Perspectives for Using CO2 as a Feedstock for Biomanufacturing of Fuels and Chemicals
Abstract
:1. Introduction
2. State of the Art of Current Technologies
2.1. One-Step Strategy—Direct Conversion
2.1.1. Natural CO2 Fixation Pathways
Common Natural CO2 Fixation Cycles
Less Common Natural CO2 Fixation Cycles
2.1.2. Synthetic CO2 Fixation Pathways
2.1.3. Host Selection and Reducing Power
CO2-Fixing Autotrophs and Synthetic Hosts
Energy Supplies for Microbial CO2 Fixation
2.1.4. Microbial Electrosynthesis
2.2. Two-Step Strategy—Fixing CO2 into C1/C2 Chemicals via Electrochemical Catalysis and Converting C1/C2 Chemicals into Bioproducts via Biomanufacturing
2.2.1. Using CO2-Derived C1 Chemicals for Biomanufacturing
Carbon Monoxide
Methane
Methanol
Formate
2.2.2. Using CO2-Derived C2 Chemicals for Biomanufacturing
Acetate
Ethanol
2.2.3. Biomanufacturing with Syngas via Gas Fermentation
2.2.4. Current Attempts to Industrialize Microbial CO2 Fixation
3. Challenges and Future Perspectives
3.1. Challenges for Biomanufacturing with Direct Fixation of CO2
- (1)
- Only low-energy utilization efficiency can be achieved when light is used as the energy source to fix CO2. Green plants, algae, and certain bacteria are capable of using sunlight via the photosynthesis process to capture and fix CO2 into carbohydrates, but at low-energy efficiency, with less than 1% of the sunlight energy stored in the biosynthesized chemicals [5,265].
- (2)
- Energy-intensive chemicals such as H2 gas can be used to fix CO2 and provide the reducing power to convert CO2 into the desired carbohydrate products, but there are concerns of extra material cost, technical challenges of using gas for fermentation, increased process complexity, and operating safety due to the use of H2 gas or similar energy-intensive materials.
- (3)
- A very limited number of microbial hosts, genetic manipulation methods and tools, and pathway engineering strategies are available for more generalized applications of direct CO2 fixation and conversion. Many synthetic pathways for direct CO2 fixation face major challenges, such as enzymes with toxicity to host cells or with non-compatible optimum temperatures. Innovations such as the allyl-CoA carboxylase/reductase, which boasts an activity rate 37 times that of the CBB cycle, show promise in addressing this [26]. Introducing mechanisms to concentrate carbon also seems to be a viable strategy to enhance the carbon flux in these pathways. With synthetic biology’s progress, exploring and designing novel pathways might be the key. Predictions even suggest that certain pathways, like those using phosphoenolpyruvate carboxylase, could potentially offer two to three times the carbon-fixation rate of the Calvin cycle [56].
- (4)
- Microbial electrosynthesis (MES) can be used to produce certain fuels or valuable organic acids [92,93,94] by utilizing a biofilm on an electrode as a catalyst to directly reduce CO2 to the products [23], but the species of the microorganisms and the categories of the fuels and chemicals that can be produced are very limited. Acetate is the current major product and its production titer and yield are still too low, which significantly increases the downstream recovery cost [266]. In addition, there is strict requirement for the materials that can be used for cathode. More challenges for further process design and scale-up are expected for large-scale applications in future [266].
3.2. Challenges for Biomanufacturing with CO2-Derived C1/C2 Chemicals
- (1)
- Mass transfer challenges limits the microbial fermentation productivity when the CO2-derived C1 gases, such as CO or CH4, are used as the substrate. Metabolic engineering strategies for using appropriate microorganisms to metabolize the C1 gases are also to be established and further optimized. In addition, safety concerns are also another challenge that may limit the use of CO for biomanufacturing.
- (2)
- Though formic acid and acetic acid can be used as the substrate for biomanufacturing, most current electrochemical catalysis processes can only fix CO2 into the form of formate or acetate salts in aqueous solution, which need to be further treated with acid and base and go through a complicated purification process to obtain the acid products so that they can be fed into the bioreactor for microbial fermentation. Progress has been achieved in electrochemically fixing CO2 into nearly pure formic acid [267], but the productivity needs to be further improved for large-scale application. Comparing to the electrochemical reduction in CO2 into formic acid, converting CO2 into acetic acid at high yield is still a challenge [268].
- (3)
- Direct feeding too much formic acid or acetic acid into a bioreactor may cause sudden acidic pH spikes in fermentation and kill the microbial cells. Therefore, new formic/acetic acid feeding strategies should be developed to avoid/minimize pH spikes in a bioreactor while providing enough substrate(s) for cell growth and product formation [269,270].
- (4)
- Methanol and ethanol can be used as fermentation substrates with high-energy densities, but high concentrations of the alcohol substrates may cause toxicity to the microbial cells. In addition, further metabolic engineering strategies for efficient assimilation of methanol and/or ethanol should be explored for significantly higher product yield.
3.3. Future Perspectives for Biomanufacturing with CO2
- (1)
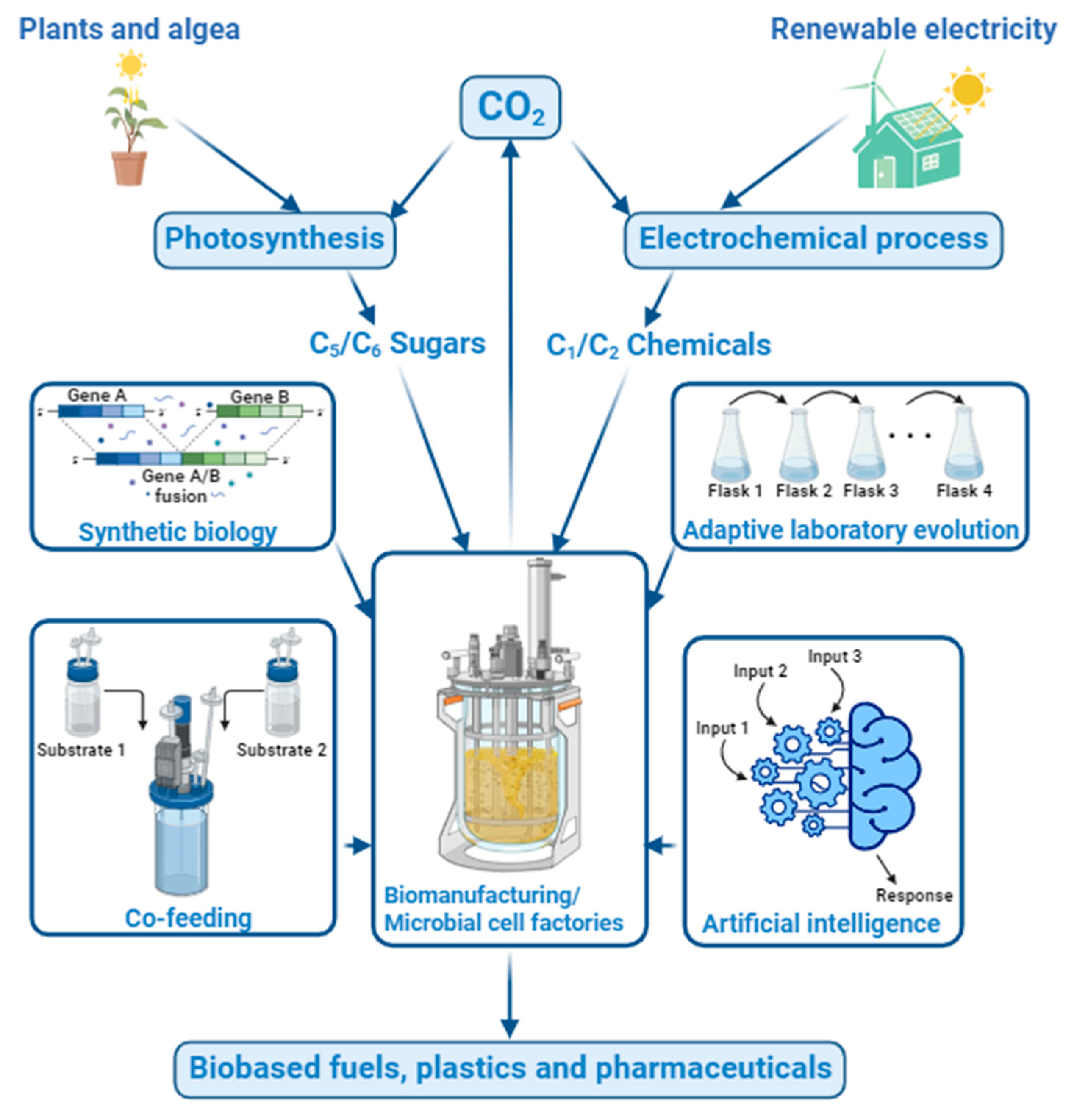
- Using advanced synthetic biology to create new microbial cell factories to utilize CO2 and CO2-derived chemicals for high-yield biomanufacturing: Researchers are now at the forefront of devising more efficient synthetic systems. This involves engineering pivotal enzymes and transferring whole or partial carbon-fixation pathways into heterotrophic cells, enabling them to perform carbon fixation. A testament to these efforts includes the creation of pathways like the MCG pathway and the CETCH cycle using different carboxylases [26]. Although the enhancement in carbon-fixation rate remains modest, these innovations may lead to designing more adept systems. Host selection also is a challenge for keeping CO2 fixation sustainable. For example, most CO2-fixing microbes cannot tolerate high CO2 concentrations, necessitating research into strains that can endure and efficiently process higher levels of CO2 or CO2-derived substrates. Adaptive laboratory evolution (ALE) methods may be applied to help develop more robust production strains that are suitable for large-scale applications.
- (2)
- Using artificial intelligence (AI) to guide the discoveries of new strains, metabolic pathways, enzymes, and fermentation process controls that may lead to complete bioconversion of CO2 or CO2-derived substrates [271,272,273]: This may also help discover new valuable products that may be produced from the pathways using CO2 or having CO2 as the major intermediates. More advanced process, such as continuous biomanufacturing with extremely high yield and productivity, can also be developed [8].
- (3)
- Exploring a cofeeding strategy that uses a mixed C1 and C2 substrates for biomanufacturing: Current electrochemical reduction in CO2 focuses on maximizing the production of a single C1/C2 product at high yield and selectivity. However, the microbial cells may be capable of using a mixed C1 and C2 feed for producing a desired fermentation product. This may help relieve the burden in the electrochemical catalysis system and significantly reduce its cost. More strain engineering and fermentation process development work should be conducted to use a medium or feed with mixed C1/C2 substrates, including methanol, formic acid, ethanol, and acetic acid, for various biomanufacturing purposes. A joint research effort between the electrochemists, biologists, and chemical engineers are expected to achieve the goal.
- (4)
- Developing an advanced process control strategy based on online monitoring/measurements of dissolved CO2 in an aqueous medium, exhausted CO2 in off-gas flow, and the cellular redox levels: Technologies for measuring dissolved CO2 in liquid and gas-phase CO2 have been well established and become commercially available. Monitoring redox cofactor (NAD/NADH, NADP/NADPH, FAD/FADH2) balance has also been investigated and demonstrated a capability for advanced fermentation control to further improve the biomanufacturing yield [274,275,276]. In particular, a nutrient-induced metabolic shift for high productivity and low-waste generation has been demonstrated in cultures of various cell lines and products. However, as the cells rapidly respond to culture conditions, it is crucial to closely monitor their metabolism for a controlled balance between the target metabolic pathway and unfavorable consequences. In particular, during biosynthesis of bioproducts from CO2-derived C1/C2 substrates, additional reduction power (NADH, NADPH, FADH2) has to be supplied to produce compounds whose degree of reduction is higher than that of the substrate [277,278]. Therefore, adjusting the metabolic status and pathways for improved NADH/NADPH in microbial cells is an effective method to enhance the biosynthesis of many bioproducts [277,279,280]. Moreover, other parameters like temperature (to consider O2 and CO2 solubility), pH (regarding the host optimal pH), dissolved oxygen, and total inorganic carbon should be optimized for reaching higher yields [281,282].
- (5)
- Developing a novel biomanufacturing platform that can produce fuels and chemicals from sugars at zero or near-zero life cycle carbon emissions via in situ CO2 recycling: Most microbial fermentation processes that use C5/C6 sugars as substrates have nearly 50% or more carbon loss due to the need for metabolizing a portion of the sugar substrate into CO2 to generate energy (ATP) and cofactors for cell growth and biosynthesis. To date, there has been very rare research aiming for biomanufacturing with direct recycling of the exhausted CO2. The capturing and fixation of CO2 into C1/C2 chemicals can be achieved via similar electrochemical catalysis processes [116,283]. There are several trials to combine electrochemical reduction in CO2 and the fermentation of its reduced products. However, there is still a long way to go for the optimization of this combined system to work effectively [284]. The developed new biomanufacturing platform should employ newly engineered strains that can co-utilize C5/C6 sugars and CO2-derived C1/C2 chemicals for producing the desired fermentation products as shown in Figure 7. Recycling the exhausted CO2 back to fermentation not only avoids/minimizes the CO2 release from the biomanufacturing processes, but also maximizes the use of the renewable feedstocks for significantly higher product yield.
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lemaire, O.N.; Jespersen, M.; Wagner, T. CO2-Fixation Strategies in Energy Extremophiles: What Can We Learn from Acetogens? Front. Microbiol. 2020, 11, 486. [Google Scholar] [CrossRef]
- Hossain, M.F. Extreme Level of CO2 Accumulation into the Atmosphere Due to the Unequal Global Carbon Emission and Sequestration. Water Air Soil Pollut. 2022, 233, 105. [Google Scholar] [CrossRef]
- Bhat, P. Carbon Needs to Cost at Least $100/tonne Now to Reach Net Zero by 2050: Reuters Poll. 2021. Available online: https://www.reuters.com/business/cop/carbon-needs-cost-least-100tonne-now-reach-net-zero-by-2050-2021-10-25/ (accessed on 23 November 2023).
- Baylin-Stern, A.; Berghout, N. Is Carbon Capture Too Expensive? 2021. Available online: https://www.iea.org/commentaries/is-carbon-capture-too-expensive (accessed on 23 November 2023).
- Claassens, N.J.; Sousa, D.Z.; Dos Santos, V.A.; de Vos, W.M.; van der Oost, J. Harnessing the power of microbial autotrophy. Nat. Rev. Microbiol. 2016, 14, 692–706. [Google Scholar] [CrossRef]
- Park, J.O.; Liu, N.; Holinski, K.M.; Emerson, D.F.; Qiao, K.; Woolston, B.M.; Xu, J.; Lazar, Z.; Islam, M.A.; Vidoudez, C. Synergistic substrate cofeeding stimulates reductive metabolism. Nat. Metab. 2019, 1, 643–651. [Google Scholar] [CrossRef]
- Liao, J.C.; Mi, L.; Pontrelli, S.; Luo, S. Fuelling the future: Microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 2016, 14, 288–304. [Google Scholar] [CrossRef]
- Xie, D. Continuous biomanufacturing with microbes—Upstream progresses and challenges. Curr. Opin. Biotechnol. 2022, 78, 102793. [Google Scholar] [CrossRef]
- Zhang, C.; Ottenheim, C.; Weingarten, M.; Ji, L. Microbial Utilization of Next-Generation Feedstocks for the Biomanufacturing of Value-Added Chemicals and Food Ingredients. Front. Bioeng. Biotechnol. 2022, 10, 874612. [Google Scholar] [CrossRef]
- Schrader, J.; Schilling, M.; Holtmann, D.; Sell, D.; Filho, M.V.; Marx, A.; Vorholt, J.A. Methanol-based industrial biotechnology: Current status and future perspectives of methylotrophic bacteria. Trends Biotechnol. 2009, 27, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, D.; Merkel, M.; Lilge, L.; Henkel, M.; Hausmann, R. From acetate to bio-based products: Underexploited potential for industrial biotechnology. Trends Biotechnol. 2021, 39, 397–411. [Google Scholar] [CrossRef]
- Peter, S.C. Reduction of CO2 to Chemicals and Fuels: A Solution to Global Warming and Energy Crisis. ACS Energy Lett. 2018, 3, 1557–1561. [Google Scholar] [CrossRef]
- Gomez Vidales, A.; Bruant, G.; Omanovic, S.; Tartakovsky, B. Carbon dioxide conversion to C1-C2 compounds in a microbial electrosynthesis cell with in situ electrodeposition of nickel and iron. Electrochim. Acta 2021, 383, 138349. [Google Scholar] [CrossRef]
- Bae, J.; Jin, S.; Kang, S.; Cho, B.-K.; Oh, M.-K. Recent progress in the engineering of C1-utilizing microbes. Curr. Opin. Biotechnol. 2022, 78, 102836. [Google Scholar] [CrossRef]
- Iguchi, H.; Yurimoto, H.; Sakai, Y. Interactions of Methylotrophs with Plants and Other Heterotrophic Bacteria. Microorganisms 2015, 3, 137–151. [Google Scholar] [CrossRef]
- Chou, A.; Lee, S.H.; Zhu, F.; Clomburg, J.M.; Gonzalez, R. An orthogonal metabolic framework for one-carbon utilization. Nat. Metab. 2021, 3, 1385–1399. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Liao, J.C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds. Nat. Commun. 2018, 9, 3992. [Google Scholar] [CrossRef]
- Chen, F.Y.-H.; Jung, H.-W.; Tsuei, C.-Y.; Liao, J.C. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell 2020, 182, 933–946. e914. [Google Scholar] [CrossRef]
- Antonovsky, N.; Gleizer, S.; Noor, E.; Zohar, Y.; Herz, E.; Barenholz, U.; Zelcbuch, L.; Amram, S.; Wides, A.; Tepper, N.; et al. Sugar Synthesis from CO2 in Escherichia coli. Cell 2016, 166, 115–125. [Google Scholar] [CrossRef]
- Xia, P.-F.; Zhang, G.-C.; Walker, B.; Seo, S.-O.; Kwak, S.; Liu, J.-J.; Kim, H.; Ort, D.R.; Wang, S.-G.; Jin, Y.-S. Recycling Carbon Dioxide during Xylose Fermentation by Engineered Saccharomyces cerevisiae. ACS Synth. Biol. 2017, 6, 276–283. [Google Scholar] [CrossRef]
- Xi, Y.-l.; Chen, K.-q.; Li, J.; Fang, X.-j.; Zheng, X.-y.; Sui, S.-S.; Jiang, M.; Wei, P. Optimization of culture conditions in CO2 fixation for succinic acid production using Actinobacillus succinogenes. J. Ind. Microbiol. Biotechnol. 2011, 38, 1605. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Q.; Cheng, C.-L.; Nagarajan, D.; Chang, J.-S.; Hu, J.; Lee, D.-J. Carbon capture and utilization of fermentation CO2: Integrated ethanol fermentation and succinic acid production as an efficient platform. Appl. Energy 2017, 206, 364–371. [Google Scholar] [CrossRef]
- Salehizadeh, H.; Yan, N.; Farnood, R. Recent advances in microbial CO2 fixation and conversion to value-added products. Chem. Eng. J. 2020, 390, 124584. [Google Scholar] [CrossRef]
- Morales, M.; Sánchez, L.; Revah, S. The impact of environmental factors on carbon dioxide fixation by microalgae. FEMS Microbiol. Lett. 2018, 365, fnx262. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Abraham, J.; Jordan, N.; Lindsay, M.; Chauhan, N. Synthetic, Photosynthetic, and Chemical Strategies to Enhance Carbon Dioxide Fixation. C 2022, 8, 18. [Google Scholar] [CrossRef]
- Schwander, T.; Schada von Borzyskowski, L.; Burgener, S.; Cortina, N.S.; Erb, T.J. A synthetic pathway for the fixation of carbon dioxide in vitro. Science 2016, 354, 900–904. [Google Scholar] [CrossRef]
- Santos Correa, S.; Schultz, J.; Lauersen, K.J.; Soares Rosado, A. Natural carbon fixation and advances in synthetic engineering for redesigning and creating new fixation pathways. J. Adv. Res. 2023, 47, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.R.; Ahn, Y.-J.; Lee, S.Y. Bacterial conversion of CO2 to organic compounds. J. CO2 Util. 2022, 58, 101929. [Google Scholar] [CrossRef]
- Erb, T.J.; Zarzycki, J. A short history of Rubisco: The rise and fall (?) of Nature’s predominant CO2 fixing enzyme. Curr. Opin. Biotechnol. 2018, 49, 100–107. [Google Scholar] [CrossRef]
- Flamholz, A.I.; Prywes, N.; Moran, U.; Davidi, D.; Bar-On, Y.M.; Oltrogge, L.M.; Alves, R.; Savage, D.; Milo, R. Revisiting Trade-offs between Rubisco Kinetic Parameters. Biochemistry 2019, 58, 3365–3376. [Google Scholar] [CrossRef]
- Li, Z.; Xin, X.; Xiong, B.; Zhao, D.; Zhang, X.; Bi, C. Engineering the Calvin–Benson–Bassham cycle and hydrogen utilization pathway of Ralstonia eutropha for improved autotrophic growth and polyhydroxybutyrate production. Microb. Cell Factories 2020, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Davidi, D.; Shamshoum, M.; Guo, Z.; Bar-On, Y.M.; Prywes, N.; Oz, A.; Jablonska, J.; Flamholz, A.; Wernick, D.G.; Antonovsky, N.; et al. Highly active rubiscos discovered by systematic interrogation of natural sequence diversity. EMBO J. 2020, 39, e104081. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Park, B.-R.; Jeong, D.; Park, J.; Ko, J.K.; Kim, S.-J.; Kim, J.-S.; Jin, Y.-S.; Kim, S.R. Functional expression of RuBisCO reduces CO2 emission during fermentation by engineered Saccharomyces cerevisiae. Process Biochem. 2023, 134, 286–293. [Google Scholar] [CrossRef]
- Martin, W.F. Hydrogen, metals, bifurcating electrons, and proton gradients: The early evolution of biological energy conservation. FEBS Lett. 2012, 586, 485–493. [Google Scholar] [CrossRef]
- Pu, X.; Han, Y. Promotion of Carbon Dioxide Biofixation through Metabolic and Enzyme Engineering. Catalysts 2022, 12, 399. [Google Scholar] [CrossRef]
- Jang, Y.-S.; Kim, W.J.; Im, J.A.; Palaniswamy, S.; Yao, Z.; Lee, H.L.; Yoon, Y.R.; Seong, H.J.; Papoutsakis, E.T.; Lee, S.Y. Efforts to install a heterologous Wood-Ljungdahl pathway in Clostridium acetobutylicum enable the identification of the native tetrahydrofolate (THF) cycle and result in early induction of solvents. Metab. Eng. 2023, 77, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Orsi, E.; Nikel, P.I.; Nielsen, L.K.; Donati, S. Synergistic investigation of natural and synthetic C1-trophic microorganisms to foster a circular carbon economy. Nat. Commun. 2023, 14, 6673. [Google Scholar] [CrossRef] [PubMed]
- Claassens, N.J.; Satanowski, A.; Bysani, V.R.; Dronsella, B.; Orsi, E.; Rainaldi, V.; Yilmaz, S.; Wenk, S.; Lindner, S.N. Engineering the reductive glycine pathway: A promising synthetic metabolism approach for c1-assimilation. In One-Carbon Feedstocks for Sustainable Bioproduction; Springer: Cham, Switzerland, 2022; pp. 299–350. [Google Scholar]
- Gonzales, J.N.; Matson, M.M.; Atsumi, S. Nonphotosynthetic Biological CO2 Reduction. Biochemistry 2019, 58, 1470–1477. [Google Scholar] [CrossRef]
- Song, Y.; Lee, J.S.; Shin, J.; Lee, G.M.; Jin, S.; Kang, S.; Lee, J.-K.; Kim, D.R.; Lee, E.Y.; Kim, S.C. Functional cooperation of the glycine synthase-reductase and Wood–Ljungdahl pathways for autotrophic growth of Clostridium drakei. Proc. Natl. Acad. Sci. USA 2020, 117, 7516–7523. [Google Scholar] [CrossRef]
- Bruinsma, L.; Wenk, S.; Claassens, N.J.; Martins dos Santos, V.A.P. Paving the way for synthetic C1-Metabolism in Pseudomonas putida through the reductive glycine pathway. Metab. Eng. 2023, 76, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Giraldo, N.; Rainaldi, V.; Machens, F.; Collas, F.; Kubis, A.; Kensy, F.; Bar-Even, A.; Lindner, S.N. Optimizing E. coli as a formatotrophic platform for bioproduction via the reductive glycine pathway. Front. Bioeng. Biotechnol. 2023, 11, 1091899. [Google Scholar] [CrossRef]
- Steffens, L.; Pettinato, E.; Steiner, T.M.; Eisenreich, W.; Berg, I.A. Tracking the Reversed Oxidative Tricarboxylic Acid Cycle in Bacteria. Bio Protoc 2022, 12, e4364. [Google Scholar] [CrossRef]
- Chen, C.-H.; Tseng, I.-T.; Lo, S.-C.; Yu, Z.-R.; Pang, J.-J.; Chen, Y.-H.; Huang, C.-C.; Li, S.-Y. Manipulating ATP Supply Improve in situ CO2 recycling by reductive TCA cycle in engineered Escherichia coli. 3 Biotech 2020, 10, 125. [Google Scholar] [CrossRef]
- Fuchs, G. Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu. Rev. Microbiol. 2011, 65, 631–658. [Google Scholar] [CrossRef]
- Erb Tobias, J. Carboxylases in Natural and Synthetic Microbial Pathways. Appl. Environ. Microbiol. 2011, 77, 8466–8477. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, M.; Chen, Y.; Wang, Z.; Nielsen, J.; Liu, Z. Engineering yeast mitochondrial metabolism for 3-hydroxypropionate production. Biotechnol. Biofuels Bioprod. 2023, 16, 64. [Google Scholar] [CrossRef]
- Min, Z.; Zhang, X.; Wu, W.; Xin, Y.; Liu, M.; Wang, K.; Zhang, X.; He, Y.; Fan, C.; Wang, Z.; et al. Crystal Structure of an Intramolecular Mesaconyl-coenzyme A Transferase From the 3-Hydroxypropionic Acid Cycle of Roseiflexus castenholzii. Front. Microbiol. 2022, 13, 923367. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, K.; Chen, Y.; Tan, T.; Nielsen, J. Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat. Catal. 2020, 3, 274–288. [Google Scholar] [CrossRef]
- Liu, L.; Schubert, D.M.; Könneke, M.; Berg, I.A. (S)-3-Hydroxybutyryl-COA Dehydrogenase From the Autotrophic 3-Hydroxypropionate/4-Hydroxybutyrate Cycle in Nitrosopumilus maritimus. Front. Microbiol. 2021, 12, 712030. [Google Scholar] [CrossRef]
- McLean, R.; Schwander, T.; Diehl, C.; Cortina, N.S.; Paczia, N.; Zarzycki, J.; Erb, T.J. Exploring alternative pathways for the in vitro establishment of the HOPAC cycle for synthetic CO2 fixation. Sci. Adv. 2023, 9, eadh4299. [Google Scholar] [CrossRef]
- Satanowski, A.; Dronsella, B.; Noor, E.; Vögeli, B.; He, H.; Wichmann, P.; Erb, T.J.; Lindner, S.N.; Bar-Even, A. Awakening a latent carbon fixation cycle in Escherichia coli. Nat. Commun. 2020, 11, 5812. [Google Scholar] [CrossRef]
- Cai, T.; Sun, H.; Qiao, J.; Zhu, L.; Zhang, F.; Zhang, J.; Tang, Z.; Wei, X.; Yang, J.; Yuan, Q. Cell-free chemoenzymatic starch synthesis from carbon dioxide. Science 2021, 373, 1523–1527. [Google Scholar] [CrossRef]
- Xiao, L.; Liu, G.; Gong, F.; Zhu, H.; Zhang, Y.; Cai, Z.; Li, Y. A minimized synthetic carbon fixation cycle. ACS Catal. 2021, 12, 799–808. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, H.; Xu, Y.; Meng, H.; Zeng, A.-P. Turn air-captured CO2 with methanol into amino acid and pyruvate in an ATP/NAD (P) H-free chemoenzymatic system. Nat. Commun. 2023, 14, 2772. [Google Scholar] [CrossRef]
- Bar-Even, A.; Noor, E.; Lewis, N.E.; Milo, R. Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 8889–8894. [Google Scholar] [CrossRef]
- Liu, Z.; Shi, S.; Ji, Y.; Wang, K.; Tan, T.; Nielsen, J. Opportunities of CO2-based biorefineries for production of fuels and chemicals. Green Carbon 2023, 1, 75–84. [Google Scholar] [CrossRef]
- Humphreys, C.M.; Minton, N.P. Advances in metabolic engineering in the microbial production of fuels and chemicals from C1 gas. Curr. Opin. Biotechnol. 2018, 50, 174–181. [Google Scholar] [CrossRef]
- Kato, Y.; Inabe, K.; Hidese, R.; Kondo, A.; Hasunuma, T. Metabolomics-based engineering for biofuel and bio-based chemical production in microalgae and cyanobacteria: A review. Bioresour. Technol. 2022, 344, 126196. [Google Scholar] [CrossRef]
- Farrokh, P.; Sheikhpour, M.; Kasaeian, A.; Asadi, H.; Bavandi, R. Cyanobacteria as an eco-friendly resource for biofuel production: A critical review. Biotechnol. Prog. 2019, 35, e2835. [Google Scholar] [CrossRef]
- Ramey, C.J.; Barón-Sola, A.n.; Aucoin, H.R.; Boyle, N.R. Genome engineering in cyanobacteria: Where we are and where we need to go. ACS Synth. Biol. 2015, 4, 1186–1196. [Google Scholar] [CrossRef]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Factories 2018, 17, 1–21. [Google Scholar] [CrossRef]
- Gimpel, J.A.; Henríquez, V.; Mayfield, S.P. In metabolic engineering of eukaryotic microalgae: Potential and challenges come with great diversity. Front. Microbiol. 2015, 6, 1376. [Google Scholar] [CrossRef]
- Liu, C.; Colón, B.C.; Ziesack, M.; Silver, P.A.; Nocera, D.G. Water splitting–biosynthetic system with CO2 reduction efficiencies exceeding photosynthesis. Science 2016, 352, 1210–1213. [Google Scholar] [CrossRef]
- Li, H.; Opgenorth, P.H.; Wernick, D.G.; Rogers, S.; Wu, T.-Y.; Higashide, W.; Malati, P.; Huo, Y.-X.; Cho, K.M.; Liao, J.C. Integrated electromicrobial conversion of CO2 to higher alcohols. Science 2012, 335, 1596. [Google Scholar] [CrossRef]
- Nybo, S.E.; Khan, N.E.; Woolston, B.M.; Curtis, W.R. Metabolic engineering in chemolithoautotrophic hosts for the production of fuels and chemicals. Metab. Eng. 2015, 30, 105–120. [Google Scholar] [CrossRef]
- Lu, J.; Brigham, C.J.; Gai, C.S.; Sinskey, A.J. Studies on the production of branched-chain alcohols in engineered Ralstonia eutropha. Appl. Microbiol. Biotechnol. 2012, 96, 283–297. [Google Scholar] [CrossRef]
- Bi, C.; Su, P.; Müller, J.; Yeh, Y.-C.; Chhabra, S.R.; Beller, H.R.; Singer, S.W.; Hillson, N.J. Development of a broad-host synthetic biology toolbox for Ralstonia eutropha and its application to engineering hydrocarbon biofuel production. Microb. Cell Factories 2013, 12, 1–10. [Google Scholar] [CrossRef]
- Kernan, T.; Majumdar, S.; Li, X.; Guan, J.; West, A.C.; Banta, S. Engineering the iron-oxidizing chemolithoautotroph Acidithiobacillus ferrooxidans for biochemical production. Biotechnol. Bioeng. 2016, 113, 189–197. [Google Scholar] [CrossRef]
- Lee, H.; Bae, J.; Jin, S.; Kang, S.; Cho, B.-K. Engineering Acetogenic Bacteria for Efficient One-Carbon Utilization. Front. Microbiol. 2022, 13, 865168. [Google Scholar] [CrossRef]
- Liew, F.E.; Nogle, R.; Abdalla, T.; Rasor, B.J.; Canter, C.; Jensen, R.O.; Wang, L.; Strutz, J.; Chirania, P.; De Tissera, S. Carbon-negative production of acetone and isopropanol by gas fermentation at industrial pilot scale. Nat. Biotechnol. 2022, 40, 335–344. [Google Scholar] [CrossRef]
- Yoon, J.; Oh, M.-K. Strategies for Biosynthesis of C1 Gas-derived Polyhydroxyalkanoates: A review. Bioresour. Technol. 2022, 344, 126307. [Google Scholar] [CrossRef] [PubMed]
- Gleizer, S.; Ben-Nissan, R.; Bar-On, Y.M.; Antonovsky, N.; Noor, E.; Zohar, Y.; Jona, G.; Krieger, E.; Shamshoum, M.; Bar-Even, A. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell 2019, 179, 1255–1263. e1212. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Kim, S.-J.; Kim, S.-K.; Seo, S.-O.; Park, S.; Shin, J.; Kim, J.-S.; Park, B.-R.; Jin, Y.-S.; Chang, P.-S.; et al. Yeast metabolic engineering for carbon dioxide fixation and its application. Bioresour. Technol. 2021, 346, 126349. [Google Scholar]
- Liu, C.; Gallagher, J.J.; Sakimoto, K.K.; Nichols, E.M.; Chang, C.J.; Chang, M.C.; Yang, P. Nanowire–bacteria hybrids for unassisted solar carbon dioxide fixation to value-added chemicals. Nano Lett. 2015, 15, 3634–3639. [Google Scholar] [CrossRef]
- Hu, P.; Chakraborty, S.; Kumar, A.; Woolston, B.; Liu, H.; Emerson, D.; Stephanopoulos, G. Integrated bioprocess for conversion of gaseous substrates to liquids. Proc. Natl. Acad. Sci. USA 2016, 113, 3773–3778. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liu, L.; Chen, X. Bacterial photosynthesis: State-of-the-art in light-driven carbon fixation in engineered bacteria. Curr. Opin. Microbiol. 2022, 69, 102174. [Google Scholar] [CrossRef]
- Johnson, M.P. Photosynthesis. Essays Biochem. 2016, 60, 255–273. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, F.; Meng, H.; Zhang, Y.; Li, Y. Introducing extra NADPH consumption ability significantly increases the photosynthetic efficiency and biomass production of cyanobacteria. Metab. Eng. 2016, 38, 217–227. [Google Scholar] [CrossRef]
- Nürnberg, D.J.; Morton, J.; Santabarbara, S.; Telfer, A.; Joliot, P.; Antonaru, L.A.; Ruban, A.V.; Cardona, T.; Krausz, E.; Boussac, A. Photochemistry beyond the red limit in chlorophyll f–containing photosystems. Science 2018, 360, 1210–1213. [Google Scholar] [CrossRef]
- Work, V.H.; D’Adamo, S.; Radakovits, R.; Jinkerson, R.E.; Posewitz, M.C. Improving photosynthesis and metabolic networks for the competitive production of phototroph-derived biofuels. Curr. Opin. Biotechnol. 2012, 23, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Sakimoto, K.K.; Wong, A.B.; Yang, P. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production. Science 2016, 351, 74–77. [Google Scholar] [CrossRef]
- Teng, Y.; Xu, Y.; Wang, X.; Christie, P. Function of biohydrogen metabolism and related microbial communities in environmental bioremediation. Front. Microbiol. 2019, 10, 106. [Google Scholar] [CrossRef]
- Lubitz, W.; Ogata, H.; Rudiger, O.; Reijerse, E. Hydrogenases. Chem. Rev. 2014, 114, 4081–4148. [Google Scholar] [CrossRef]
- Sargent, F. The model [NiFe]-hydrogenases of Escherichia coli. Adv. Microb. Physiol. 2016, 68, 433–507. [Google Scholar]
- Lamont, C.M.; Sargent, F. Design and characterisation of synthetic operons for biohydrogen technology. Arch. Microbiol. 2017, 199, 495–503. [Google Scholar] [CrossRef] [PubMed]
- White, G.F.; Edwards, M.J.; Gomez-Perez, L.; Richardson, D.J.; Butt, J.N.; Clarke, T.A. Mechanisms of bacterial extracellular electron exchange. Adv. Microb. Physiol. 2016, 68, 87–138. [Google Scholar] [PubMed]
- Van Kessel, M.A.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.; Kartal, B.; Jetten, M.S.; Lücker, S. Complete nitrification by a single microorganism. Nature 2015, 528, 555–559. [Google Scholar] [CrossRef]
- Kuypers, M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Wang, R.; Lin, J.-Q.; Liu, X.-M.; Pang, X.; Zhang, C.-J.; Yang, C.-L.; Gao, X.-Y.; Lin, C.-M.; Li, Y.-Q.; Li, Y. Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 2019, 9, 3290. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, I.; Coates, J. Microbial phosphite oxidation and its potential role in the global phosphorus and carbon cycles. Adv. Appl. Microbiol. 2017, 98, 93–117. [Google Scholar]
- Butler, C.S.; Lovley, D.R. How to sustainably feed a microbe: Strategies for biological production of carbon-based commodities with renewable electricity. Front. Microbiol. 2016, 7, 1879. [Google Scholar] [CrossRef]
- Chen, H.; Simoska, O.; Lim, K.; Grattieri, M.; Yuan, M.; Dong, F.; Lee, Y.S.; Beaver, K.; Weliwatte, S.; Gaffney, E.M. Fundamentals, applications, and future directions of bioelectrocatalysis. Chem. Rev. 2020, 120, 12903–12993. [Google Scholar] [CrossRef]
- Bajracharya, S.; Srikanth, S.; Mohanakrishna, G.; Zacharia, R.; Strik, D.P.; Pant, D. Biotransformation of carbon dioxide in bioelectrochemical systems: State of the art and future prospects. J. Power Sources 2017, 356, 256–273. [Google Scholar] [CrossRef]
- Liu, Z.; Xue, X.; Cai, W.; Cui, K.; Patil, S.A.; Guo, K. Recent progress on microbial electrosynthesis reactor designs and strategies to enhance the reactor performance. Biochem. Eng. J. 2023, 190, 108745. [Google Scholar] [CrossRef]
- Bhagchandanii, D.D.; Babu, R.P.; Sonawane, J.M.; Khanna, N.; Pandit, S.; Jadhav, D.A.; Khilari, S.; Prasad, R. A comprehensive understanding of electro-fermentation. Fermentation 2020, 6, 92. [Google Scholar] [CrossRef]
- Zhen, G.; Lu, X.; Kumar, G.; Bakonyi, P.; Xu, K.; Zhao, Y. Microbial electrolysis cell platform for simultaneous waste biorefinery and clean electrofuels generation: Current situation, challenges and future perspectives. Prog. Energy Combust. Sci. 2017, 63, 119–145. [Google Scholar] [CrossRef]
- Das, S.; Ghangrekar, M. Value added product recovery and carbon dioxide sequestration from biogas using microbial electrosynthesis. Indian J. Exp. Biol. 2018, 56, 484–492. [Google Scholar]
- Gong, Y.; Ebrahim, A.; Feist, A.M.; Embree, M.; Zhang, T.; Lovley, D.; Zengler, K. Sulfide-driven microbial electrosynthesis. Environ. Sci. Technol. 2013, 47, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Leang, C.; Ueki, T.; Nevin, K.P.; Lovley, D.R. A genetic system for Clostridium ljungdahlii: A chassis for autotrophic production of biocommodities and a model homoacetogen. Appl. Environ. Microbiol. 2013, 79, 1102–1109. [Google Scholar] [CrossRef]
- Mohanakrishna, G.; Vanbroekhoven, K.; Pant, D. Impact of dissolved carbon dioxide concentration on the process parameters during its conversion to acetate through microbial electrosynthesis. React. Chem. Eng. 2018, 3, 371–378. [Google Scholar] [CrossRef]
- Liu, H.; Song, T.; Fei, K.; Wang, H.; Xie, J. Microbial electrosynthesis of organic chemicals from CO2 by Clostridium scatologenes ATCC 25775 T. Bioresour. Bioprocess. 2018, 5, 1–10. [Google Scholar] [CrossRef]
- Bajracharya, S.; Yuliasni, R.; Vanbroekhoven, K.; Buisman, C.J.; Strik, D.P.; Pant, D. Long-term operation of microbial electrosynthesis cell reducing CO2 to multi-carbon chemicals with a mixed culture avoiding methanogenesis. Bioelectrochemistry 2017, 113, 26–34. [Google Scholar] [CrossRef]
- Gildemyn, S.; Verbeeck, K.; Slabbinck, R.; Andersen, S.J.; Prévoteau, A.; Rabaey, K. Integrated Production, Extraction, and concentration of Acetic Acid from CO2 through Microbial Electrosynthesis. Environ. Sci. Technol. Lett. 2015, 2, 325–328. [Google Scholar] [CrossRef]
- Sciarria, T.P.; Batlle-Vilanova, P.; Colombo, B.; Scaglia, B.; Balaguer, M.D.; Colprim, J.; Puig, S.; Adani, F. Bio-electrorecycling of carbon dioxide into bioplastics. Green Chem. 2018, 20, 4058–4066. [Google Scholar] [CrossRef]
- Jourdin, L.; Raes, S.M.; Buisman, C.J.; Strik, D.P. Critical biofilm growth throughout unmodified carbon felts allows continuous bioelectrochemical chain elongation from CO2 up to caproate at high current density. Front. Energy Res. 2018, 6, 7. [Google Scholar] [CrossRef]
- Batlle-Vilanova, P.; Ganigue, R.; Ramió-Pujol, S.; Baneras, L.; Jiménez, G.; Hidalgo, M.; Balaguer, M.D.; Colprim, J.; Puig, S. Microbial electrosynthesis of butyrate from carbon dioxide: Production and extraction. Bioelectrochemistry 2017, 117, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Chu, N.; Hao, W.; Wu, Q.; Liang, Q.; Jiang, Y.; Liang, P.; Ren, Z.J.; Zeng, R.J. Microbial Electrosynthesis for Producing Medium Chain Fatty Acids. Engineering 2022, 16, 141–153. [Google Scholar] [CrossRef]
- Yu, L.; Yuan, Y.; Tang, J.; Zhou, S. Thermophilic Moorella thermoautotrophica-immobilized cathode enhanced microbial electrosynthesis of acetate and formate from CO2. Bioelectrochemistry 2017, 117, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Agostino, V.; Rosenbaum, M.A. Sulfate-reducing electroautotrophs and their applications in bioelectrochemical systems. Front. Energy Res. 2018, 6, 55. [Google Scholar] [CrossRef]
- Deutzmann, J.S.; Spormann, A.M. Enhanced microbial electrosynthesis by using defined co-cultures. ISME J. 2017, 11, 704–714. [Google Scholar] [CrossRef]
- Jiang, Y.; Chu, N.; Zhang, W.; Ma, J.; Zhang, F.; Liang, P.; Zeng, R.J. Zinc: A promising material for electrocatalyst-assisted microbial electrosynthesis of carboxylic acids from carbon dioxide. Water Res. 2019, 159, 87–94. [Google Scholar] [CrossRef]
- Lee, S.Y.; Oh, Y.-K.; Lee, S.; Fitriana, H.N.; Moon, M.; Kim, M.-S.; Lee, J.; Min, K.; Park, G.W.; Lee, J.-P.; et al. Recent developments and key barriers to microbial CO2 electrobiorefinery. Bioresour. Technol. 2021, 320, 124350. [Google Scholar] [CrossRef]
- Karthikeyan, R.; Singh, R.; Bose, A. Microbial electron uptake in microbial electrosynthesis: A mini-review. J. Ind. Microbiol. Biotechnol. 2019, 46, 1419–1426. [Google Scholar] [CrossRef]
- Liu, N.; Santala, S.; Stephanopoulos, G. Mixed carbon substrates: A necessary nuisance or a missed opportunity. Curr. Opin. Biotechnol. 2020, 62, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Costentin, C.; Robert, M.; Savéant, J.-M. Catalysis of the electrochemical reduction of carbon dioxide. Chem. Soc. Rev. 2013, 42, 2423–2436. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.C.; Yuan, Z.; Virdis, B. Towards carbon neutral chemicals production: Opportunities for combining fermentation with electrochemical processes. Curr. Opin. Electrochem. 2023, 37, 101177. [Google Scholar] [CrossRef]
- Cotton, C.A.R.; Claassens, N.J.; Benito-Vaquerizo, S.; Bar-Even, A. Renewable methanol and formate as microbial feedstocks. Curr. Opin. Biotechnol. 2020, 62, 168–180. [Google Scholar] [CrossRef]
- Mustafa, A.; Lougou, B.G.; Shuai, Y.; Wang, Z.; Tan, H. Current technology development for CO2 utilization into solar fuels and chemicals: A review. J. Energy Chem. 2020, 49, 96–123. [Google Scholar] [CrossRef]
- Rayne, S. Thermal Carbon Dioxide Splitting: A Summary of the Peer-Reviewed Scientific Literature. Nat. Preced. 2008. [Google Scholar] [CrossRef]
- Kamkeng, A.D.N.; Wang, M.; Hu, J.; Du, W.; Qian, F. Transformation technologies for CO2 utilisation: Current status, challenges and future prospects. Chem. Eng. J. 2021, 409, 128138. [Google Scholar] [CrossRef]
- Bae, J.; Song, Y.; Lee, H.; Shin, J.; Jin, S.; Kang, S.; Cho, B.-K. Valorization of C1 gases to value-added chemicals using acetogenic biocatalysts. Chem. Eng. J. 2022, 428, 131325. [Google Scholar] [CrossRef]
- Liew, F.; Martin, M.E.; Tappel, R.C.; Heijstra, B.D.; Mihalcea, C.; Köpke, M. Gas Fermentation—A Flexible Platform for commercial Scale Production of Low-Carbon-Fuels and Chemicals from Waste and Renewable Feedstocks. Front. Microbiol. 2016, 7, 694. [Google Scholar] [CrossRef]
- Herranz, J.; Pătru, A.; Fabbri, E.; Schmidt, T.J. Co-electrolysis of CO2 and H2O: From electrode reactions to cell-level development. Curr. Opin. Electrochem. 2020, 23, 89–95. [Google Scholar] [CrossRef]
- King, G.M.; Weber, C.F. Distribution, diversity and ecology of aerobic co-oxidizing bacteria. Nat. Rev. Microbiol. 2007, 5, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Can, M.; Armstrong, F.A.; Ragsdale, S.W. Structure, Function, and Mechanism of the Nickel Metalloenzymes, Co Dehydrogenase, and Acetyl-COA Synthase. Chem. Rev. 2014, 114, 4149–4174. [Google Scholar] [CrossRef] [PubMed]
- Oelgeschlager, E.; Rother, M. Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch Microbiol 2008, 190, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Takors, R.; Kopf, M.; Mampel, J.; Bluemke, W.; Blombach, B.; Eikmanns, B.; Bengelsdorf, F.R.; Weuster-Botz, D.; Dürre, P. Using gas mixtures of CO, CO2 and H2 as microbial substrates: The do’s and don’ts of successful technology transfer from laboratory to production scale. Microb. Biotechnol. 2018, 11, 606–625. [Google Scholar] [CrossRef]
- Grenz, S.; Baumann, P.T.; Rückert, C.; Nebel, B.A.; Siebert, D.; Schwentner, A.; Eikmanns, B.J.; Hauer, B.; Kalinowski, J.; Takors, R.; et al. Exploiting Hydrogenophaga pseudoflava for aerobic syngas-based production of chemicals. Metab. Eng. 2019, 55, 220–230. [Google Scholar] [CrossRef]
- Diender, M.; Stams, A.J.; Sousa, D.Z. Pathways and Bioenergetics of Anaerobic Carbon Monoxide Fermentation. Front. Microbiol. 2015, 6, 1275. [Google Scholar] [CrossRef]
- Roberts, D.L.; James-Hagstrom, J.E.; Garvin, D.K.; Gorst, C.M.; Runquist, J.A.; Baur, J.R.; Haase, F.C.; Ragsdale, S.W. Cloning and expression of the gene cluster encoding key proteins involved in acetyl-COA synthesis in Clostridium thermoaceticum: Co dehydrogenase, the corrinoid/Fe-S protein, and methyltransferase. Proc. Natl. Acad. Sci. USA 1989, 86, 32–36. [Google Scholar] [CrossRef]
- Fast Alan, G.; Papoutsakis Eleftherios, T. Functional Expression of the Clostridium ljungdahlii Acetyl-coenzyme A Synthase in Clostridium acetobutylicum as Demonstrated by a Novel In Vivo CO Exchange Activity En Route to Heterologous Installation of a Functional Wood-Ljungdahl Pathway. Appl. Environ. Microbiol. 2018, 84, e02307–e02317. [Google Scholar] [CrossRef]
- Kang, H.; Park, B.; Oh, S.; Pathiraja, D.; Kim, J.-Y.; Jung, S.; Jeong, J.; Cha, M.; Park, Z.-Y.; Choi, I.-G.; et al. Metabolism perturbation Causedby the overexpression of carbon monoxide dehydrogenase/Acetyl-COA synthase gene complex accelerated gas to acetate conversion rate of Eubacterium limosumKIST612. Bioresour. Technol. 2021, 341, 125879. [Google Scholar] [CrossRef]
- Jin, S.; Kang, S.; Bae, J.; Lee, H.; Cho, B.-K. Development of CO gas conversion system using high CO tolerance biocatalyst. Chem. Eng. J. 2022, 449, 137678. [Google Scholar] [CrossRef]
- Karakurt, I.; Aydin, G.; Aydiner, K. Sources and mitigation of methane emissions by sectors: A critical review. Renew. Energy 2012, 39, 40–48. [Google Scholar] [CrossRef]
- Krupp, F. Reducing Methane Will Help Hit the Brakes on Runaway Global Warming. Available online: https://www.edf.org/blog/2022/05/16/reducing-methane-will-help-hit-brakes-runaway-global-warming (accessed on 23 November 2023).
- Semrau, J.D.; DiSpirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, K.K.; Goswami, G.; Das, D. Biotransformation of Methane and Carbon Dioxide Into High-Value Products by Methanotrophs: Current State of Art and Future Prospects. Front. Microbiol. 2021, 12, 636486. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-y.; Kim, C.-g. Application and development of methanotrophs in environmental engineering. J. Mater. Cycles Waste Manag. 2019, 21, 415–422. [Google Scholar] [CrossRef]
- Ge, X.; Yang, L.; Sheets, J.P.; Yu, Z.; Li, Y. Biological conversion of methane to liquid fuels: Status and opportunities. Biotechnol. Adv. 2014, 32, 1460–1475. [Google Scholar] [CrossRef]
- Boetius, A.; Ravenschlag, K.; Schubert, C.J.; Rickert, D.; Widdel, F.; Gieseke, A.; Amann, R.; Jørgensen, B.B.; Witte, U.; Pfannkuche, O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 2000, 407, 623–626. [Google Scholar] [CrossRef]
- Beal, E.J.; House, C.H.; Orphan, V.J. Manganese-and iron-dependent marine methane oxidation. Science 2009, 325, 184–187. [Google Scholar] [CrossRef]
- Haroon, M.F.; Hu, S.; Shi, Y.; Imelfort, M.; Keller, J.; Hugenholtz, P.; Yuan, Z.; Tyson, G.W. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 2013, 500, 567–570. [Google Scholar] [CrossRef]
- Ettwig, K.F.; Butler, M.K.; Le Paslier, D.; Pelletier, E.; Mangenot, S.; Kuypers, M.M.; Schreiber, F.; Dutilh, B.E.; Zedelius, J.; de Beer, D. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 2010, 464, 543–548. [Google Scholar] [CrossRef]
- Lee, O.K.; Hur, D.H.; Nguyen, D.T.N.; Lee, E.Y. Metabolic engineering of methanotrophs and its application to production of chemicals and biofuels from methane. Biofuels Bioprod. Biorefining 2016, 10, 848–863. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Nguyen, A.D.; Nguyen, T.T.; Nguyen, L.T.; Lee, O.K.; Lee, E.Y. Biological conversion of methane to chemicals and fuels: Technical challenges and issues. Appl. Microbiol. Biotechnol. 2018, 102, 3071–3080. [Google Scholar] [CrossRef] [PubMed]
- Dürre, P.; Eikmanns, B.J. C1-carbon sources for chemical and fuel production by microbial gas fermentation. Curr. Opin. Biotechnol. 2015, 35, 63–72. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Lee, E.Y. Engineered Methanotrophy: A Sustainable Solution for Methane-Based Industrial Biomanufacturing. Trends Biotechnol. 2021, 39, 381–396. [Google Scholar] [CrossRef] [PubMed]
- Zilly, F.E.; Acevedo, J.P.; Augustyniak, W.; Deege, A.; Häusig, U.W.; Reetz, M.T. Tuning a P450 Enzyme for Methane Oxidation. Angew. Chem. Int. Ed. 2011, 50, 2720–2724. [Google Scholar] [CrossRef] [PubMed]
- Meinhold, P.; Peters, M.W.; Chen, M.M.Y.; Takahashi, K.; Arnold, F.H. Direct conversion of Ethane to Ethanol by Engineered Cytochrome P450 BM3. ChemBioChem 2005, 6, 1765–1768. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Smith, S.M.; Rawat, S.; Yatsunyk, L.A.; Stemmler, T.L.; Rosenzweig, A.C. Oxidation of methane by a biological dicopper centre. Nature 2010, 465, 115–119. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, J.; Kwon, Y.W.; Park, D.; Yu, Y.; Jang, Y.E.; Lee, B.-R.; Jo, E.; Lee, E.J.; Heo, Y. Biological conversion of methane to methanol through genetic reassembly of native catalytic domains. Nat. Catal. 2019, 2, 342–353. [Google Scholar] [CrossRef]
- Bennett, R.K.; Nyaradzo, D.; Michael, D.; Stephanie, J.; Kelley, H.; Baolong, Z.; Noah, H.; Derek, G.; Elizabeth, C.; Eleftherios, T.P. Expression of soluble methane monooxygenase in Escherichia coli enables methane conversion. bioRxiv 2021, 2021.2008.2005.455234. [Google Scholar] [CrossRef]
- Raili, K.; Markku, H.; Pekka, O. Methanol-Managing greenhouse gas emissions in the production chain by optimizing the resource base. AIMS Energy 2018, 6, 1074–1102. [Google Scholar] [CrossRef]
- Du, X.-L.; Jiang, Z.; Su, D.S.; Wang, J.-Q. Research Progress on the Indirect Hydrogenation of Carbon Dioxide to Methanol. ChemSusChem 2016, 9, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, W.B.; Sandoval, N.R.; Bennett, R.K.; Fast, A.G.; Papoutsakis, E.T. Synthetic methylotrophy: Engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization. Curr. Opin. Biotechnol. 2015, 33, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Y.; Lan, E.I. Chemical Production from Methanol Using Natural and Synthetic Methylotrophs. Biotechnol. J. 2020, 15, 1900356. [Google Scholar] [CrossRef] [PubMed]
- Pfeifenschneider, J.; Brautaset, T.; Wendisch, V.F. Methanol as carbon substrate in the bio-economy: Metabolic engineering of aerobic methylotrophic bacteria for production of value-added chemicals. Biofuels Bioprod. Biorefining 2017, 11, 719–731. [Google Scholar] [CrossRef]
- Antoniewicz, M.R. Synthetic methylotrophy: Strategies to assimilate methanol for growth and chemicals production. Curr. Opin. Biotechnol. 2019, 59, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Yurimoto, H.; Sakai, Y. Methylotrophic yeasts: Current understanding of their C1-metabolism and its regulation by sensing methanol for survival on plant leaves. Curr. Issues Mol. Biol. 2019, 33, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Amaro, R.; Nicaud, J.M. Metabolic Engineering for Expanding the Substrate Range of Yarrowia lipolytica. Trends Biotechnol. 2016, 34, 798–809. [Google Scholar] [CrossRef]
- Lv, X.; Yu, W.; Zhang, C.; Ning, P.; Li, J.; Liu, Y.; Du, G.; Liu, L. C1-based biomanufacturing: Advances, challenges and perspectives. Bioresour. Technol. 2023, 367, 128259. [Google Scholar] [CrossRef]
- Zhan, C.; Li, X.; Yang, Y.; Nielsen, J.; Bai, Z.; Chen, Y. Strategies and challenges with the microbial conversion of methanol to high-value chemicals. Biotechnol. Bioeng. 2021, 118, 3655–3668. [Google Scholar] [CrossRef]
- Kremp, F.; Müller, V. Methanol and methyl group conversion in acetogenic bacteria: Biochemistry, physiology and application. FEMS Microbiol. Rev. 2021, 45, fuaa040. [Google Scholar] [CrossRef]
- Gregory, G.J.; Bennett, R.K.; Papoutsakis, E.T. Recent advances toward the bioconversion of methane and methanol in synthetic methylotrophs. Metab. Eng. 2022, 71, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Bae, J.; Shin, J.; Jin, S.; Kang, S.; Lee, H.; Cho, S.; Cho, B.-K. Systems Biology on Acetogenic Bacteria for Utilizing C1 Feedstocks. In One-Carbon Feedstocks for Sustainable Bioproduction; Springer: Berlin/Heidelberg, Germany, 2022; pp. 57–90. [Google Scholar]
- Bennett, R.K.; Steinberg, L.M.; Chen, W.; Papoutsakis, E.T. Engineering the bioconversion of methane and methanol to fuels and chemicals in native and synthetic methylotrophs. Curr. Opin. Biotechnol. 2018, 50, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qin, R.; Guo, Y.; Ma, C.; Wang, X.; Chen, K.; Ouyang, P. Engineering the native methylotrophs for the bioconversion of methanol to value-added chemicals: Current status and future perspectives. Green Chem. Eng. 2023, 4, 199–211. [Google Scholar] [CrossRef]
- Schultenkämper, K.; Gütle, D.D.; López, M.G.; Keller, L.B.; Zhang, L.; Einsle, O.; Jacquot, J.-P.; Wendisch, V.F. Interrogating the Role of the Two Distinct Fructose-Bisphosphate Aldolases of Bacillus methanolicus by Site-Directed Mutagenesis of Key Amino Acids and Gene Repression by CRISPR Interference. Front. Microbiol. 2021, 12, 669220. [Google Scholar] [CrossRef]
- Lim, C.K.; Villada, J.C.; Chalifour, A.; Duran, M.F.; Lu, H.; Lee, P.K. Designing and engineering Methylorubrum extorquens AM1 for itaconic acid production. Front. Microbiol. 2019, 10, 1027. [Google Scholar] [CrossRef]
- Cai, P.; Wu, X.; Deng, J.; Gao, L.; Shen, Y.; Yao, L.; Zhou, Y.J. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris. Proc. Natl. Acad. Sci. USA 2022, 119, e2201711119. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Li, Y.; Yu, W.; Zhou, Y.J. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol. Nat. Metab. 2022, 4, 932–943. [Google Scholar] [CrossRef]
- Gao, B.; Zhao, N.; Deng, J.; Gu, Y.; Jia, S.; Hou, Y.; Lv, X.; Liu, L. Constructing a methanol-dependent Bacillus subtilis by engineering the methanol metabolism. J. Biotechnol. 2022, 343, 128–137. [Google Scholar] [CrossRef]
- Zhu, T.; Zhao, T.; Bankefa, O.E.; Li, Y. Engineering unnatural methylotrophic cell factories for methanol-based biomanufacturing: Challenges and opportunities. Biotechnol. Adv. 2020, 39, 107467. [Google Scholar] [CrossRef]
- Sanford, P.A.; Woolston, B.M. Synthetic or natural? Metabolic engineering for assimilation and valorization of methanol. Curr. Opin. Biotechnol. 2022, 74, 171–179. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Park, J.Y.; Hwang, I.Y.; Hamilton, R.; Kalyuzhnaya, M.G.; Kim, D.; Lee, E.Y. Genome-scale evaluation of core one-carbon metabolism in gammaproteobacterial methanotrophs grown on methane and methanol. Metab. Eng. 2020, 57, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.E.; Meyer, F.; Litsanov, B.; Kiefer, P.; Potthoff, E.; Heux, S.; Quax, W.J.; Wendisch, V.F.; Brautaset, T.; Portais, J.-C. Engineering Escherichia coli for methanol conversion. Metab. Eng. 2015, 28, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Price, J.V.; Chen, L.; Whitaker, W.B.; Papoutsakis, E.; Chen, W. Scaffoldless engineered enzyme assembly for enhanced methanol utilization. Proc. Natl. Acad. Sci. USA 2016, 113, 12691–12696. [Google Scholar] [CrossRef] [PubMed]
- Woolston, B.M.; King, J.R.; Reiter, M.; Van Hove, B.; Stephanopoulos, G. Improving formaldehyde consumption drives methanol assimilation in engineered E. coli. Nat. Commun. 2018, 9, 2387. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Keller, P.; Hartl, J.; Gröninger, O.G.; Kiefer, P.; Vorholt, J.A. Methanol-essential growth of Escherichia coli. Nat. Commun. 2018, 9, 1508. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, M.I.; Gonzalez-Garcia, R.A.; Valgepea, K.; Plan, M.R.; Scott, C.; Pretorius, I.S.; Marcellin, E.; Paulsen, I.T.; Williams, T.C. Adaptive laboratory evolution of native methanol assimilation in Saccharomyces cerevisiae. Nat. Commun. 2020, 11, 5564. [Google Scholar] [CrossRef]
- Wang, G.; Olofsson-Dolk, M.; Hansson, F.G.; Donati, S.; Li, X.; Chang, H.; Cheng, J.; Dahlin, J.; Borodina, I. Engineering Yeast Yarrowia lipolytica for Methanol Assimilation. ACS Synth. Biol. 2021, 10, 3537–3550. [Google Scholar] [CrossRef]
- Jiang, W.; Hernandez Villamor, D.; Peng, H.; Chen, J.; Liu, L.; Haritos, V.; Ledesma-Amaro, R. Metabolic engineering strategies to enable microbial utilization of C1 feedstocks. Nat. Chem. Biol. 2021, 17, 845–855. [Google Scholar] [CrossRef]
- Yishai, O.; Lindner, S.N.; Gonzalez de la Cruz, J.; Tenenboim, H.; Bar-Even, A. The formate bio-economy. Curr. Opin. Chem. Biol. 2016, 35, 1–9. [Google Scholar] [CrossRef]
- Bar-Even, A.; Noor, E.; Flamholz, A.; Milo, R. Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes. Biochim. Biophys. Acta (BBA)-Bioenerg. 2013, 1827, 1039–1047. [Google Scholar] [CrossRef]
- Bar-Even, A.; Flamholz, A.; Noor, E.; Milo, R. Thermodynamic constraints shape the structure of carbon fixation pathways. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2012, 1817, 1646–1659. [Google Scholar] [CrossRef] [PubMed]
- Grunwald, S.; Mottet, A.; Grousseau, E.; Plassmeier, J.K.; Popović, M.K.; Uribelarrea, J.-L.; Gorret, N.; Guillouet, S.E.; Sinskey, A. Kinetic and stoichiometric characterization of organoautotrophic growth of Ralstonia eutropha on formic acid in fed-batch and continuous cultures. Microb. Biotechnol. 2015, 8, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Xu, S.; Liu, Z.; Fu, X.; Zhao, H.; Shi, S. Challenges and opportunities in C1-based biomanufacturing. Bioresour. Technol. 2022, 364, 128095. [Google Scholar] [CrossRef]
- Claassens, N.J.; He, H.; Bar-Even, A. Synthetic Methanol and Formate Assimilation Via Modular Engineering and Selection Strategies. Curr. Issues Mol. Biol. 2019, 33, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Bar-Even, A. Formate Assimilation: The Metabolic Architecture of Natural and Synthetic Pathways. Biochemistry 2016, 55, 3851–3863. [Google Scholar] [CrossRef]
- Claassens, N.J. Reductive Glycine Pathway: A Versatile Route for One-Carbon Biotech. Trends Biotechnol. 2021, 39, 327–329. [Google Scholar] [CrossRef]
- Claassens, N.J.; Bordanaba-Florit, G.; Cotton, C.A.; De Maria, A.; Finger-Bou, M.; Friedeheim, L.; Giner-Laguarda, N.; Munar-Palmer, M.; Newell, W.; Scarinci, G. Replacing the Calvin cycle with the reductive glycine pathway in Cupriavidus necator. Metab. Eng. 2020, 62, 30–41. [Google Scholar] [CrossRef]
- Sánchez-Andrea, I.; Guedes, I.A.; Hornung, B.; Boeren, S.; Lawson, C.E.; Sousa, D.Z.; Bar-Even, A.; Claassens, N.J.; Stams, A.J.M. The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nat. Commun. 2020, 11, 5090. [Google Scholar] [CrossRef]
- Hong, Y.; Arbter, P.; Wang, W.; Rojas, L.N.; Zeng, A.-P. Introduction of glycine synthase enables uptake of exogenous formate and strongly impacts the metabolism in Clostridium pasteurianum. Biotechnol. Bioeng. 2021, 118, 1366–1380. [Google Scholar] [CrossRef]
- Yishai, O.; Goldbach, L.; Tenenboim, H.; Lindner, S.N.; Bar-Even, A. Engineered assimilation of exogenous and endogenous formate in Escherichia coli. ACS Synth. Biol. 2017, 6, 1722–1731. [Google Scholar] [CrossRef]
- Yishai, O.; Bouzon, M.; Döring, V.; Bar-Even, A. In vivo assimilation of one-carbon via a synthetic reductive glycine pathway in Escherichia coli. ACS Synth. Biol. 2018, 7, 2023–2028. [Google Scholar] [CrossRef] [PubMed]
- Bang, J.; Lee, S.Y. Assimilation of formic acid and CO2 by engineered Escherichia coli equipped with reconstructed one-carbon assimilation pathways. Proc. Natl. Acad. Sci. USA 2018, 115, E9271–E9279. [Google Scholar] [CrossRef] [PubMed]
- Bang, J.; Hwang, C.H.; Ahn, J.H.; Lee, J.A.; Lee, S.Y. Escherichia coli is engineered to grow on CO2 and formic acid. Nat. Microbiol. 2020, 5, 1459–1463. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, Y.; Hirano, S.; Matson, M.M.; Atsumi, S.; Kondo, A. Electrical-biological hybrid system for CO2 reduction. Metab. Eng. 2018, 47, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lindner, S.N.; Aslan, S.; Yishai, O.; Wenk, S.; Schann, K.; Bar-Even, A. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat. Chem. Biol. 2020, 16, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Machens, F.; Messerschmidt, K.; Bar-Even, A. core Catalysis of the Reductive Glycine Pathway Demonstrated in Yeast. ACS Synth. Biol. 2019, 8, 911–917. [Google Scholar]
- Neuendorf, C.S.; Vignolle, G.A.; Derntl, C.; Tomin, T.; Novak, K.; Mach, R.L.; Birner-Grünberger, R.; Pflügl, S. A quantitative metabolic analysis reveals Acetobacterium woodii as a flexible and robust host for formate-based bioproduction. Metab. Eng. 2021, 68, 68–85. [Google Scholar] [CrossRef]
- Moon, J.; Dönig, J.; Kramer, S.; Poehlein, A.; Daniel, R.; Müller, V. Formate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 2021, 23, 4214–4227. [Google Scholar] [CrossRef]
- Wood, J.C.; Marcellin, E.; Plan, M.R.; Virdis, B. High methanol-to-formate ratios induce butanol production in Eubacterium limosum. Microb. Biotechnol. 2022, 15, 1542–1549. [Google Scholar] [CrossRef]
- Nicholls, P. Formate as an inhibitor of cytochrome c oxidase. Biochem. Biophys. Res. Commun. 1975, 67, 610–616. [Google Scholar] [CrossRef]
- Warnecke, T.; Gill, R.T. Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Microb. Cell Factories 2005, 4, 25. [Google Scholar] [CrossRef]
- Overkamp, K.M.; Kötter, P.; van der Hoek, R.; Schoondermark-Stolk, S.; Luttik, M.A.; van Dijken, J.P.; Pronk, J.T. Functional analysis of structural genes for NAD+-dependent formate dehydrogenase in Saccharomyces cerevisiae. Yeast 2002, 19, 509–520. [Google Scholar] [CrossRef]
- Litty, D.; Müller, V. Butyrate production in the acetogen Eubacterium limosum is dependent on the carbon and energy source. Microb. Biotechnol. 2021, 14, 2686–2692. [Google Scholar] [CrossRef]
- Du, C.; Li, Y.; Xiang, R.; Yuan, W. Formate dehydrogenase improves the resistance to formic acid and acetic acid simultaneously in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2022, 23, 3406. [Google Scholar] [CrossRef] [PubMed]
- Babel, W. The Auxiliary Substrate concept: From simple considerations to heuristically valuable knowledge. Eng. Life Sci. 2009, 9, 285–290. [Google Scholar] [CrossRef]
- Nielsen, J.; Keasling, J.D. Engineering Cellular Metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.G.; Lee, J.H.; Noh, M.H.; Jung, G.Y. Rediscovering Acetate Metabolism: Its Potential Sources and Utilization for Biobased Transformation into Value-Added Chemicals. J. Agric. Food Chem. 2018, 66, 3998–4006. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lama, S.; Agrawal, D.; Kumar, V.; Park, S. Acetate as a potential feedstock for the production of value-added chemicals: Metabolism and applications. Biotechnol. Adv. 2021, 49, 107736. [Google Scholar] [CrossRef] [PubMed]
- Mutyala, S.; Kim, J.R. Recent advances and challenges in the bioconversion of acetate to value-added chemicals. Bioresour. Technol. 2022, 364, 128064. [Google Scholar] [CrossRef]
- Wang, S.; Sun, X.; Yuan, Q. Strategies for enhancing microbial tolerance to inhibitors for biofuel production: A review. Bioresour. Technol. 2018, 258, 302–309. [Google Scholar] [CrossRef]
- Kutscha, R.; Pflugl, S. Microbial Upgrading of Acetate into Value-Added Products-Examining Microbial Diversity, Bioenergetic constraints and Metabolic Engineering Approaches. Int. J. Mol. Sci. 2020, 21, 8777. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Lama, S.; Jiang, J.; Sankaranarayanan, M.; Park, S. Use of acetate for the production of 3-hydroxypropionic acid by metabolically-engineered Pseudomonas denitrificans. Bioresour. Technol. 2020, 307, 123194. [Google Scholar] [CrossRef] [PubMed]
- Seong, W.; Han, G.H.; Lim, H.S.; Baek, J.I.; Kim, S.-J.; Kim, D.; Kim, S.K.; Lee, H.; Kim, H.; Lee, S.-G.; et al. Adaptive laboratory evolution of Escherichia coli lacking cellular byproduct formation for enhanced acetate utilization through compensatory ATP consumption. Metab. Eng. 2020, 62, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Wu, B.; Liu, L.; Li, J.; Zhu, Q.; He, M.; Hu, G. Metabolic engineering using acetate as a promising building block for the production of bio-based chemicals. Eng. Microbiol. 2022, 2, 100036. [Google Scholar] [CrossRef]
- Deng, Y.; Ma, N.; Zhu, K.; Mao, Y.; Wei, X.; Zhao, Y. Balancing the carbon flux distributions between the TCA cycle and glyoxylate shunt to produce glycolate at high yield and titer in Escherichia coli. Metab. Eng. 2018, 46, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Lama, S.; Kim, Y.; Nguyen, D.T.; Im, C.H.; Sankaranarayanan, M.; Park, S. Production of 3-hydroxypropionic acid from acetate using metabolically-engineered and glucose-grown Escherichia coli. Bioresour. Technol. 2021, 320, 124362. [Google Scholar] [CrossRef]
- Chang, Z.; Dai, W.; Mao, Y.; Cui, Z.; Zhang, Z.; Wang, Z.; Ma, H.; Chen, T. Enhanced 3-hydroxypropionic acid production from acetate via the malonyl-COA pathway in corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2022, 9, 808258. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, S.; Jia, X. Production of medium chain length polyhydroxyalkanoate from acetate by engineered Pseudomonas putida KT2440. J. Ind. Microbiol. Biotechnol. 2019, 46, 793–800. [Google Scholar] [CrossRef]
- Wei, N.; Quarterman, J.; Kim, S.R.; Cate, J.H.; Jin, Y.-S. Enhanced biofuel production through coupled acetic acid and xylose Consumption by engineered yeast. Nat. Commun. 2013, 4, 2580. [Google Scholar] [CrossRef]
- Huang, X.-f.; Shen, Y.; Luo, H.-j.; Liu, J.-n.; Liu, J. Enhancement of extracellular lipid production by oleaginous yeast through preculture and sequencing batch culture strategy with acetic acid. Bioresour. Technol. 2018, 247, 395–401. [Google Scholar] [CrossRef]
- Gong, G.; Zhang, X.; Tan, T. Simultaneously enhanced intracellular lipogenesis and β-carotene biosynthesis of Rhodotorula glutinis by light exposure with sodium acetate as the substrate. Bioresour. Technol. 2020, 295, 122274. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yan, W.; Qian, X.; Chen, M.; Zhang, X.; Xin, F.; Zhang, W.; Jiang, M.; Ochsenreither, K. Increased lipid production in Yarrowia lipolytica from acetate through metabolic engineering and cosubstrate fermentation. ACS Synth. Biol. 2021, 10, 3129–3138. [Google Scholar] [CrossRef]
- Kövilein, A.; Umpfenbach, J.; Ochsenreither, K. Acetate as substrate for l-malic acid production with Aspergillus oryzae DSM 1863. Biotechnol. Biofuels 2021, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Ding, Y.; Liu, M.; Xian, M.; Zhao, G. Comparison of glucose, acetate and ethanol as carbon resource for production of poly (3-hydroxybutyrate) and other acetyl-COA derivatives. Front. Bioeng. Biotechnol. 2020, 8, 833. [Google Scholar] [CrossRef]
- Jo, M.; Noh, M.H.; Lim, H.G.; Kang, C.W.; Im, D.-K.; Oh, M.-K.; Jung, G.Y. Precise tuning of the glyoxylate cycle in Escherichia coli for efficient tyrosine production from acetate. Microb. Cell Factories 2019, 18, 1–9. [Google Scholar] [CrossRef]
- Xu, J.; Liu, N.; Qiao, K.; Vogg, S.; Stephanopoulos, G. Application of metabolic controls for the maximization of lipid production in semicontinuous fermentation. Proc. Natl. Acad. Sci. USA 2017, 114, E5308–E5316. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, Y.; Desai, S.H.; Atsumi, S. Two-dimensional isobutyl acetate production pathways to improve carbon yield. Nat. Commun. 2015, 6, 7488. [Google Scholar] [CrossRef]
- Yang, J.; Nie, Q. Engineering Escherichia coli to convert acetic acid to β-caryophyllene. Microb. Cell Factories 2016, 15, 1–9. [Google Scholar] [CrossRef]
- Lam, F.H.; Turanlı-Yıldız, B.; Liu, D.; Resch, M.G.; Fink, G.R.; Stephanopoulos, G. Engineered yeast tolerance enables efficient production from toxified lignocellulosic feedstocks. Sci. Adv. 2021, 7, eabf7613. [Google Scholar] [CrossRef]
- Hahn-Hägerdal, B.; Galbe, M.; Gorwa-Grauslund, M.-F.; Lidén, G.; Zacchi, G. Bio-ethanol–the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006, 24, 549–556. [Google Scholar] [CrossRef]
- Liang, H.; Ma, X.; Ning, W.; Liu, Y.; Sinskey, A.J.; Stephanopoulos, G.; Zhou, K. Constructing an ethanol utilization pathway in Escherichia coli to produce acetyl-COA derived compounds. Metab. Eng. 2021, 65, 223–231. [Google Scholar] [CrossRef]
- Liu, Y.; Bai, C.; Liu, Q.; Xu, Q.; Qian, Z.; Peng, Q.; Yu, J.; Xu, M.; Zhou, X.; Zhang, Y.; et al. Engineered ethanol-driven biosynthetic system for improving production of acetyl-COA derived drugs in Crabtree-negative yeast. Metab. Eng. 2019, 54, 275–284. [Google Scholar] [CrossRef]
- Cao, Y.; Mu, H.; Guo, J.; Liu, H.; Zhang, R.; Liu, W.; Xian, M.; Liu, H. Metabolic engineering of Escherichia coli for the utilization of ethanol. J. Biol. Res. 2020, 27, 1. [Google Scholar] [CrossRef]
- Westfall, P.J.; Pitera, D.J.; Lenihan, J.R.; Eng, D.; Woolard, F.X.; Regentin, R.; Horning, T.; Tsuruta, H.; Melis, D.J.; Owens, A. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc. Natl. Acad. Sci. USA 2012, 109, E111–E118. [Google Scholar] [CrossRef] [PubMed]
- Felenbok, B.; Flipphi, M.; Nikolaev, I. Ethanol catabolism in Aspergillus nidulans: A model system for studying gene regulation. Prog. Nucleic Acid. Res. Mol. Biol. 2001, 69, 149–204. [Google Scholar] [PubMed]
- Ricci, L.; Seifert, A.; Bernacchi, S.; Fino, D.; Pirri, C.F.; Re, A. Leveraging substrate flexibility and product selectivity of acetogens in two-stage systems for chemical production. Microb. Biotechnol. 2023, 16, 218–237. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Rouboa, A.; Silva, V.; Monteiro, E.; Bouziane, K. Influence of the biomass gasification processes on the final composition of syngas. Energy Procedia 2013, 36, 596–606. [Google Scholar] [CrossRef]
- Molino, A.; Chianese, S.; Musmarra, D. Biomass gasification technology: The state of the art overview. J. Energy Chem. 2016, 25, 10–25. [Google Scholar] [CrossRef]
- Ahmad, A.A.; Zawawi, N.A.; Kasim, F.H.; Inayat, A.; Khasri, A. Assessing the gasification performance of biomass: A review on biomass gasification process conditions, optimization and economic evaluation. Renew. Sustain. Energy Rev. 2016, 53, 1333–1347. [Google Scholar] [CrossRef]
- Griffin, D.W.; Schultz, M.A. Fuel and chemical products from biomass syngas: A comparison of gas fermentation to thermochemical onversion routes. Environ. Prog. Sustain. Energy 2012, 31, 219–224. [Google Scholar] [CrossRef]
- Daniell, J.; Köpke, M.; Simpson, S.D. Commercial biomass syngas fermentation. Energies 2012, 5, 5372–5417. [Google Scholar] [CrossRef]
- Akhtar, A.; Krepl, V.; Ivanova, T. A combined overview of combustion, pyrolysis, and gasification of biomass. Energy Fuels 2018, 32, 7294–7318. [Google Scholar] [CrossRef]
- Phillips, J.R.; Huhnke, R.L.; Atiyeh, H.K. Syngas fermentation: A microbial conversion process of gaseous substrates to various products. Fermentation 2017, 3, 28. [Google Scholar] [CrossRef]
- Lemgruber, R.d.S.P.; Valgepea, K.; Tappel, R.; Behrendorff, J.B.; Palfreyman, R.W.; Plan, M.; Hodson, M.P.; Simpson, S.D.; Nielsen, L.K.; Köpke, M. Systems-level engineering and characterisation of Clostridium autoethanogenum through heterologous production of poly-3-hydroxybutyrate (PHB). Metab. Eng. 2019, 53, 14–23. [Google Scholar] [CrossRef]
- Pavan, M.; Reinmets, K.; Garg, S.; Mueller, A.P.; Marcellin, E.; Köpke, M.; Valgepea, K. Advances in systems metabolic engineering of autotrophic carbon oxide-fixing biocatalysts towards a circular economy. Metab. Eng. 2022, 71, 117–141. [Google Scholar] [CrossRef]
- Pala, L.P.R.; Wang, Q.; Kolb, G.; Hessel, V. Steam gasification of biomass with subsequent syngas adjustment using shift reaction for syngas production: An Aspen Plus model. Renew. Energy 2017, 101, 484–492. [Google Scholar] [CrossRef]
- Gunes, B. A critical review on biofilm-based reactor systems for enhanced syngas fermentation processes. Renew. Sustain. Energy Rev. 2021, 143, 110950. [Google Scholar] [CrossRef]
- Stoll, I.K.; Boukis, N.; Sauer, J. Syngas Fermentation to Alcohols: Reactor Technology and Application Perspective. Chem. Ing. Tech. 2020, 92, 125–136. [Google Scholar] [CrossRef]
- Köpke, M.; Simpson, S.D. Pollution to products: Recycling of ‘above ground’carbon by gas fermentation. Curr. Opin. Biotechnol. 2020, 65, 180–189. [Google Scholar] [CrossRef]
- Marcellin, E.; Angenent, L.T.; Nielsen, L.K.; Molitor, B. Recycling Carbon for Sustainable Protein Production Using Gas Fermentation. Curr. Opin. Biotechnol. 2022, 76, 102723. [Google Scholar] [CrossRef]
- Onyeaka, H.; Ekwebelem, O.C. A review of recent advances in engineering bacteria for enhanced CO2 capture and utilization. Int. J. Environ. Sci. Technol. 2023, 20, 4635–4648. [Google Scholar] [CrossRef] [PubMed]
- Fackler, N.; Heijstra, B.D.; Rasor, B.J.; Brown, H.; Martin, J.; Ni, Z.; Shebek, K.M.; Rosin, R.R.; Simpson, S.D.; Tyo, K.E.; et al. Stepping on the Gas to a Circular Economy: Accelerating Development of Carbon-Negative Chemical Production from Gas Fermentation. Annu. Rev. Chem. Biomol. Eng. 2021, 12, 439–470. [Google Scholar] [CrossRef]
- Nesbitt, E.R. Using waste carbon feedstocks to produce chemicals. Ind. Biotechnol. 2020, 16, 147–163. [Google Scholar] [CrossRef]
- Haas, T.; Krause, R.; Weber, R.; Demler, M.; Schmid, G. Technical photosynthesis involving CO2 electrolysis and fermentation. Nat. Catal. 2018, 1, 32–39. [Google Scholar] [CrossRef]
- Air Protein. Available online: https://www.airprotein.com/ (accessed on 19 November 2023).
- Deep Branch Biotechnology. Available online: https://deepbranch.com/ (accessed on 19 November 2023).
- Kiverdi. Available online: https://www.kiverdi.om/ (accessed on 19 November 2023).
- Solar Foods. Available online: https://solarfoods.com/science/ (accessed on 19 November 2023).
- NovoNutrients. Available online: https://www.novonutrients.com/ (accessed on 19 November 2023).
- Burlacot, A.; Dao, O.; Auroy, P.; Cuiné, S.; Li-Beisson, Y.; Peltier, G. Alternative photosynthesis pathways drive the algal CO2-concentrating mechanism. Nature 2022, 605, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Diels, L.; Pant, D.; Patil, S.A.; Ghangrekar, M.M. Review—Microbial Electrosynthesis: A Way Towards The Production of Electro-ommodities Through Carbon Sequestration with Microbes as Biocatalysts. J. Electrochem. Soc. 2020, 167, 155510. [Google Scholar] [CrossRef]
- Fan, L.; Xia, C.; Zhu, P.; Lu, Y.; Wang, H. Electrochemical CO2 reduction to high-concentration pure formic acid solutions in an all-solid-state reactor. Nat. Commun. 2020, 11, 3633. [Google Scholar] [CrossRef]
- Ahmad, W.; Koley, P.; Dwivedi, S.; Lakshman, R.; Shin, Y.K.; van Duin, A.C.T.; Shrotri, A.; Tanksale, A. Aqueous phase conversion of CO2 into acetic acid over thermally transformed MIL-88B catalyst. Nat. Commun. 2023, 14, 2821. [Google Scholar] [CrossRef]
- Vlaeminck, E.; Quataert, K.; Uitterhaegen, E.; De Winter, K.; Soetaert, W.K. Advanced PHB fermentation strategies with CO2-derived organic acids. J. Biotechnol. 2022, 343, 102–109. [Google Scholar] [CrossRef]
- Cao, M.; Tran, V.G.; Qin, J.; Olson, A.; Mishra, S.; Schultz, J.C.; Huang, C.; Xie, D.; Zhao, H. Metabolic engineering of oleaginous yeast Rhodotorula toruloides for overproduction of triacetic acid lactone. Biotechnol. Bioeng. 2022, 119, 2529–2540. [Google Scholar] [CrossRef]
- Lawson, C.E.; Martí, J.M.; Radivojevic, T.; Jonnalagadda, S.V.R.; Gentz, R.; Hillson, N.J.; Peisert, S.; Kim, J.; Simmons, B.A.; Petzold, C.J.; et al. Machine learning for metabolic engineering: A review. Metab. Eng. 2021, 63, 34–60. [Google Scholar] [CrossRef]
- Helmy, M.; Smith, D.; Selvarajoo, K. Systems biology approaches integrated with artificial intelligence for optimized metabolic engineering. Metab. Eng. Commun. 2020, 11, e00149. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Duncan, J.D.; Setati, M.E.; Divol, B. Redox cofactor metabolism in Saccharomyces cerevisiae and its impact on the production of alcoholic fermentation end-products. Food Res. Int. 2023, 163, 112276. [Google Scholar] [CrossRef] [PubMed]
- Bloem, A.; Sanchez, I.; Dequin, S.; Camarasa, C. Metabolic Impact of Redox Cofactor Perturbations on the Formation of Aroma Compounds in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2016, 82, 174–183. [Google Scholar] [CrossRef]
- Liu, C.-G.; Xue, C.; Lin, Y.-H.; Bai, F.-W. Redox potential control and applications in microaerobic and anaerobic fermentations. Biotechnol. Adv. 2013, 31, 257–265. [Google Scholar] [CrossRef]
- Hou, J.; Scalcinati, G.; Oldiges, M.; Vemuri, G.N. Metabolic impact of increased NADH availability in Saccharomyces cerevisiae. Appl Env. Microbiol 2010, 76, 851–859. [Google Scholar] [CrossRef]
- Qiao, K.; Wasylenko, T.M.; Zhou, K.; Xu, P.; Stephanopoulos, G. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat. Biotechnol. 2017, 35, 173–177. [Google Scholar] [CrossRef]
- Kim, J.-E.; Jang, I.-S.; Sung, B.H.; Kim, S.C.; Lee, J.Y. Rerouting of NADPH synthetic pathways for increased protopanaxadiol production in Saccharomyces cerevisiae. Sci. Rep. 2018, 8, 15820. [Google Scholar] [CrossRef]
- Qiao, K.; Imam Abidi, S.H.; Liu, H.; Zhang, H.; Chakraborty, S.; Watson, N.; Kumaran Ajikumar, P.; Stephanopoulos, G. Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 2015, 29, 56–65. [Google Scholar] [CrossRef]
- Kajla, S.; Kumari, R.; Nagi, G.K. Microbial CO2 fixation and biotechnology in reducing industrial CO2 emissions. Arch. Microbiol. 2022, 204, 149. [Google Scholar] [CrossRef] [PubMed]
- Tharak, A.; Katakojwala, R.; Kajla, S.; Venkata Mohan, S. Chemolithoautotrophic reduction of CO2 to acetic acid in gas and gas-electro fermentation systems: Enrichment, microbial dynamics, and sustainability assessment. Chem. Eng. J. 2023, 454, 140200. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, S.-X.; Gandionco, K.A.; Bond, A.M.; Zhang, J. Electrocatalytic carbon dioxide reduction: From fundamental principles to catalyst design. Mater. Today Adv. 2020, 7, 100074. [Google Scholar] [CrossRef]
- Stöckl, M.; Claassens, N.J.; Lindner, S.N.; Klemm, E.; Holtmann, D. Coupling electrochemical CO2 reduction to microbial product generation–identification of the gaps and opportunities. Curr. Opin. Biotechnol. 2022, 74, 154–163. [Google Scholar] [CrossRef] [PubMed]
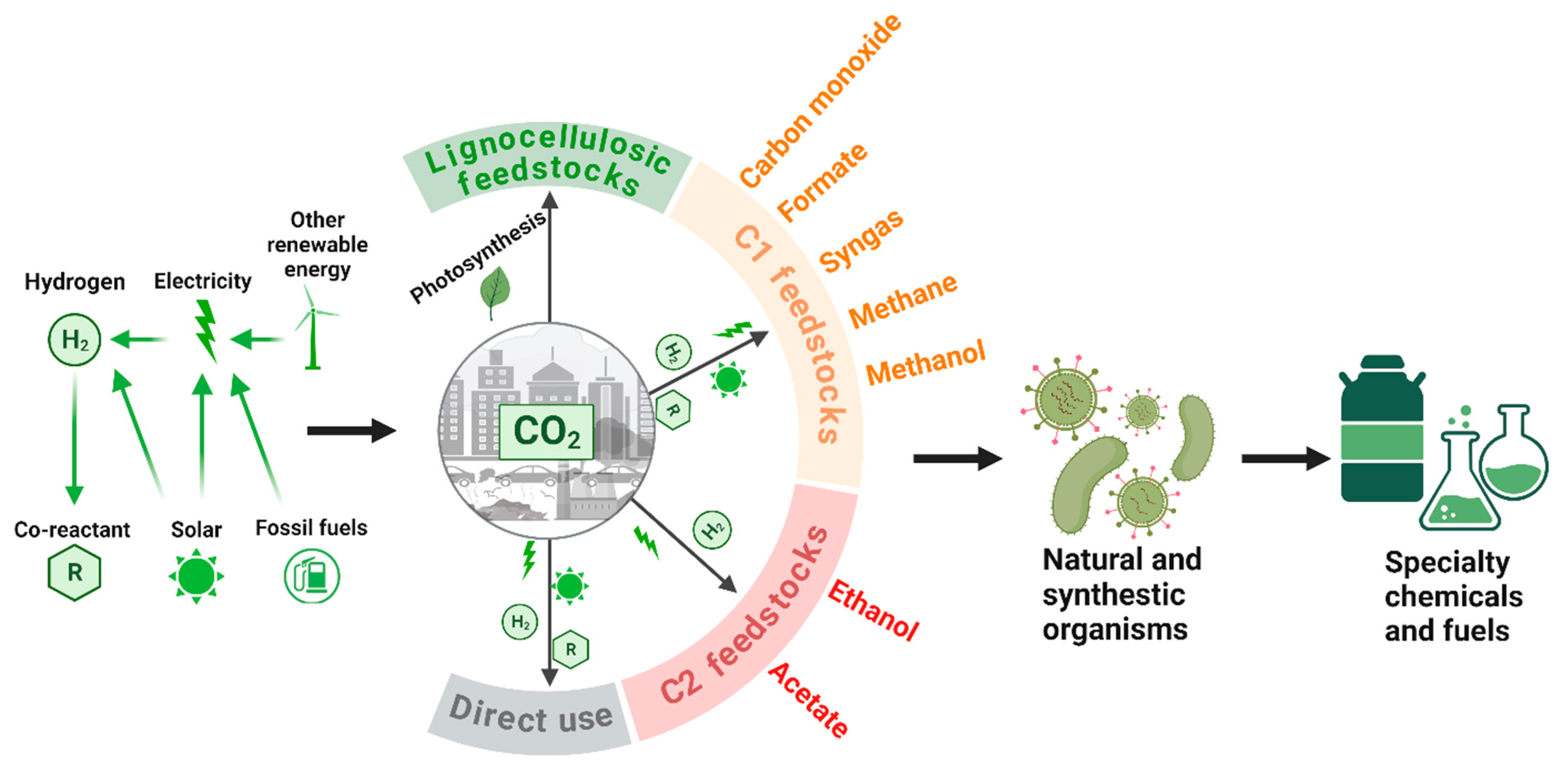


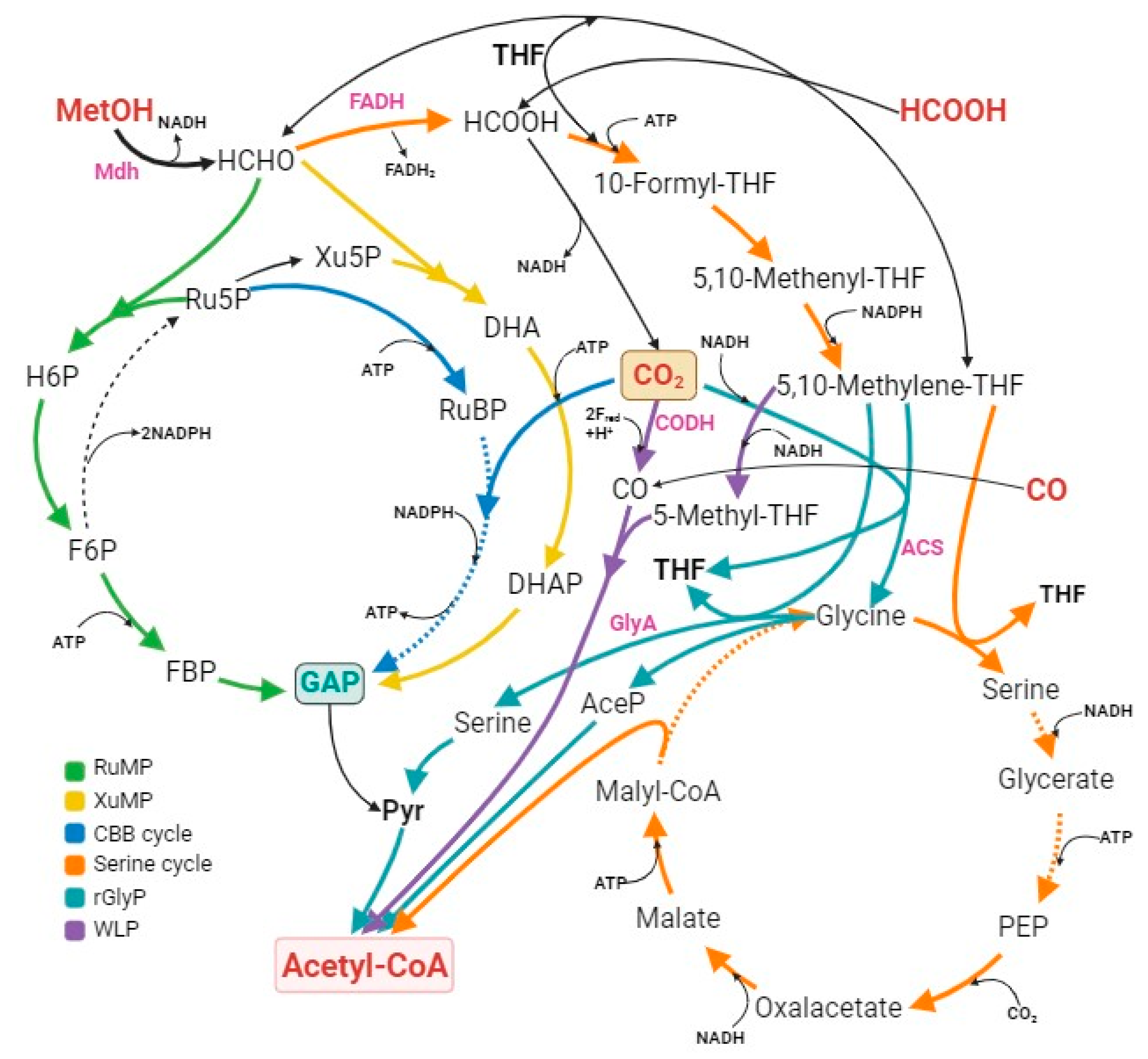
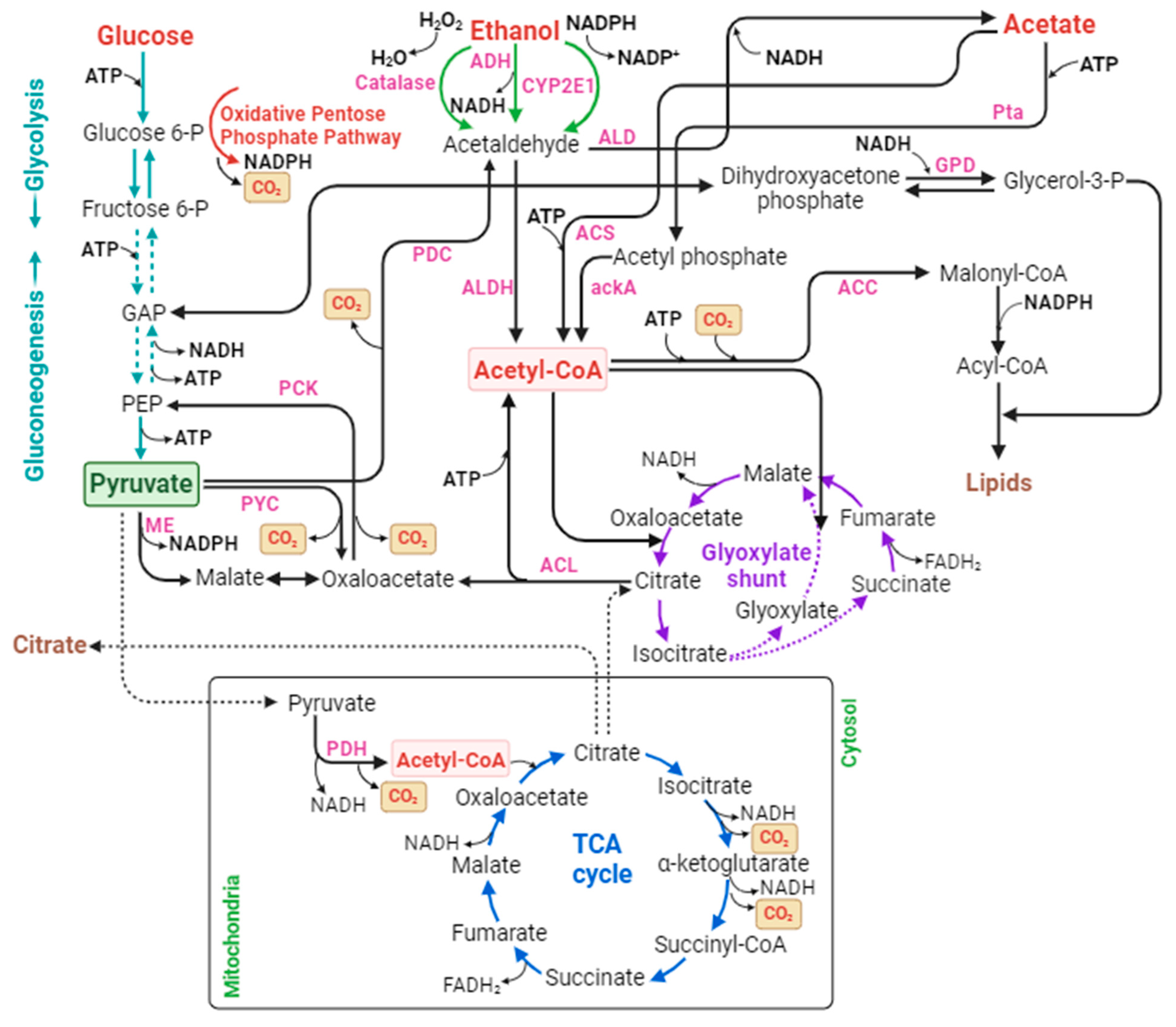

| Methods | Major Steps and Overall Reaction of CO2 Fixation | |
|---|---|---|
| One-step/Direct CO2 fixation and conversion |
| |
| Two-step CO2 fixation and conversion | Step 1 (electrochemical catalysis): CO2 + H2O + electricity → C1/C2 chemicals | Step 2 (biomanufacturing): C1/C2 → biofuels and chemicals |
|
| |
| Substrate | Key Enzyme | Major Biochemical Reactions | Eq. ATP/Substrate | ||
|---|---|---|---|---|---|
| Reaction 1 | Reaction 2 | Reaction 3 | |||
| CO2 | N/A | CO2 + RuBP + 2NADPH + 2ADP + 2Pi → 2GAP + 2NADP + 2ATP | N/A | N/A | 3.3 |
| CO | N/A | CO + 5-Methyl-THF → AcCoA | N/A | N/A | 6.0 |
| Methane (CH4) | N/A | CH4 + O2 + NADH → HCHO + NAD | HCHO + Xu5P + ATP → 2GAP + ADP + Pi | N/A | 8.7 |
| Methanol (CH3OH or MeOH) | RuMp | MeOH + NADH → HCHO + NAD | HCHO + Ru5P + ATP → 2GAP + ADP + Pi | 2GAP + 8ADP + Pi + 8NAD → 2AcCoA + 8ATP + 8NADH + 2CO2 | 8.7 |
| XuMp | HCHO + Xu5P + ATP → 2GAP + ADP + Pi | N/A | 9.2 | ||
| Serine | HCHO + FAD + 3ATP + 2NADPH + 2NADH + Glycine + CO2 → AcCoA + FADH2 + 3ADP + 2NADP + 2NAD + Glyoxylate | N/A | −6.0 | ||
| Formate (HCOOH) | CBB | HCOOH + NAD → CO2 + NADH | RuBP + CO2 + 2NADPH + 2ADP + Pi → 2GAP + 2NADP + 2ATP | 2GAP + 8ADP + Pi + 8NAD → 2AcCoA + 8ATP + 8NADH + 2CO2 | 9.2 |
| HCOOH + ATP → 10-Formyl-THF + ADP + Pi | 10-Formyl-THF + NADPH + NADH + CO2 + FADH2 → AcCoA + NADP + NAD + FAD | N/A | 5.0 | ||
| Acetate (CH3COOH or OAc) | Pta/ackA | OAc + ATP → ADP + AcP | AcP + CoA → AcCoA + pi | N/A | 11 |
| ACS | OAc + ATP + CoA → AcCoA + AMP + PPi | N/A | N/A | 11 | |
| Ethanol (CH3CH2OH or EtOH) | CYP2E1 | EtOH + NADPH + H + O2 → MeCHO + NADP + H2O | MeCHO + NADH → NAD + OAc | OAc + ATP → ADP + AcP; AcP + CoA → AcCoA + pi | 5 |
| OAc + ATP + CoA → AcCoA + AMP + PPi | 5 | ||||
| ADH | EtOH + NAD → MeCHO + NADH | OAc + ATP → ADP + AcP; AcP + CoA → AcCoA + pi | 11 | ||
| OAc + ATP + CoA → AcCoA + AMP + Ppi | 11 | ||||
| Catalase | EtOH + H2O2 → MeCHO + H2O | OAc + ATP → ADP + AcP; AcP + CoA → AcCoA + pi | 8 | ||
| OAc + ATP + CoA → AcCoA + AMP + PPi | 8 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurt, E.; Qin, J.; Williams, A.; Zhao, Y.; Xie, D. Perspectives for Using CO2 as a Feedstock for Biomanufacturing of Fuels and Chemicals. Bioengineering 2023, 10, 1357. https://doi.org/10.3390/bioengineering10121357
Kurt E, Qin J, Williams A, Zhao Y, Xie D. Perspectives for Using CO2 as a Feedstock for Biomanufacturing of Fuels and Chemicals. Bioengineering. 2023; 10(12):1357. https://doi.org/10.3390/bioengineering10121357
Chicago/Turabian StyleKurt, Elif, Jiansong Qin, Alexandria Williams, Youbo Zhao, and Dongming Xie. 2023. "Perspectives for Using CO2 as a Feedstock for Biomanufacturing of Fuels and Chemicals" Bioengineering 10, no. 12: 1357. https://doi.org/10.3390/bioengineering10121357
APA StyleKurt, E., Qin, J., Williams, A., Zhao, Y., & Xie, D. (2023). Perspectives for Using CO2 as a Feedstock for Biomanufacturing of Fuels and Chemicals. Bioengineering, 10(12), 1357. https://doi.org/10.3390/bioengineering10121357







