A Comprehensive Review of Computer-Aided Models for Breast Cancer Diagnosis Using Histopathology Images
Abstract
:1. Introduction
1.1. Scope of the Review
- (a)
- Which histopathological image datasets are widely used in breast CAD systems?
- (b)
- What are the preprocessing methods and their impact on the CAD systems?
- (c)
- What are the employed segmentation and feature extraction methods?
- (d)
- What are the most common performance metrics used?
- (e)
- What are the trending methodologies and associated challenges in the field?
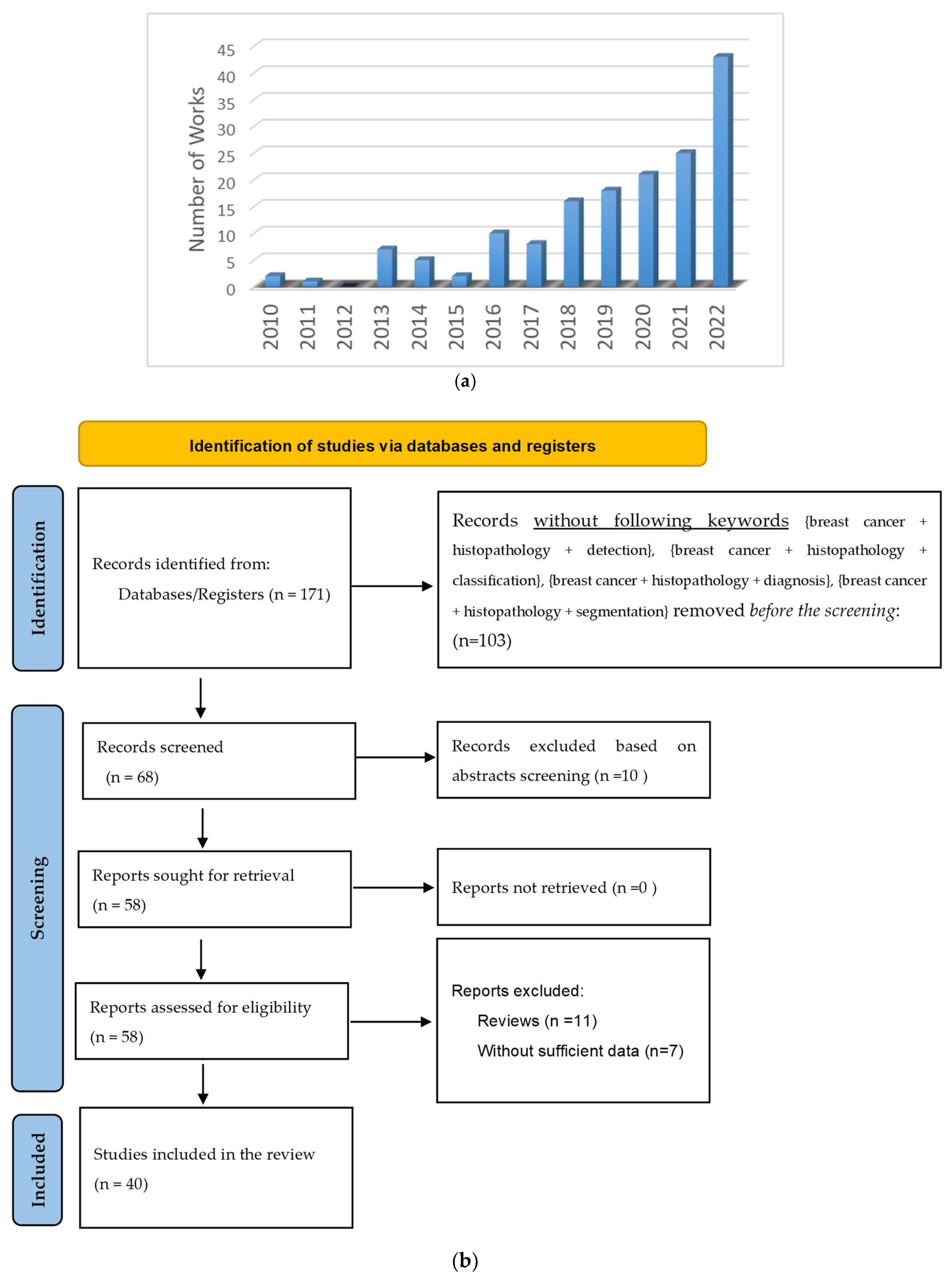
1.2. Article Selection Criteria
2. Basics and Background
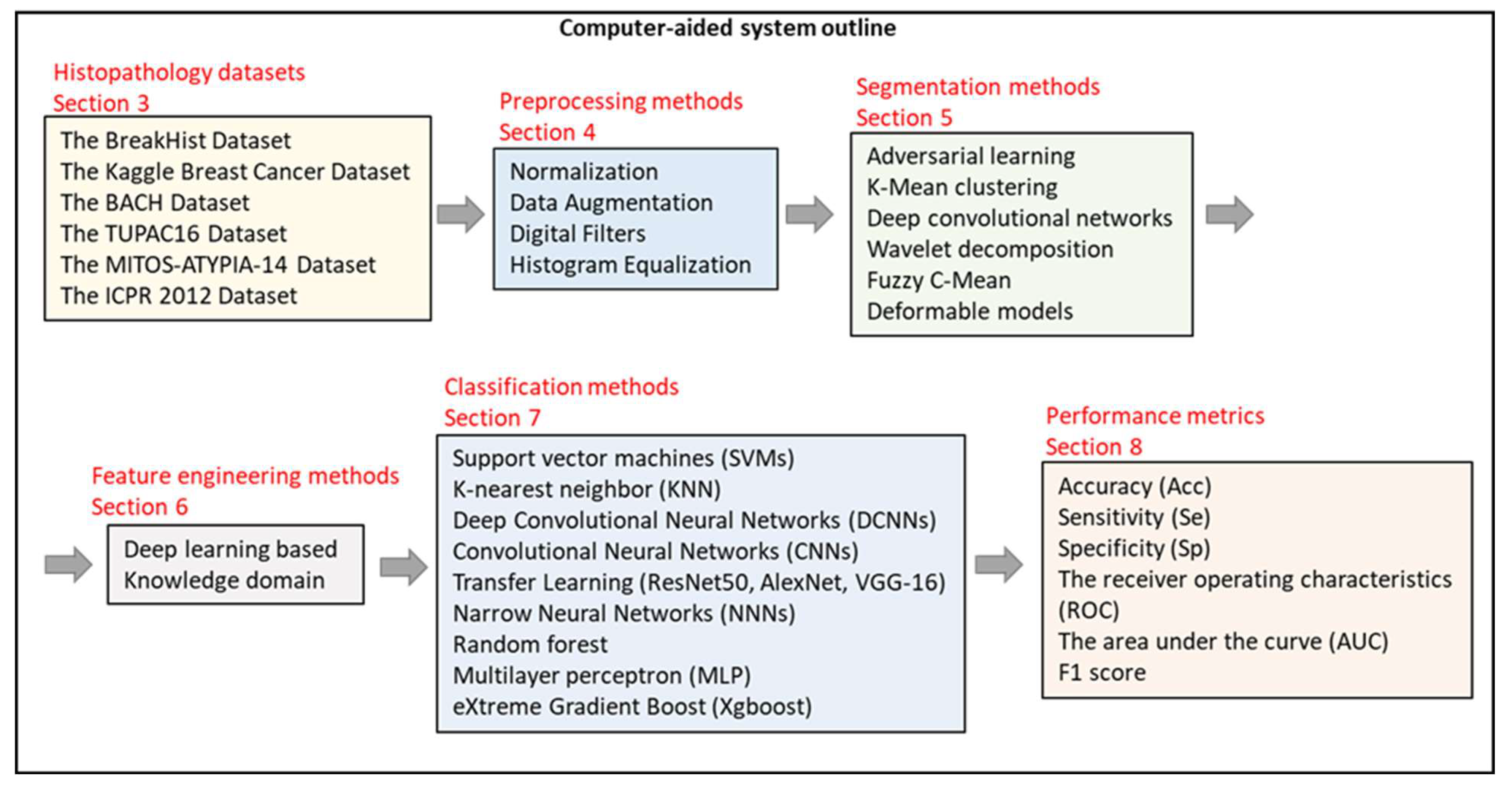
3. Histopathology Image Datasets
3.1. The BreakHis Dataset
3.2. The Kaggle Breast Cancer
3.3. The ICIAR 2018 Grand Challenge on Breast Cancer Histology Images (BACH) Dataset
3.4. The TUPAC16 Dataset
3.5. The MITOS-ATYPIA-14 Dataset
3.6. The ICPR 2012 Dataset
4. Preprocessing Methods
4.1. Normalization
4.2. Data Augmentation
4.3. Digital Filters
4.4. Histogram Equalization
5. Segmentation Methods
6. Feature Engineering Methods
7. Classification/Detection/Diagnosis Algorithms
8. Performance Evaluation Metrics
- TP represents the image correctly classified as malignant,
- TN represents the image correctly classified as benign,
- FP represents the image falsely classified as malignant, and
- FN represents the image falsely classified as benign.
9. Discussions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cancer Facts & Figures 2022. American Cancer Society. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2022.html (accessed on 16 August 2022).
- Stump-Sutliff, K.A. Breast Cancer: What Are the Survival Rates? WebMD. Available online: https://www.webmd.com/breast-cancer/guide/breast-cancer-survival-rates (accessed on 16 August 2022).
- U.S. Breast Cancer Statistics. Breastcancer.org. 13 January 2022. Available online: https://www.breastcancer.org/symptoms/understand_bc/statistics (accessed on 16 August 2022).
- Breast Cancer—Metastatic: Statistics|cancer.net. Available online: https://www.cancer.net/cancer-types/breast-cancer-metastatic/statistics (accessed on 17 August 2022).
- Veta, M.; Pluim, J.P.; van Diest, P.J.; Viergever, M.A. Breast Cancer Histopathology Image Analysis: A Review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Łukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanisławek, A. Breast Cancer—Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef]
- Angahar, T.L. An overview of breast cancer epidemiology, risk factors, pathophysiology, and cancer risks reduction. MOJ Biol. Med. 2017, 1, 92–96. [Google Scholar] [CrossRef]
- Nassif, A.B.; Talib, M.A.; Nasir, Q.; Afadar, Y.; Elgendy, O. Breast cancer detection using artificial intelligence techniques: A systematic literature review. Artif. Intell. Med. 2022, 127, 102276. [Google Scholar] [CrossRef] [PubMed]
- Yassin, N.I.; Omran, S.; El Houby, E.M.; Allam, H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: A systematic review. Comput. Methods Programs Biomed. 2018, 156, 25–45. [Google Scholar] [CrossRef]
- Petrelli, F.; Viale, G.; Cabiddu, M.; Barni, S. Prognostic value of different cut-off levels of Ki-67 in breast cancer: A systematic review and meta-analysis of 64,196 patients. Breast Cancer Res. Treat. 2015, 153, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Luporsi, E.; Andre, F.; Spyratos, F.; Martin, P.M.; Jacquemier, J.; Penault-Llorca, F.; Tubiana-Mathieu, N.; Sigal-Zafrani, B.; Arnould, L.; Gompel, A.; et al. Ki-67: Level of evidence and methodological considerations for its role in the clinical management of breast cancer: Analytical and critical review. Breast Cancer Res. Treat. 2012, 132, 895–915. [Google Scholar] [CrossRef]
- Saxena, S.; Gyanchandani, M. Machine learning methods for computer-aided breast cancer diagnosis using histopathology: A narrative review. J. Med. Imaging Radiat. Sci. 2020, 51, 182–193. [Google Scholar] [CrossRef]
- Zhou, X.; Li, C.; Rahaman, M.M.; Yao, Y.; Ai, S.; Sun, C.; Wang, Q.; Zhang, Y.; Li, M.; Li, X. A comprehensive review for breast histopathology image analysis using classical and deep neural networks. IEEE Access 2020, 8, 90931–90956. [Google Scholar] [CrossRef]
- Abhisheka, B.; Biswas, S.K.; Purkayastha, B. A comprehensive review on breast cancer detection, classification and segmentation using deep learning. Arch. Comput. Methods Eng. 2023, 30, 5023–5052. [Google Scholar] [CrossRef]
- Kaushal, C.; Bhat, S.; Koundal, D.; Singla, A. Recent trends in computer-assisted diagnosis (CAD) system for breast cancer diagnosis using histopathological images. IRBM 2019, 40, 211–227. [Google Scholar] [CrossRef]
- PRISMA Transparent Reporting of Systematic Reviews and Meta-Analyses. 2017. Available online: http://www.prismastatement.org/ (accessed on 18 October 2023).
- Accardi, T. Mammography Matters: Screening for Breast Cancer—Then and Now. Available online: https://www.radiologytoday.net/archive/rt0517p7.shtml#:~:text=Although%20the%20concept%20of%20mammography,Society%20to%20officially%20recommend%20it (accessed on 24 August 2022).
- Mammograms. National Cancer Institute. Available online: https://www.cancer.gov/types/breast/mammograms-fact-sheet (accessed on 24 August 2022).
- Sree, S.V. Breast Imaging: A survey. World J. Clin. Oncol. 2011, 2, 171. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, P.J. The history of breast ultrasound. J. Ultrasound Med. 2004, 23, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Breast Ultrasound. Johns Hopkins Medicine. 8 August 2021. Available online: https://www.hopkinsmedicine.org/health/treatment-tests-and-therapies/breast-ultrasound#:~:text=A%20breast%20ultrasound%20is%20most,some%20early%20signs%20of%20cancer (accessed on 24 August 2022).
- What Is a Breast Ultrasound?: Breast Cancer Screening. American Cancer Society. Available online: https://www.cancer.org/cancer/breast-cancer/screening-tests-and-early-detection/breast-ultrasound.html (accessed on 24 August 2022).
- Kelly, K.M.; Dean, J.; Comulada, W.S.; Lee, S.-J. Breast cancer detection using automated whole breast ultrasound and mammography in radiographically dense breasts. Eur. Radiol. 2009, 20, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Heywang, S.H.; Hahn, D.; Schmidt, H.; Krischke, I.; Eiermann, W.; Bassermann, R.; Lissner, J. MR imaging of the breast using gadolinium-DTPA. J. Comput. Assist. Tomogr. 1986, 10, 199–204. [Google Scholar] [CrossRef] [PubMed]
- What Is a Breast MRI: Breast Cancer Screening. American Cancer Society. Available online: https://www.cancer.org/cancer/breast-cancer/screening-tests-and-early-detection/breast-mri-scans.html (accessed on 24 August 2022).
- History of Breast Biopsy. Siemens Healthineers. Available online: https://www.siemens-healthineers.com/mammography/news/history-of-breast-biopsy.html (accessed on 24 August 2022).
- Breast Biopsy: Biopsy Procedure for Breast Cancer. American Cancer Society. Available online: https://www.cancer.org/cancer/breast-cancer/screening-tests-and-early-detection/breast-biopsy.html (accessed on 24 August 2022).
- Breast Biopsy. Johns Hopkins Medicine. 8 August 2021. Available online: https://www.hopkinsmedicine.org/health/treatment-tests-and-therapies/breast-biopsy (accessed on 24 August 2022).
- Versaggi, S.L.; De Leucio, A. Breast Biopsy. Available online: https://www.ncbi.nlm.nih.gov/books/NBK559192/ (accessed on 24 August 2022).
- Barkana, B.D.; Saricicek, I. Classification of Breast Masses in Mammograms using 2D Homomorphic Transform Features and Supervised Classifiers. J. Med. Imaging Health Inform. 2017, 7, 1566–1571. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef]
- Bukun, Breast Cancer Histopathological Database (BreakHis), Kaggle. 10 March 2020. Available online: https://www.kaggle.com/ambarish/breakhis (accessed on 5 February 2022).
- Mooney, P. Breast Histopathology Images. Kaggle. 19 December 2017. Available online: https://www.kaggle.com/paultimothymooney/breast-histopathology-images (accessed on 5 February 2022).
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M.; et al. BACH: Grand challenge on breast cancer histology images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef]
- ICIAR 2018—Grand Challenge. Grand Challenge. Available online: https://iciar2018-challenge.grand-challenge.org/Dataset/ (accessed on 5 February 2022).
- Wahab, N.; Khan, A. Multifaceted fused-CNN based scoring of breast cancer whole-slide histopathology images. Appl. Soft Comput. 2020, 97, 106808. [Google Scholar] [CrossRef]
- Veta, M.; Heng, Y.J.; Stathonikos, N.; Bejnordi, B.E.; Beca, F.; Wollmann, T.; Rohr, K.; Shah, M.A.; Wang, D.; Rousson, M.; et al. Predicting breast tumor proliferation from whole-slide images: The TUPAC16 challenge. Med. Image Anal. 2019, 56, 43, Erratum in Med Image Anal. 2019, 54, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Mitos-ATYPIA-14—Grand Challenge. Available online: https://mitos-atypia-14.grand-challenge.org/Dataset/ (accessed on 30 August 2022).
- Ludovic, R.; Daniel, R.; Nicolas, L.; Maria, K.; Humayun, I.; Jacques, K.; Frédérique, C.; Catherine, G. Mitosis detection in breast cancer histological images An ICPR 2012 contest. J. Pathol. Inform. 2013, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, R. Breast cancer histopathological image classification using stochastic dilated residual ghost model. Int. J. Inf. Retr. Res. 2022, 12, 1–24. [Google Scholar]
- Al Noumah, W.; Jafar, A.; Al Joumaa, K. Using parallel pre-trained types of DCNN model to predict breast cancer with color normalization. BMC Res. Notes 2022, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Boumaraf, S.; Liu, X.; Zheng, Z.; Ma, X.; Ferkous, C. A new transfer learning-based approach to magnification dependent and independent classification of breast cancer in histopathological images. Biomed. Signal Process. Control 2021, 63, 102192. [Google Scholar] [CrossRef]
- Kate, V.; Shukla, P. A new approach to breast cancer analysis through histopathological images using MI, MD binary, and eight class classifying techniques. J. Ambient. Intell. Human Comput. 2021. [Google Scholar] [CrossRef]
- Hameed, Z.; Zahia, S.; Garcia-Zapirain, B.; Javier Aguirre, J.; María Vanegas, A. Breast Cancer Histopathology Image Classification Using an Ensemble of Deep Learning Models. Sensors 2020, 20, 4373. [Google Scholar] [CrossRef]
- Vo, D.M.; Nguyen, N.-Q.; Lee, S.-W. Classification of breast cancer histology images using incremental boosting convolution networks. Inf. Sci. 2019, 482, 123–138. [Google Scholar] [CrossRef]
- Kausar, T.; Wang, M.J.; Idrees, M.; Lu, Y. HWDCNN: Multiclass recognition in breast histopathology with Haar wavelet decomposed image based convolution neural network. Biocybern. Biomed. Eng. 2019, 39, 967–982. [Google Scholar] [CrossRef]
- Li, X.; Radulovic, M.; Kanjer, K.; Plataniotis, K.N. Discriminative pattern mining for breast cancer histopathology image classification via fully convolutional autoencoder. IEEE Access 2019, 7, 36433–36445. [Google Scholar] [CrossRef]
- Rakhlin, A.; Shvets, A.; Iglovikov, V.; Kalinin, A.A. Deep convolutional neural networks for breast cancer histology image analysis. In Proceedings of the Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, 27–29 June 2018; pp. 737–744. [Google Scholar]
- Anghel, A.; Stanisavljevic, M.; Andani, S.; Papandreou, N.; Rüschoff, J.H.; Wild, P.; Gabrani, M.; Pozidis, H. A high-performance system for robust stain normalization of whole-slide images in histopathology. Front. Med. 2019, 6, 193. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.M.; Hernandez, A.A. Enhanced deep learning approach for predicting invasive ductal carcinoma from histopathology images. In Proceedings of the 2019 2nd International Conference on Artificial Intelligence and Big Data (ICAIBD), Chengdu, China, 25–28 May 2019. [Google Scholar]
- Chang, J.; Yu, J.; Han, T.; Chang, H.-J.; Park, E. A method for classifying medical images using transfer learning: A pilot study on histopathology of breast cancer. In Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), Dalian, China, 12–15 October 2017. [Google Scholar]
- Yari, Y.; Nguyen, T.V.; Nguyen, H.T. Deep learning applied for histological diagnosis of breast cancer. IEEE Access 2020, 8, 162432–162448. [Google Scholar] [CrossRef]
- Mikolajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the 2018 International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018. [Google Scholar]
- Hirra, I.; Ahmad, M.; Hussain, A.; Ashraf, M.U.; Saeed, I.A.; Qadri, S.F.; Alghamdi, A.M.; Alfakeeh, A.S. Breast cancer classification from histopathological images using patch-based Deep Learning Modeling. IEEE Access 2021, 9, 24273–24287. [Google Scholar] [CrossRef]
- Vaka, A.R.; Soni, B.; Reddy, S. Breast cancer detection by leveraging machine learning. ICT Express 2020, 6, 320–324. [Google Scholar] [CrossRef]
- Chatterjee, C.C.; Krishna, G. A novel method for IDC prediction in breast cancer histopathology images using deep residual neural networks. In Proceedings of the 2019 2nd International Conference on Intelligent Communication and Computational Techniques (ICCT), Jaipur, India, 28–29 September 2019. [Google Scholar]
- Narayanan, B.N.; Krishnaraja, V.; Ali, R. Convolutional neural network for classification of histopathology images for breast cancer detection. In Proceedings of the 2019 IEEE National Aerospace and Electronics Conference (NAECON), Dayton, OH, USA, 15–19 July 2019. [Google Scholar]
- Jiang, Y.; Chen, L.; Zhang, H.; Xiao, X. Classification of H&E stained breast cancer histopathology images based on Convolutional Neural Network. J. Phys. Conf. Ser. 2019, 1302, 032018. [Google Scholar]
- Image Segmentation: The Basics and 5 Key Techniques. Datagen. 25 October 2022. Available online: https://datagen.tech/guides/image-annotation/image-segmentation/ (accessed on 13 January 2023).
- Lin, Z.; Li, J.; Yao, Q.; Shen, H.; Wan, L. Adversarial learning with data selection for cross-domain histopathological breast cancer segmentation. Multimed. Tools Appl. 2022, 81, 5989–6008. [Google Scholar] [CrossRef]
- Li, B.; Mercan, E.; Mehta, S.; Knezevich, S.; Arnold, C.W.; Weaver, D.L.; Elmore, J.G.; Shapiro, L.G. Classifying breast histopathology images with a ductal instance-oriented pipeline. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021. [Google Scholar]
- Tan, X.J.; Mustafa, N.; Mashor, M.Y.; Rahman, K.S. Spatial neighborhood intensity constraint (SNIC) clustering framework for tumor region in breast histopathology images. Multimed. Tools Appl. 2022, 81, 18203–18222. [Google Scholar] [CrossRef]
- Sebai, M.; Wang, T.; Al-Fadhli, S.A. Partmitosis: A partially supervised deep learning framework for mitosis detection in breast cancer histopathology images. IEEE Access 2020, 8, 45133–45147. [Google Scholar] [CrossRef]
- Priego-Torres, B.M.; Sanchez-Morillo, D.; Fernandez-Granero, M.A.; Garcia-Rojo, M. Automatic segmentation of whole-slide H&E stained breast histopathology images using a deep convolutional neural network architecture. Expert Syst. Appl. 2020, 151, 113387. [Google Scholar]
- Belsare, A.D.; Mushrif, M.M.; Pangarkar, M.A.; Meshram, N. Breast histopathology image segmentation using spatio-colour-texture based graph partition method. J. Microsc. 2015, 262, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Hu, X.; Li, Y.; Liu, Q.; Zhu, X. Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process. 2016, 122, 1–13. [Google Scholar] [CrossRef]
- Kaushal, C.; Singla, A. Automated segmentation technique with self-driven post-processing for histopathological breast cancer images. CAAI Trans. Intell. Technol. 2020, 5, 294–300. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Tang, K.; Zhao, Y.; Lu, Z.; Feng, Q. DDTNet: A dense dual-task network for tumor-infiltrating lymphocyte detection and segmentation in histopathological images of breast cancer. Med. Image Anal. 2022, 78, 102415. [Google Scholar] [CrossRef]
- Wahab, N.; Khan, A.; Lee, Y.S. Transfer learning based deep CNN for segmentation and detection of mitoses in breast cancer histopathological images. Microscopy 2019, 68, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.U.; Akhtar, S.; Zakwan, M.; Mahmood, M.H. Novel architecture with selected feature vector for effective classification of mitotic and non-mitotic cells in breast cancer histology images. Biomed. Signal Process. Control 2022, 71, 103212. [Google Scholar] [CrossRef]
- Jiang, H.; Li, S.; Li, H. Parallel ‘same’ and ‘valid’ convolutional block and input-collaboration strategy for Histopathological Image Classification. Appl. Soft Comput. 2022, 117, 108417. [Google Scholar] [CrossRef]
- Karthiga, R.; Narashimhan, K. Deep Convolutional Neural Network for computer-aided detection of breast cancer using histopathology images. J. Phys. Conf. Ser. 2021, 1767, 012042. [Google Scholar] [CrossRef]
- Labrada, A.; Barkana, B.D. Breast cancer diagnosis from histopathology images using supervised algorithms. In Proceedings of the 2022 IEEE 35th International Symposium on Computer-Based Medical Systems (CBMS), Shenzen, China, 21–23 July 2022. [Google Scholar]
- Wang, P.; Wang, J.; Li, Y.; Li, P.; Li, L.; Jiang, M. Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing. Biomed. Signal Process. Control 2021, 65, 102341. [Google Scholar] [CrossRef]
- Yari, Y.; Nguyen, H.; Nguyen, T.V. Accuracy improvement in binary and multiclass classification of breast histopathology images. In Proceedings of the 2020 IEEE Eighth International Conference on Communications and Electronics (ICCE), Phu Quoc Island, Vietnam, 13–15 January 2021. [Google Scholar]
- Yang, H.; Kim, J.-Y.; Kim, H.; Adhikari, S.P. Guided Soft Attention Network for classification of breast cancer histopathology images. IEEE Trans. Med. Imaging 2020, 39, 1306–1315. [Google Scholar] [CrossRef]
- Kode, H.; Barkana, B.D. Deep Learning- and Expert Knowledge-Based Feature Extraction and Performance Evaluation in Breast Histopathology Images. Cancers 2023, 15, 3075. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.; Bhushan, M. Classifying breast cancer using transfer learning models based on histopathological images. Neural Comput. Appl. 2023, 35, 14243–14257. [Google Scholar] [CrossRef]
- Boumaraf, S.; Liu, X.; Wan, Y.; Zheng, Z.; Ferkous, C.; Ma, X.; Li, Z.; Bardou, D. Conventional machine learning versus deep learning for magnification dependent histopathological breast cancer image classification: A comparative study with visual explanation. Diagnostics 2021, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, E.D.; Filho, A.O.C.; Silva, R.R.V.; Araújo, F.H.D.; Diniz, J.O.B.; Silva, A.C.; Paiva, A.C.; Gattass, M. Breast cancer diagnosis from histopathological images using textural features and CBIR. Artif. Intell. Med. 2020, 105, 101845. [Google Scholar] [CrossRef] [PubMed]

| Dataset Name | URL |
|---|---|
| The Breast Cancer Histopathological Image Classification (BreakHis) | https://www.kaggle.com/ambarish/breakhis (accessed on 28 April 2023.) |
| The Kaggle Breast Cancer Histopathology Images | https://www.kaggle.com/paultimothymooney/breast-histopathology-images (accessed on 28 April 2023.) |
| The ICIAR 2018 Grand Challenge on Breast Cancer Histology images (BACH) | https://iciar2018-challenge.grand-challenge.org/Dataset/ (accessed on 28 April 2023.) |
| Tumor Proliferation Assessment Challenge 2016 (TUPAC16) | https://github.com/DeepPathology/TUPAC16_AlternativeLabels (accessed on 28 April 2023.) |
| MITOS-ATYPIA-14 challenge | https://mitos-atypia-14.grand-challenge.org/Dataset/ (accessed on 28 April 2023.) |
| International Conference on Pattern Recognition (ICPR 2012) dataset | http://ludo17.free.fr/mitos_2012/download.html (accessed on 28 April 2023.) |
| Magnification | Benign | Malignant | Total |
|---|---|---|---|
| ×40 | 625 | 1370 | 1995 |
| ×100 | 644 | 1437 | 2081 |
| ×200 | 623 | 1390 | 2013 |
| ×400 | 588 | 1232 | 1820 |
| Total images | 2480 | 5429 | 7909 |
| Magnification | Benign | Malignant | Total |
|---|---|---|---|
| ×40 | 198,738 | 78,786 | 277,524 |
| Magnification | Normal | Benign | In Situ Carcinoma | Invasive Carcinoma | Total |
|---|---|---|---|---|---|
| ×200 | 100 | 100 | 100 | 100 | 400 |
| Score 1 | Score 2 | Score 3 | PAM50 Score (Mean ± STD) | |
|---|---|---|---|---|
| Training | 236 (47%) | 117(23%) | 147(30%) | −0.166 ± 0.446 |
| Testing | 147 (46%) | 77(24%) | 97(30%) | −0.192 ± 0.400 |
| Magnification | Number of Frames | Information |
|---|---|---|
| ×20 | 284 | Nuclear atypia score as a number 1, 2, or 3 |
| ×40 | 1136 | Atypia scoring regarding the size of nuclei, size of nucleoli, the density of chromatin, thickness of the nuclear membrane, regularity of the nuclear contour, and anisonucleosis. |
| Data Sets | Both Scanners | Multispectral Microscope |
|---|---|---|
| Training: 35 HPF | 226 | 224 |
| Evaluation: 15 HPF | 100 | 98 |
| Total | 326 | 322 |
| Work Year | Dataset | Preprocessing | Segmentation | Features | Classifier | Performance |
|---|---|---|---|---|---|---|
| 2023 [77] | BreakHis | - | - | Seven transfer learning models, VGG16, Darknet19, DarkNet53, LENET, ResNet50, Inception, and Xception | - | 2-class: VGG16: 67.51% Darknet19: 80.57% DarkNet53: 70.59% LENET: 75.99% ResNet50: 81.85% Inception: 80.5% Xception: 83.09% |
| 2023 [78] | BreakHis | - | - | Convolutional Neural Network, (2) a transfer learning architecture VGG16 | Neural Network (64 units), Random Forest, Multilayer Perceptron, Decision Tree, Support Vector Machines, K-Nearest Neighbors, and Narrow Neural Network (10 units) | Magnification: 400× CNN achieved up to 85% for the Neural Network and Random Forest, the VGG16 method achieved up to 86% for the Neural Network |
| 2022 [68] | Two public datasets and a new dataset: Bca-lym, Post-NAT-BRCA, TCGA-lym | - | Dense dual-task network (DDTNet) | Spatial and context cues, the multi-scale features with lymphocyte location information | All networks using Pytorch 1.1.0 and a NVIDIA GeForce RTX 2080 Ti GPU | Segmentation performance: Bca-lym dataset: Dice: 85.6% Post-NAT-BRCA dataset: Dice: 83.6% TCGA-lym dataset: Dice: 77.8% |
| 2022 [40] | BreakHis BreCaHAD | Contrast-limited adaptive histogram equalization; Data augmentation | - | Ghost features | Stochastic Dilated Residual Ghost (SDRG) Model including ghost unit, stochastic downsampling, stochastic up-sampling units, and other convolution layers | BreakHis (x40) Original (93.13 ± 4.36) Augmented (98.41 ± 1.00) BreCaHAD Original (95.23 ± 4.38) Augmented (98.60 ± 0.99) |
| 2022 [41] | BreakHis | Stain color normalization by Vahadane method; Random Zoom Augmentation with value 2, Random Rotation Augmentation with a value of 90° and Horizontal and Vertical Flip Augmentation | - | - | Three pre-trained deep convolutional neural networks work in parallel (xception, NASNet, and eptoin_resnet_V2) | The range of threshold values: 50–97% The range of accuracy depending on the threshold value: 96–98% |
| 2022 [60] | Private dataset | Color augmentation, HE-stained and IHC-stained | Segmentation networks: Deeplab_v2, Linknet, Pspnet | - | Domain adaptation framework: Adversarial learning, Target domain data selection, Model refinement, Atrous Spatial Pyramid Pooling | Dice on HE: 87.9% Dice on IHC: 84.6% |
| 2022 [62] | Private dataset of a total of 200 images at 10× magnification | Histogram matching algorithm for color normalization | Spatial neighborhood intensity constraint (SNIC) and knowledge-based clustering framework | Spatial information | K-Mean clustering algorithm | 91.2% |
| 2022 [70] | MITOS 2012 AMIDA 2013 MITOS 2014 TUPAC 2016 | - | - | Three features vector sets Extended Local Pattern features, GLCM features from grayscale, GLCM features from V channel of HSV image | SVM, Random Forest Naïve Bayes Majority voting | MITOS 2012 Majority voting: F score: 95.64% MITOS 2014 Majority voting: F score: 86.38% AMIDA 13 Majority voting: F score: 73.09% TUPAC 16 Majority voting: F score: 78.25% |
| 2022 [71] | DS1, DS2, DS3 | - | - | Step-by-step valid convolutions | Input-collaborative PSV ConvNet | DS2: 90.4–93% |
| 2022 [73] | BreakHis | Histogram Equalization | Otsu’s thresholding method using Red Channel | Geometrical Features Directional Features Intensity-based features | Decision Tree: Fine tree Linear SVM Fine KNN Narrow Neural Network (NNN) | 2 class: NNN: 96.9% |
| 2021 [72] | BreakHis | - | - | DCNN | Alexnet, VGG-16 Transfer learning methods, DCNN | 2-class: 40×: 94% 100×: 95.45% 200×: 98.36% 400×: 85.71% |
| 2021 [42] | BreakHis | Global contrast normalization; Three-fold data augmentation on training data | - | ResNet-18 | Transfer learning based on block-wise fine-tuning strategy | MI classification: Binary: 98.42% Eight-class: 92.03% MD classification: Binary: 98.84% Eight-class: 92.15% |
| 2021 [54] | The HUP 239 images, CINJ 40 images and TCGS 195, CWRU 110 images | Reduced image size, RGB to grayscale conversion, smoothing by Gaussian Filter | - | Unsupervised pre-training and supervised fine-tuning phase | Patch-based deep learning method called Pa-DBN-BC, Deep Belief Network (DBN), Logistic regressions | Overall: 86% |
| 2021 [74] | BreaKHis | - | - | Convolution and capsule features Integrated sematic and special features | Deep feature fusion and enhanced routing, FE-BkCapsNet | 2-class: 40×: 92.71% 100×: 94.52% 200×: 94.03% 400×: 93.54% |
| 2021 [79] | BreaKHis | Color normalization technique | - | Feature Extraction-Based CML Approaches, Zernike moments, Haralick, and color histogram features | Conventional machine learning (CML) and deep learning (DL)-based methods | 2-class: DL: 94.05–98.13% CML: 85.65–89.32% 8-class: DL: 76.77–88.95% CML: 63.55–69.69% |
| 2020 [67] | Two small datasets: 50 images of 11 patients; 30 H&E marked 40× magnified images | Median filter, Bottom + Top Hat filter | Identifying thresholds based on the energy curve, finding the best threshold using the entropy | Area, major axis length, minor axis length | - | Dataset 1: 93.1% Dataset 2: 93.5% |
| 2020 [76] | BACH dataset | Data augmentation by color normalization, vertical and horizontal mirroring, random rotations, addition of random noise and random change in intensity of the images | - | CNN-based feature extraction network | Region Guided Soft Attention | 90.25% |
| 2020 [80] | BACH 2018 | - | - | Indexes based on phylogenetic diversity. | SVM, Random Forest MLP XGBoost | 4-class: 95% |
| 2020 [55] | Private dataset of 8009 histopathology images from over 683 patients with different magnification levels | Gaussian filtering technique for noise removal, data augmentation by rotation | Histo-sigmoid-based fuzzy clustering | - | Deep Neural Network with Support Value (DNNS) | 97.21% |
| 2020 [44] | Private dataset | Data augmentation | - | Multi-level and multiscale deep features | Ensemble of fine-tuned VGG16 and fined tuned VGG19 | Up to 95.29% |
| 2020 [52] | BreakHis | Data augmentation, random horizontal flip, color jitter, random rotation, and crop | - | Feature maps | Deep transfer learning-based models: DensNet and ResNet, ResNet101, VGG19, AlexNet, and SqueezeNet | 2-class: BreakHis (40×): 100% BreakHis (100×): 100% BreakHis (200×): 98.08% BreakHis (400×): 98.99% Multi-class: BreakHis (40×): 97.96% BreakHis (100×): 97.14% BreakHis (200×): 95.19% BreakHis (400×): 94.95% |
| 2020 [61] | Private dataset consists of 428 images from 240 breast biopsies | - | Ductal Instance-Oriented Pipeline (DIOP) segmentation model: a duct-level instance segmentation model, tissue-level semantic segmentation model, three levels of features | Histogram features Co-occurrence features Structural features | Random forest model, 3-degree polynomial SVM SVM-RBF Multilayer perception with four hidden layers | 2-class: Invasive vs. non invasive: 95% Atypia and DCIS vs Benign: 79% DCIS vs. Atypia: 90% Multi-class: 70% |
| 2020 [63] | ICPR 2012 MITOSIS Dataset, 2014 ICPR dataset, and the AMIDA13 dataset | - | Segmentation branch trained with weak and strong labels | Convolution features | Pre-trained and fine-tuned Partially supervised framework based on two parallel, deep fully convolutional networks | 2012 ICPR MITOSIS dataset F-scores: 0.788 2014 ICPR dataset: F-scores: 0.575 AMIDA13 dataset: F-scores: 0.698 |
| 2020 [64] | Dataset of 640 H&E-stained breast histopathology images | Data augmentation by random zooming, cropping, horizontal and vertical flips | A tile-wise segmentation strategy, (a) direct tile-wise merging; (b) tile-wise merging based on a Conditional Random Field (CRF) | - | DCNN-based architecture | Xception 65: 95.62% Mobilenet v2: 92.9% Resnet v1: 91.16% |
| 2019 [45] | -Bioimaging-2015 -BreakHis | Stain color normalization; Logarithmic transformation; Data Augmentation | - | Ensemble of DCNNs | Gradient boosting trees classifier | Bioimaging-2015 (4-class): 96.4% Bioimaginf-2015 (2-class): 99.5% BreakHis (40×): 95.1% BreakHis (100×): 96.3% BreakHis (200×): 96.9% BreakHis (400×): 93.8% |
| 2019 [46] | ICIAR 2018 BreakHis | Stain color normalization; Image decomposition via Haar wavelet; Data Augmentation | - | Deep features from Haar wavelet decomposed images by a CNN model; Incorporation of multiscale discriminant features | Three fully connected two Dropout and SoftMax layers | ICIAR 2018 (2 and 4-class): 98.2% BreakHis (Multi-class): 96.85% |
| 2019 [50] | Data Augmentation | - | Feature vectors | CNN with IDC patch-based classification | 85.41% | |
| 2019 [57] | BreakHis | Contrast enhancement by histogram Equalization, color constancy | - | CNN features | 5 Convolutional layers Fully connected and SoftMax layer | Hist. Equalization with the proposed method: AUC: 87.6% Color constancy with the proposed method: AUC: 93.5% |
| 2019 [58] | Bioimaging Challenge 2015 | Singular value decomposition (SVD), Logarithmic transformation | - | CNN based on the SE-ResNet module GoogleNet, Xception, Inception-ResNet, 3-Norm pooling method | KNN SVM | SVM-GoogleLeNet 2-class: 91.67% 4-class: 83.33% |
| 2019 [69] | TUPAC 16 MITOS12 + MITOS14 | Stain normalization Annotation Cropping | Transfer Learning-based Mitosis Segmentation (TL-Mit-Seg) | - | Hybrid-CNN based mitosis detection module (HCNN-Mit-Det); HCNN-Mit-Det-essemble; Transfer learning HCNN-Mit-Det | TUPAC 16: F-measure: 66.7% MITOS12 + MITOS14 F-measure: 65.1% |
| 2018 [48] | ICIAR 2018 | Data Augmentation: 50 random color augmentations; different image scales | - | ResNet-50, InceptionV3 and VGG-16 networks from Keras distribution | Gradient boosted trees classifier | 2-class: 93.8% 4-class: 87.2% |
| 2017 [51] | BreakHis | Data Augmentation randomly distorted images, rotated and mirrored images | - | Transfer learning Google Inception v3 | Deep convolutional neural network(CNN, ConvNet) model | 83% for benign class 89% for malignant class |
| 2015 [65] | Private dataset of 100 malignant and nonmalignant breast histology images | - | Spatial-color-texture-based graph partitioning method | Intensity-texture features Color texture features | - | |
| 2015 [66] | 68 BCH images containing more than 3600 cells. | Top-bottom hat transform | Wavelet decomposition and multiscale region growing | 4 shape-based features and 138 textural features based on color spaces, wrapper feature selection algorithm based on chain-like agent genetic algorithm (CAGA) | SVM | Normal vs. malignant: 96.19 ± 0.31% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labrada, A.; Barkana, B.D. A Comprehensive Review of Computer-Aided Models for Breast Cancer Diagnosis Using Histopathology Images. Bioengineering 2023, 10, 1289. https://doi.org/10.3390/bioengineering10111289
Labrada A, Barkana BD. A Comprehensive Review of Computer-Aided Models for Breast Cancer Diagnosis Using Histopathology Images. Bioengineering. 2023; 10(11):1289. https://doi.org/10.3390/bioengineering10111289
Chicago/Turabian StyleLabrada, Alberto, and Buket D. Barkana. 2023. "A Comprehensive Review of Computer-Aided Models for Breast Cancer Diagnosis Using Histopathology Images" Bioengineering 10, no. 11: 1289. https://doi.org/10.3390/bioengineering10111289
APA StyleLabrada, A., & Barkana, B. D. (2023). A Comprehensive Review of Computer-Aided Models for Breast Cancer Diagnosis Using Histopathology Images. Bioengineering, 10(11), 1289. https://doi.org/10.3390/bioengineering10111289








