1049 (0.1656) | 0.6168 (0.4751–0.7274) | 0.3805 | 0.0169 (0.0132–0.0206) | |
| RP240 vs. MG | 256 × 32 | 0.0653 | −0.0119 (0.0333) | 0.0170 (0.0310) | 0.9829 (0.9745–0.9886) | 0.9661 | 0.9019 (0.8729–0.9308) |
| 256 × 64 | 0.0700 | −0.0141 (0.0357) | 0.0196 (0.0330) | 0.9800 (0.9701–0.9866) | 0.9603 | 0.8976 (0.8664–0.9289) | |
| 256 × 128 | 0.0623 | −0.0113 (0.0318) | 0.0175 (0.0288) | 0.9844 (0.8815–0.9372) | 0.9690 | 0.9093 (0.8815–0.9372) | |
| RP340 vs. MG | 256 × 32 | 0.0608 | −0.0070 (0.0310) | 0.0162 (0.0273) | 0.9836 (0.9755–0.9890) | 0.9675 | 0.9447 (0.9151–0.9744) |
| 256 × 64 | 0.0581 | −0.0097 (0.0296) | 0.0157 (0.0269) | 0.9863 (0.8933–0.9460) | 0.9728 | 0.9197 (0.8933–0.9460) | |
| 256 × 128 | 0.0541 | −0.0090 (0.0276) | 0.0152 (0.0247) | 0.9880 (0.9038–0.9535) | 0.9762 | 0.9286 (0.9038–0.9535) | |
| RP480 vs. MG | 256 × 32 | 0.0487 | −0.0049 (0.0248) | 0.0149 (0.0205) | 0.9894 (0.9841–0.9929) | 0.9788 | 0.9757 (0.9511–1.0003) |
| 256 × 64 | 0.0513 | −0.0060 (0.0262) | 0.0155 (0.0219) | 0.9882 (0.9403–0.9915) | 0.9766 | 0.9659 (0.9403–0.9915) | |
| 256 × 128 | 0.0565 | −0.0084 (0.0289) | 0.0154 (0.0258) | 0.9866 (0.9031–0.9558) | 0.9734 | 0.9294 (0.9031–0.9558) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-Z.; Juroch, K.; Birch, D.G. Deep Learning-Assisted Measurements of Photoreceptor Ellipsoid Zone Area and Outer Segment Volume as Biomarkers for Retinitis Pigmentosa. Bioengineering 2023, 10, 1394. https://doi.org/10.3390/bioengineering10121394
Wang Y-Z, Juroch K, Birch DG. Deep Learning-Assisted Measurements of Photoreceptor Ellipsoid Zone Area and Outer Segment Volume as Biomarkers for Retinitis Pigmentosa. Bioengineering. 2023; 10(12):1394. https://doi.org/10.3390/bioengineering10121394
Chicago/Turabian StyleWang, Yi-Zhong, Katherine Juroch, and David Geoffrey Birch. 2023. "Deep Learning-Assisted Measurements of Photoreceptor Ellipsoid Zone Area and Outer Segment Volume as Biomarkers for Retinitis Pigmentosa" Bioengineering 10, no. 12: 1394. https://doi.org/10.3390/bioengineering10121394
APA StyleWang, Y.-Z., Juroch, K., & Birch, D. G. (2023). Deep Learning-Assisted Measurements of Photoreceptor Ellipsoid Zone Area and Outer Segment Volume as Biomarkers for Retinitis Pigmentosa. Bioengineering, 10(12), 1394. https://doi.org/10.3390/bioengineering10121394






