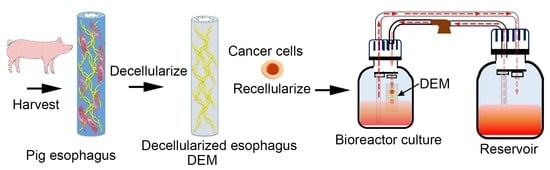
A New Model of Esophageal Cancers by Using a Detergent-Free Decellularized Matrix in a Perfusion Bioreactor
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Decellularization of Porcine Esophagus
2.3. Characterization of Decellularized Esophageal Matrix
2.3.1. Decellularization
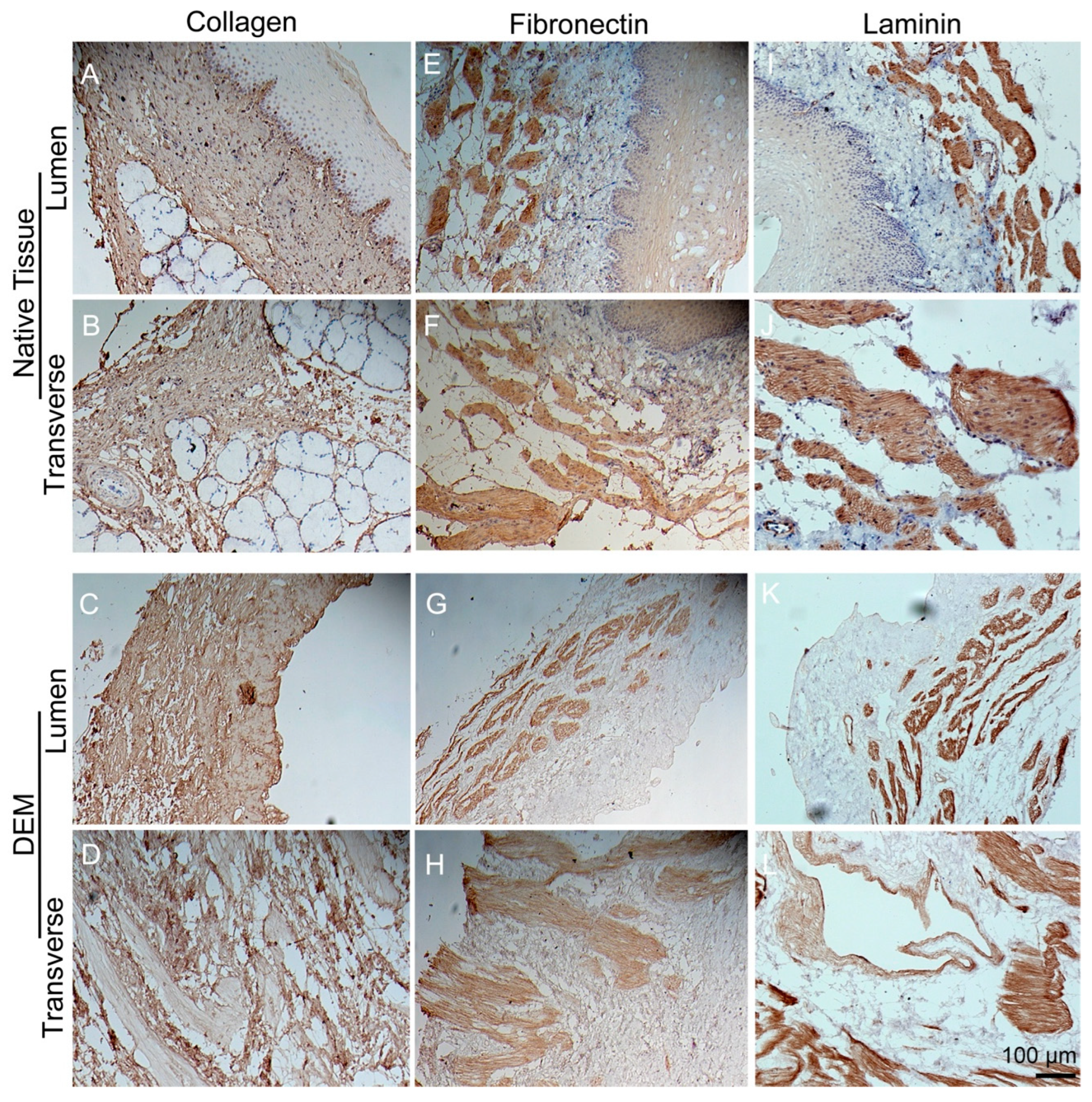
2.3.2. Matrix Proteins
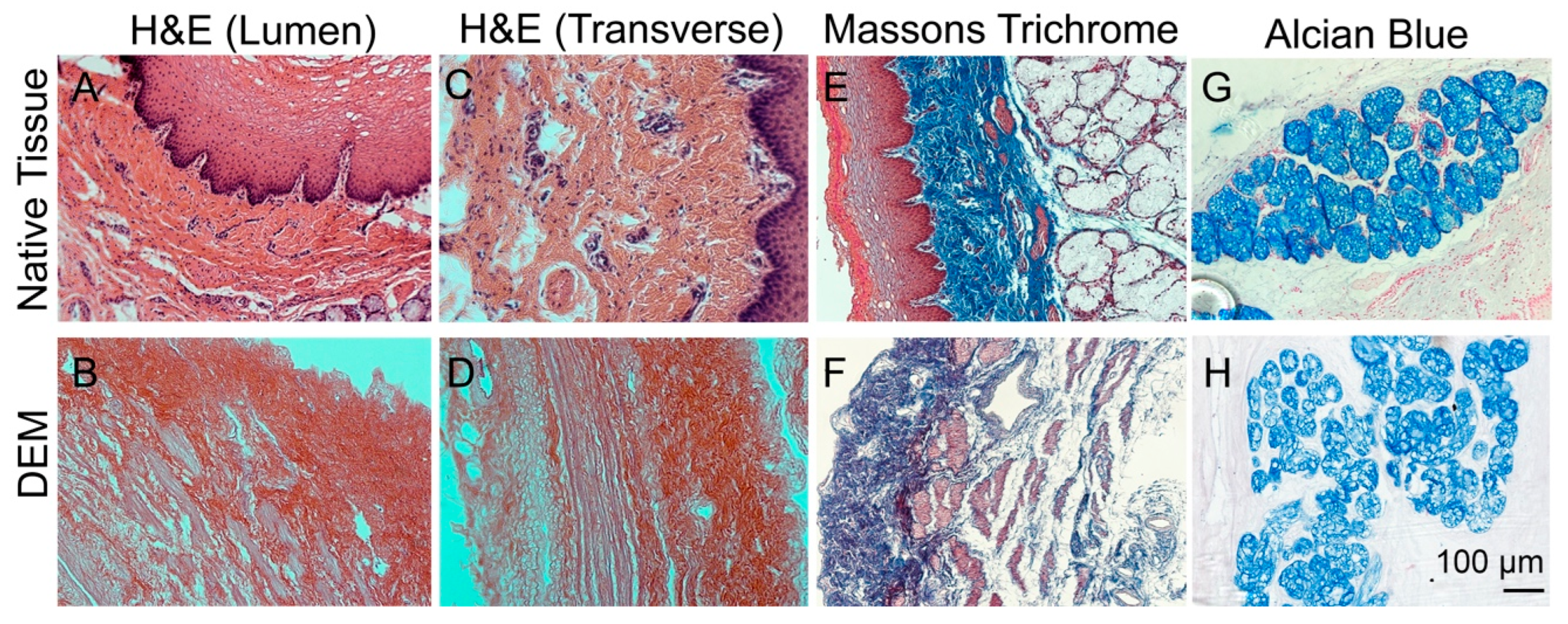
2.3.3. Masson Trichrome and Alcian Staining
2.4. Cell Viability on the Decellularized Matrix
2.5. H&E and Alcian Blue Staining
2.6. Immunohistochemistry Staining
2.7. Statistical Analysis
3. Results
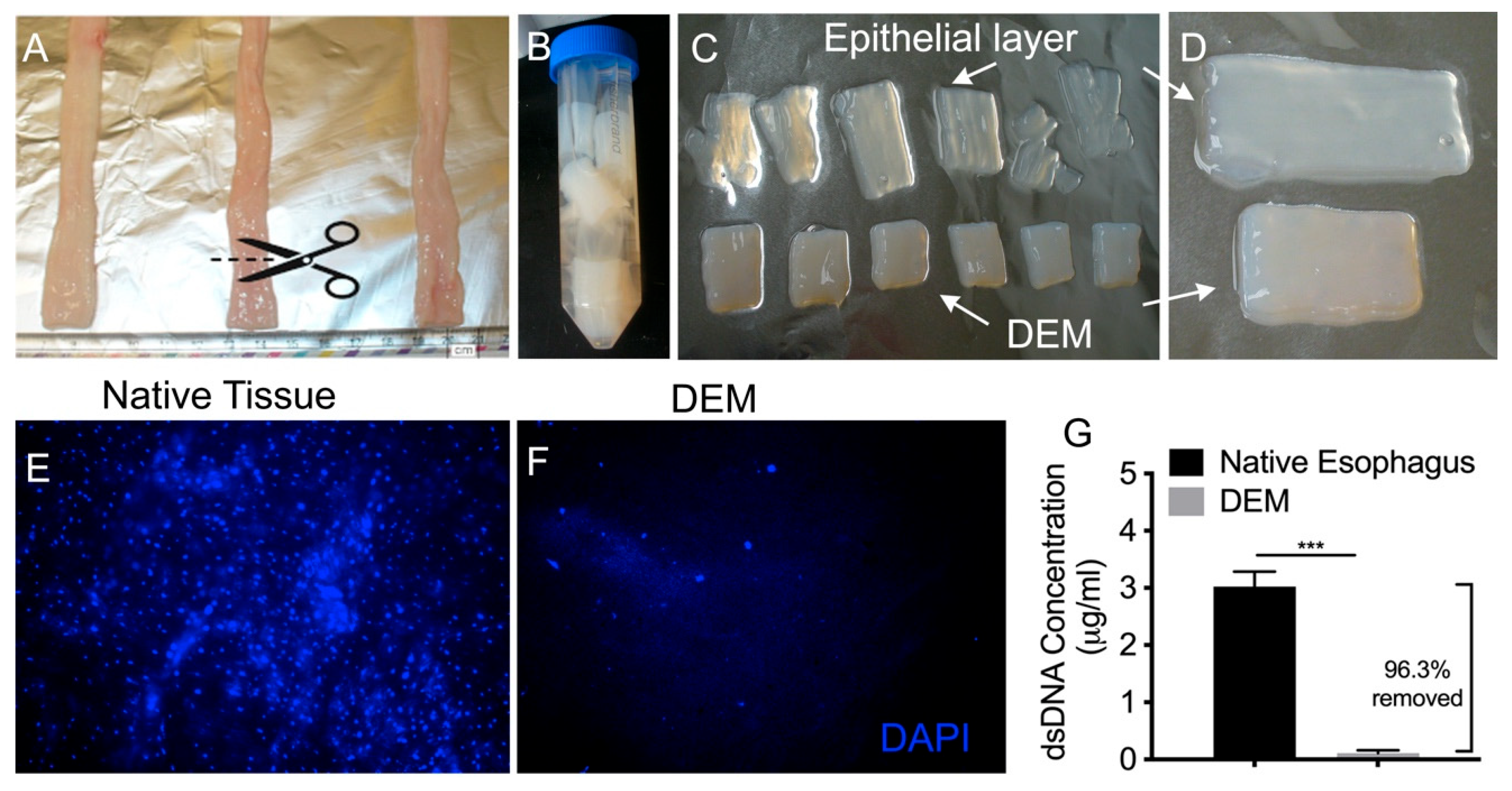
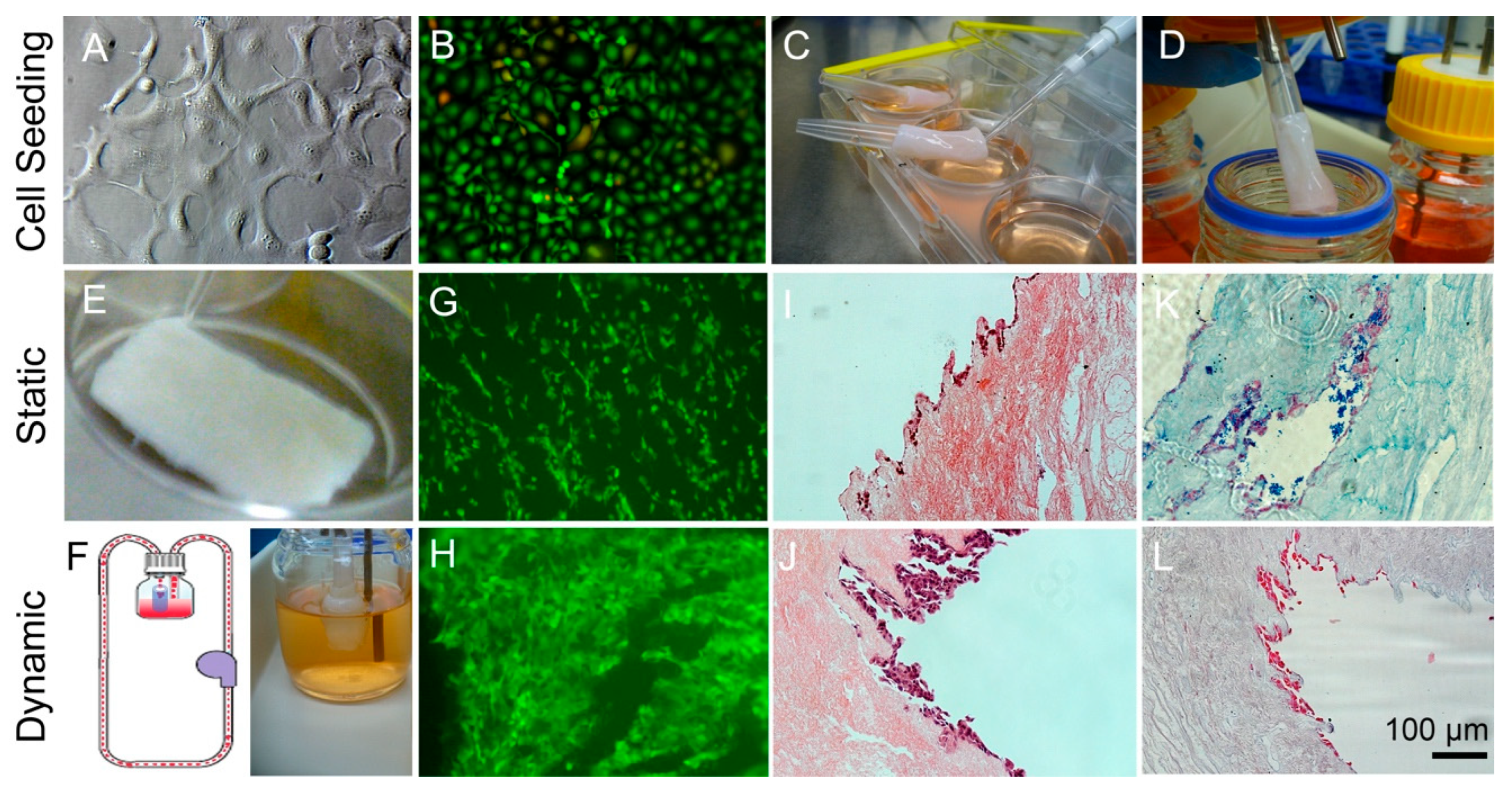
3.1. Decellularization of Esophageal Tissue
3.2. Characterization of Decellularized Esophageal Matrix
3.3. Immunohistochemistry Staining of Matrix
3.4. Compare Static Well Plate Culture to Dynamic Flow Culture Tissue Samples
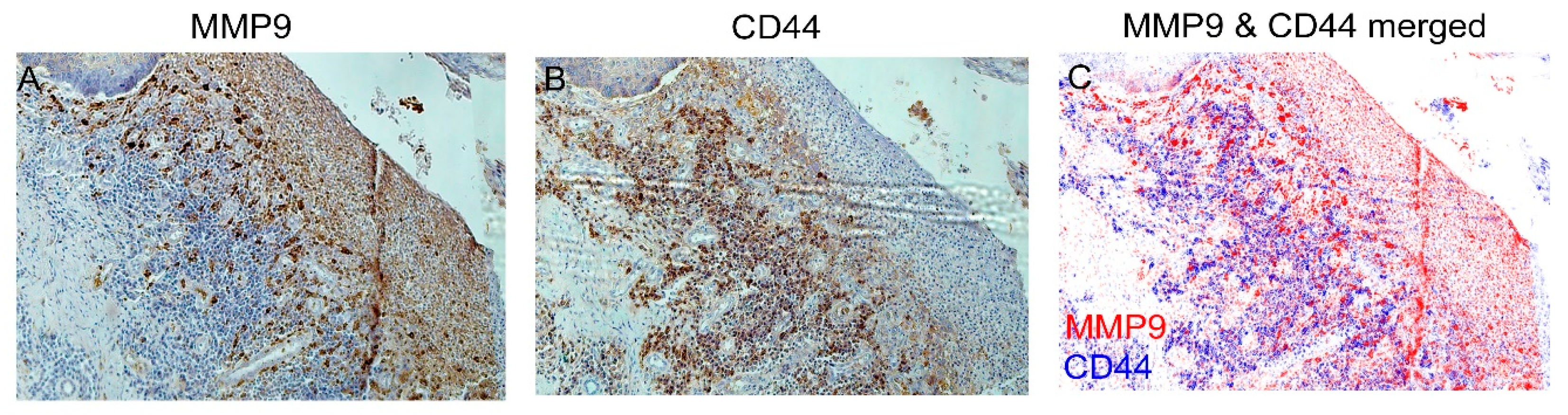
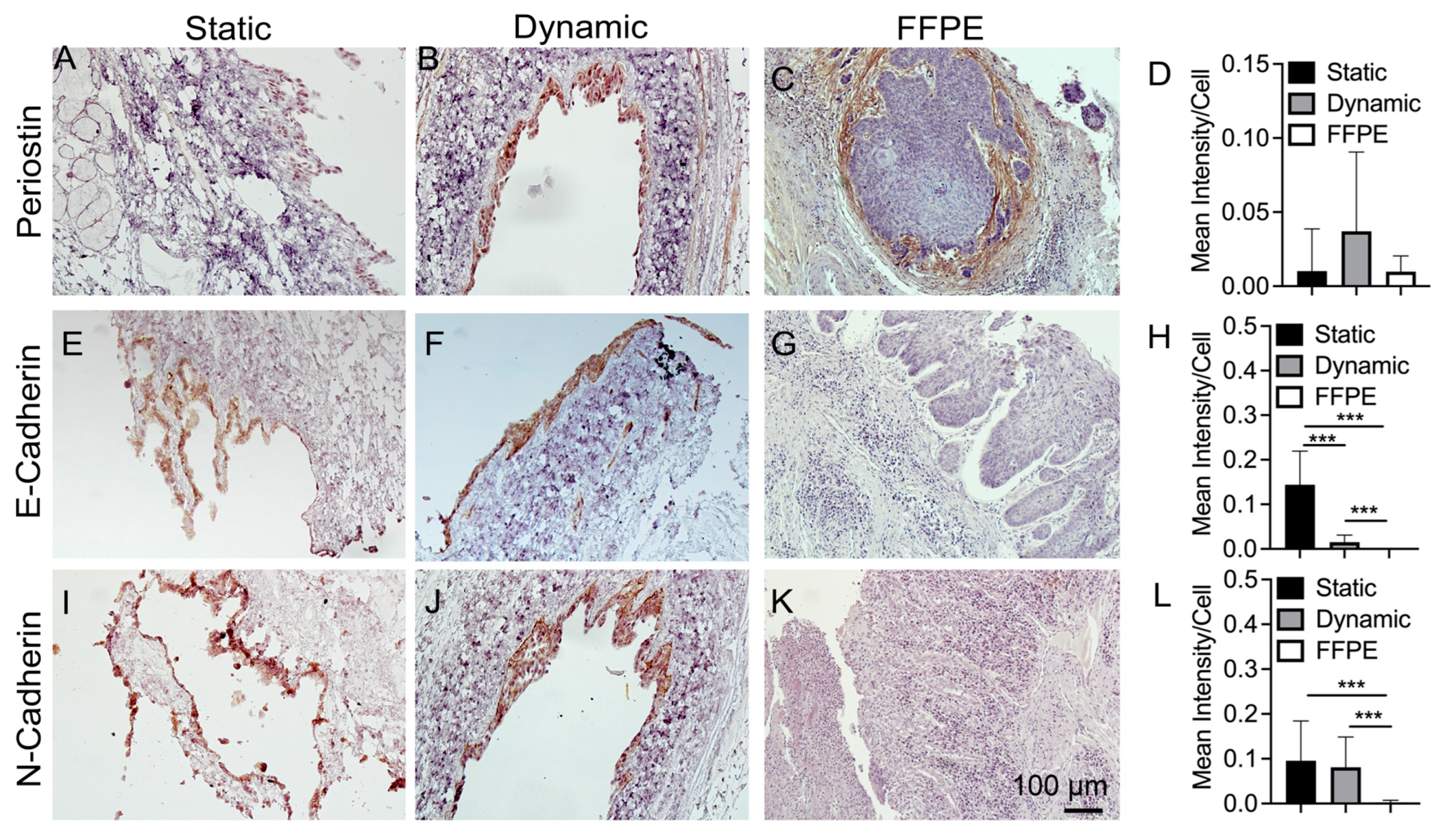
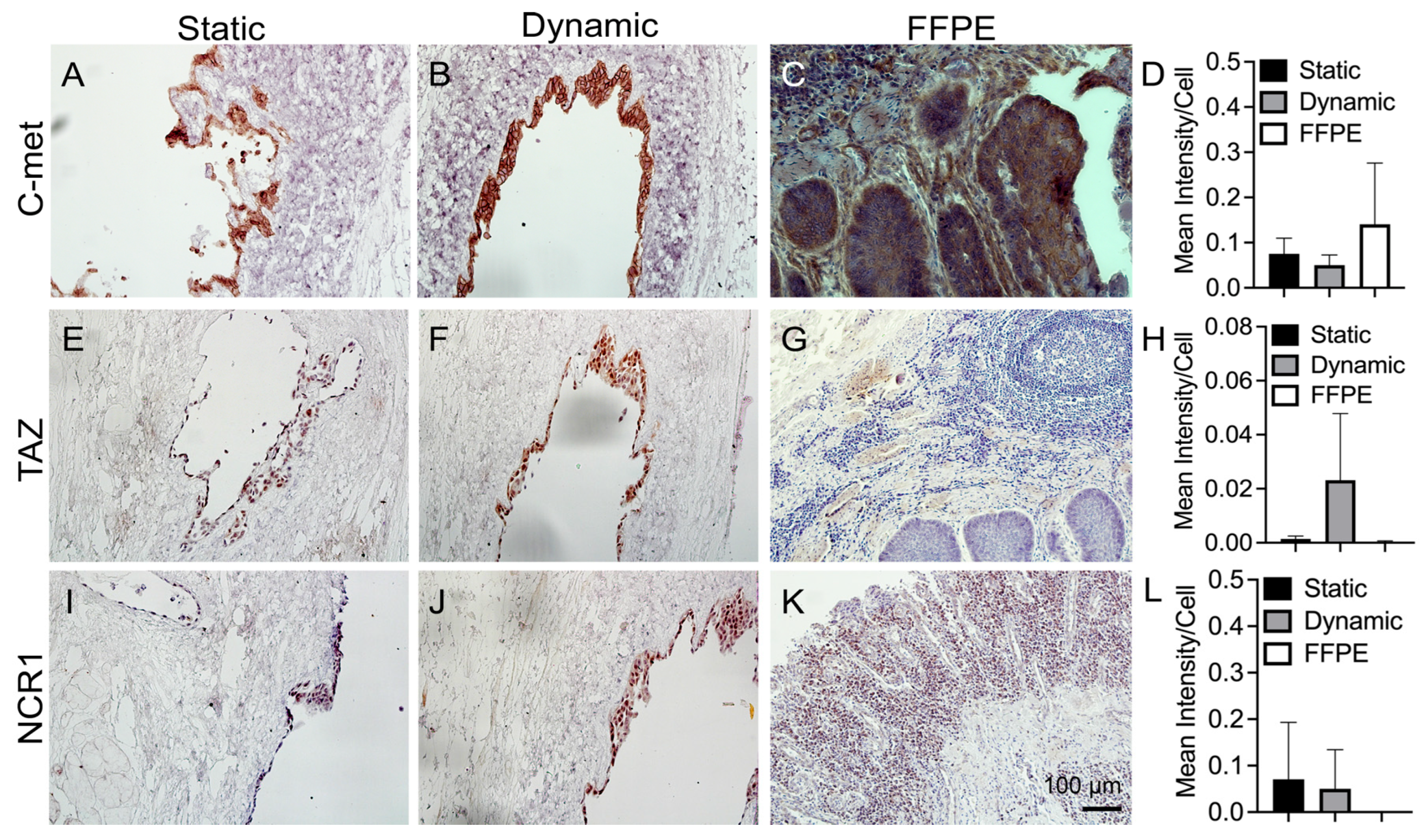
3.5. IHC Staining of Cancer Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Talukdar, F.R.; di Pietro, M.; Secrier, M.; Moehler, M.; Goepfert, K.; Lima, S.S.C.; Pinto, L.F.R.; Hendricks, D.; Parker, M.I.; Herceg, Z. Molecular landscape of esophageal cancer: Implications for early detection and personalized therapy. Ann. N. Y. Acad. Sci. 2018, 1434, 342–359. [Google Scholar] [CrossRef]
- Alsop, B.R.; Sharma, P. Esophageal Cancer. Gastroenterol. Clin. N. Am. 2016, 45, 399–412. [Google Scholar] [CrossRef]
- Vaghjiani, R.G.; Molena, D. Surgical management of esophageal cancer. Chin. Clin. Oncol. 2017, 6, 47. [Google Scholar] [CrossRef]
- Cheng, X.; Wei, L.; Huang, X.; Zheng, J.; Shao, M.; Feng, T.; Li, J.; Han, Y.; Tan, W.; Tan, W.; et al. Solute Carrier Family 39 Member 6 Gene Promotes Aggressiveness of Esophageal Carcinoma Cells by Increasing Intracellular Levels of Zinc, Activating Phosphatidylinositol 3-Kinase Signaling, and Up-regulating Genes That Regulate Metastasis. Gastroenterology 2017, 152, 1985–1997. [Google Scholar] [CrossRef]
- Liu, R.M.; Sun, D.N.; Jiao, Y.L.; Wang, P.; Zhang, J.; Wang, M.; Ma, J.; Sun, M.; Gu, B.L.; Chen, P.; et al. Macrophage migration inhibitory factor promotes tumor aggressiveness of esophageal squamous cell carcinoma via activation of Akt and inactivation of GSK3beta. Cancer Lett. 2018, 412, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Yang, J.; Guo, J.; Su, Y.; He, C.; Wu, J.; Yu, L.; Ding, W.Q.; Zhou, J. RAD18 promotes the migration and invasion of esophageal squamous cell cancer via the JNK-MMPs pathway. Cancer Lett. 2018, 417, 65–74. [Google Scholar] [CrossRef]
- Padron, J.M.; Peters, G.J. Cytotoxicity of sphingoid marine compound analogs in mono- and multilayered solid tumor cell cultures. Investig. New Drugs 2006, 24, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Weaver, V.M.; Lelievre, S.; Lakins, J.N.; Chrenek, M.A.; Jones, J.C.; Giancotti, F.; Werb, Z.; Bissell, M.J. β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2002, 2, 205–216. [Google Scholar] [CrossRef]
- Fischbach, C.; Chen, R.; Matsumoto, T.; Schmelzle, T.; Brugge, J.S.; Polverini, P.J.; Mooney, D.J. Engineering tumors with 3D scaffolds. Nat. Methods 2007, 4, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Kaminou, T.; Ohuchi, Y.; Sugiura, K.; Yata, S.; Adachi, A.; Kawai, T.; Takasugi, S.; Yamamoto, S.; Matsumoto, K.; et al. Development of a new hanging-type esophageal stent for preventing migration: A preliminary study in an animal model of esophagotracheal fistula. Cardiovasc. Interv. Radiol. 2012, 35, 1188–1194. [Google Scholar] [CrossRef]
- Baron, T.H.; Burgart, L.J.; Pochron, N.L. An internally covered (lined) self-expanding metal esophageal stent: Tissue response in a porcine model. Gastrointest. Endosc. 2006, 64, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Burden, N.; Sewell, F.; Chapman, K. Testing Chemical Safety: What Is Needed to Ensure the Widespread Application of Non-animal Approaches? PLoS Biol. 2015, 13, e1002156. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Yeong, W.Y. The future of skin toxicology testing—Three-dimensional bioprinting meets microfluidics. Int. J. Bioprint. 2019, 5, 237. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Jin, L.; Xie, N.; Luo, P.; Gao, W.; Tu, P.; Ai, X. Establishment and large-scale validation of a three-dimensional tumor model on an array chip for anticancer drug evaluation. Front. Pharmacol. 2022, 13, 1032975. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Fong, E.L.S.; Zhu, C.; Lin, Q.X.X.; Xiong, M.; Li, A.; Li, T.; Benoukraf, T.; Yu, H.; Liu, S. Hydrogel-based colorectal cancer organoid co-culture models. Acta Biomater. 2021, 132, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Paschall, A.V.; Liu, K. An Orthotopic Mouse Model of Spontaneous Breast Cancer Metastasis. J. Vis. Exp. JoVE 2016, 114, 54040. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Zhang, M.; Boughton, P.; Rose, B.; Lee, C.S.; Hong, A.M. The use of porous scaffold as a tumor model. Int. J. Biomater. 2013, 2013, 396056. [Google Scholar] [CrossRef]
- Chen, H.; Cheng, Y.; Wang, X.; Wang, J.; Shi, X.; Li, X.; Tan, W.; Tan, Z. 3D printed in vitro tumor tissue model of colorectal cancer. Theranostics 2020, 10, 12127–12143. [Google Scholar] [CrossRef] [PubMed]
- Buschhaus, J.M.; Luker, K.E.; Luker, G.D. A Facile, In Vitro 384-Well Plate System to Model Disseminated Tumor Cells in the Bone Marrow Microenvironment. Methods Mol. Biol. 2018, 1686, 201–213. [Google Scholar] [CrossRef]
- Zhuravleva, M.; Gilazieva, Z.; Grigoriev, T.E.; Shepelev, A.D.; Kh Tenchurin, T.; Kamyshinsky, R.; Krasheninnikov, S.V.; Orlov, S.; Caralogli, G.; Archipova, S.; et al. In vitro assessment of electrospun polyamide-6 scaffolds for esophageal tissue engineering. J. Biomed. Mater. Res. B Appl. Biomater. 2019, 107, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.; Blanchette, A.; Vadasz, S.; Dave, A.; Canfarotta, M.; Sayej, W.N.; Finck, C. Biomimetic and synthetic esophageal tissue engineering. Biomaterials 2015, 57, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi Sadrabadi, A.; Baei, P.; Hosseini, S.; Baghaban Eslaminejad, M. Decellularized Extracellular Matrix as a Potent Natural Biomaterial for Regenerative Medicine. Adv. Exp. Med. Biol. 2021, 1341, 27–43. [Google Scholar] [CrossRef]
- Keane, T.J.; Londono, R.; Carey, R.M.; Carruthers, C.A.; Reing, J.E.; Dearth, C.L.; D’Amore, A.; Medberry, C.J.; Badylak, S.F. Preparation and characterization of a biologic scaffold from esophageal mucosa. Biomaterials 2013, 34, 6729–6737. [Google Scholar] [CrossRef] [PubMed]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef]
- Ganz, R.A.; Utley, D.S.; Stern, R.A.; Jackson, J.; Batts, K.P.; Termin, P. Complete ablation of esophageal epithelium with a balloon-based bipolar electrode: A phased evaluation in the porcine and in the human esophagus. Gastrointest. Endosc. 2004, 60, 1002–1010. [Google Scholar] [CrossRef]
- Cornelison, R.C.; Wellman, S.M.; Park, J.H.; Porvasnik, S.L.; Song, Y.H.; Wachs, R.A.; Schmidt, C.E. Development of an apoptosis-assisted decellularization method for maximal preservation of nerve tissue structure. Acta Biomater. 2018, 77, 116–126. [Google Scholar] [CrossRef]
- Lee, J.S.; Choi, Y.S.; Cho, S.W. Decellularized Tissue Matrix for Stem Cell and Tissue Engineering. Adv. Exp. Med. Biol. 2018, 1064, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.J.; Swinehart, I.T.; Badylak, S.F. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods 2015, 84, 25–34. [Google Scholar] [CrossRef]
- Pan, M.X.; Hu, P.Y.; Cheng, Y.; Cai, L.Q.; Rao, X.H.; Wang, Y.; Gao, Y. An efficient method for decellularization of the rat liver. J. Formos. Med. Assoc. 2014, 113, 680–687. [Google Scholar] [CrossRef]
- Naderi, S.; Akbar Zadeh, M.; Mahdavi Shahri, N.; Matin, M.M.; Fereidoni, M.; Haghighitalab, A. Characterization of rat esophagus decellularized scaffold inductive effect on arrangement of human adipose derived mesenchymal Stem Cells. J. North Khorasan Univ. Med. Sci. 2013, 5, 139–145. [Google Scholar] [CrossRef]
- Beckstead, B.L.; Pan, S.; Bhrany, A.D.; Bratt-Leal, A.M.; Ratner, B.D.; Giachelli, C.M. Esophageal epithelial cell interaction with synthetic and natural scaffolds for tissue engineering. Biomaterials 2005, 26, 6217–6228. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.M.; Bissell, M.J. Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2006, 22, 287–309. [Google Scholar] [CrossRef]
- Mollica, P.A.; Booth-Creech, E.N.; Reid, J.A.; Zamponi, M.; Sullivan, S.M.; Palmer, X.L.; Sachs, P.C.; Bruno, R.D. 3D bioprinted mammary organoids and tumoroids in human mammary derived ECM hydrogels. Acta Biomater. 2019, 95, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Mayorca-Guiliani, A.E.; Willacy, O.; Madsen, C.; Rafaeva, M.; Elisabeth Heumüller, S.; Bock, F.; Sengle, G.; Koch, M.; Imhof, T.; Zaucke, F.; et al. Decellularization and antibody staining of mouse tissues to map native extracellular matrix structures in 3D. Nat. Protoc. 2019, 14, 3395–3425. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.J.; DeWard, A.; Londono, R.; Saldin, L.T.; Castleton, A.A.; Carey, L.; Nieponice, A.; Lagasse, E.; Badylak, S.F. Tissue-Specific Effects of Esophageal Extracellular Matrix. Tissue Eng. Part A 2015, 21, 2293–2300. [Google Scholar] [CrossRef] [PubMed]
- Tölg, C.; Hofmann, M.; Herrlich, P.; Ponta, H. Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res. 1993, 21, 1225–1229. [Google Scholar] [CrossRef]
- Naor, D.; Nedvetzki, S.; Golan, I.; Melnik, L.; Faitelson, Y. CD44 in cancer. Crit. Rev. Clin. Lab. Sci. 2002, 39, 527–579. [Google Scholar] [CrossRef]
- Desai, B.; Ma, T.; Zhu, J.; Chellaiah, M.A. Characterization of the expression of variant and standard CD44 in prostate cancer cells: Identification of the possible molecular mechanism of CD44/MMP9 complex formation on the cell surface. J. Cell. Biochem. 2009, 108, 272–284. [Google Scholar] [CrossRef]
- Aalinkeel, R.; Nair, B.B.; Reynolds, J.L.; Sykes, D.E.; Mahajan, S.D.; Chadha, K.C.; Schwartz, S.A. Overexpression of MMP-9 contributes to invasiveness of prostate cancer cell line LNCaP. Immunol. Investig. 2011, 40, 447–464. [Google Scholar] [CrossRef]
- Moses, M.A.; George, A.L.; Sakakibara, N.; Mahmood, K.; Ponnamperuma, R.M.; King, K.E.; Weinberg, W.C. Molecular Mechanisms of p63-Mediated Squamous Cancer Pathogenesis. Int. J. Mol. Sci. 2019, 20, 3590. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.L.; Balinth, S.; Mills, A.A. p63-related signaling at a glance. J. Cell Sci. 2020, 133, jcs228015. [Google Scholar] [CrossRef] [PubMed]
- González-González, L.; Alonso, J. Periostin: A Matricellular Protein with Multiple Functions in Cancer Development and Progression. Front. Oncol. 2018, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Morra, L.; Moch, H. Periostin expression and epithelial-mesenchymal transition in cancer: A review and an update. Virchows Arch. Int. J. Pathol. 2011, 459, 465–475. [Google Scholar] [CrossRef]
- Hu, W.W.; Chen, P.C.; Chen, J.M.; Wu, Y.M.; Liu, P.Y.; Lu, C.H.; Lin, Y.F.; Tang, C.H.; Chao, C.C. Periostin promotes epithelial-mesenchymal transition via the MAPK/miR-381 axis in lung cancer. Oncotarget 2017, 8, 62248–62260. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Safaie Qamsari, E.; Safaei Ghaderi, S.; Zarei, B.; Dorostkar, R.; Bagheri, S.; Jadidi-Niaragh, F.; Somi, M.H.; Yousefi, M. The c-Met receptor: Implication for targeted therapies in colorectal cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017, 39, 1010428317699118. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, J.; Park, S.H.; Park, J.O.; Lim, H.Y.; Kang, W.K.; Park, Y.S.; Kim, S.T. c-MET Overexpression in Colorectal Cancer: A Poor Prognostic Factor for Survival. Clin. Color. Cancer 2018, 17, 165–169. [Google Scholar] [CrossRef]
- Zhou, X.; Lei, Q.Y. Regulation of TAZ in cancer. Protein Cell 2016, 7, 548–561. [Google Scholar] [CrossRef]
- Noguchi, S.; Saito, A.; Nagase, T. YAP/TAZ Signaling as a Molecular Link between Fibrosis and Cancer. Int. J. Mol. Sci. 2018, 19, 3674. [Google Scholar] [CrossRef]
- Koch, J.; Steinle, A.; Watzl, C.; Mandelboim, O. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 2013, 34, 182–191. [Google Scholar] [CrossRef]
- White, L.J.; Taylor, A.J.; Faulk, D.M.; Keane, T.J.; Saldin, L.T.; Reing, J.E.; Swinehart, I.T.; Turner, N.J.; Ratner, B.D.; Badylak, S.F. The impact of detergents on the tissue decellularization process: A ToF-SIMS study. Acta Biomater. 2017, 50, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Roosens, A.; Somers, P.; De Somer, F.; Carriel, V.; Van Nooten, G.; Cornelissen, R. Impact of Detergent-Based Decellularization Methods on Porcine Tissues for Heart Valve Engineering. Ann. Biomed. Eng. 2016, 44, 2827–2839. [Google Scholar] [CrossRef]
- Nayakawde, N.B.; Methe, K.; Banerjee, D.; Berg, M.; Premaratne, G.U.; Olausson, M. In Vitro Regeneration of Decellularized Pig Esophagus Using Human Amniotic Stem Cells. Biores. Open Access 2020, 9, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Yosef, G.; Hayun, H.; Papo, N. Simultaneous targeting of CD44 and MMP9 catalytic and hemopexin domains as a therapeutic strategy. Biochem. J. 2021, 478, 1139–1157. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Kim, J.M.; Lee, H.J.; Seong, I.O.; Kim, K.H. Immunohistochemical expression of CD44, matrix metalloproteinase2 and matrix metalloproteinase9 in renal cell carcinomas. Urol. Oncol. 2019, 37, 742–748. [Google Scholar] [CrossRef]
- Ma, J.; Li, M.; Chai, J.; Wang, K.; Li, P.; Liu, Y.; Zhao, D.; Xu, J.; Yu, K.; Yan, Q.; et al. Expression of RSK4, CD44 and MMP-9 is upregulated and positively correlated in metastatic ccRCC. Diagn. Pathol. 2020, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Spessotto, P.; Rossi, F.M.; Degan, M.; Di Francia, R.; Perris, R.; Colombatti, A.; Gattei, V. Hyaluronan-CD44 interaction hampers migration of osteoclast-like cells by down-regulating MMP-9. J. Cell Biol. 2002, 158, 1133–1144. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; AlJohani, H.; AlQranei, M.; Majumdar, S.; Ma, T.; Chellaiah, M.A. Identification of sequence-specific interactions of the CD44-intracellular domain with RUNX2 in the transcription of matrix metalloprotease-9 in human prostate cancer cells. Cancer Drug Resist. (Alhambra Calif.) 2020, 3, 586–602. [Google Scholar] [CrossRef] [PubMed]
- Miletti-González, K.E.; Murphy, K.; Kumaran, M.N.; Ravindranath, A.K.; Wernyj, R.P.; Kaur, S.; Miles, G.D.; Lim, E.; Chan, R.; Chekmareva, M.; et al. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): Modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J. Biol. Chem. 2012, 287, 18995–19007. [Google Scholar] [CrossRef] [PubMed]
- Kii, I. Periostin Functions as a Scaffold for Assembly of Extracellular Proteins. Adv. Exp. Med. Biol. 2019, 1132, 23–32. [Google Scholar] [CrossRef]
- Heidari, P.; Esfahani, S.A.; Turker, N.S.; Wong, G.; Wang, T.C.; Rustgi, A.K.; Mahmood, U. Imaging of Secreted Extracellular Periostin, an Important Marker of Invasion in the Tumor Microenvironment in Esophageal Cancer. J. Nucl. Med. 2015, 56, 1246–1251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, M.; Jin, K.; Wang, S.; Wei, H.; Fan, C.; Wu, Y.; Li, X.; Li, X.; Li, G.; et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 2018, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Peng, Z.; Li, Z.; Lu, M.; Gao, J.; Li, Y.; Li, Y.; Shen, L. Expression and clinical significance of c-Met in advanced esophageal squamous cell carcinoma. BMC Cancer 2015, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.J.; Shi, Z.Z.; Zhao, Z.X.; Zhang, Y.; Gong, T.; Li, C.X.; Zhan, T.; Cai, Y.; Dong, J.T.; Fu, S.B.; et al. Characterization of genetic rearrangements in esophageal squamous carcinoma cell lines by a combination of M-FISH and array-CGH: Further confirmation of some split genomic regions in primary tumors. BMC Cancer 2012, 12, 367. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Ewald, A.J. Illuminating breast cancer invasion: Diverse roles for cell-cell interactions. Curr. Opin. Cell Biol. 2014, 30, 99–111. [Google Scholar] [CrossRef]
- Hofschröer, V.; Koch, A.; Ludwig, F.T.; Friedl, P.; Oberleithner, H.; Stock, C.; Schwab, A. Extracellular protonation modulates cell-cell interaction mechanics and tissue invasion in human melanoma cells. Sci. Rep. 2017, 7, 42369. [Google Scholar] [CrossRef]
- Huang, H.M.; He, X.H.; Huang, X.Y.; Wang, G.Y.; Xia, Q.X.; Du, Z.P.; Zhang, Y.F. Down-regulation of kappa opioid receptor promotes ESCC proliferation, invasion and metastasis via the PDK1-AKT signaling pathway. Cell Commun. Signal. CCS 2022, 20, 35. [Google Scholar] [CrossRef]
- Oliveira, M.N.; Pillat, M.M.; Motaln, H.; Ulrich, H.; Lah, T.T. Kinin-B1 Receptor Stimulation Promotes Invasion and is Involved in Cell-Cell Interaction of Co-Cultured Glioblastoma and Mesenchymal Stem Cells. Sci. Rep. 2018, 8, 1299. [Google Scholar] [CrossRef]
- Kodama, T.; Koma, Y.I.; Arai, N.; Kido, A.; Urakawa, N.; Nishio, M.; Shigeoka, M.; Yokozaki, H. CCL3-CCR5 axis contributes to progression of esophageal squamous cell carcinoma by promoting cell migration and invasion via Akt and ERK pathways. Lab. Investig. A J. Tech. Methods Pathol. 2020, 100, 1140–1157. [Google Scholar] [CrossRef]
- Jha, K.K.; Banga, S.; Palejwala, V.; Ozer, H.L. SV40-Mediated immortalization. Exp. Cell Res. 1998, 245, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Reddel, R.R.; De Silva, R.; Duncan, E.L.; Rogan, E.M.; Whitaker, N.J.; Zahra, D.G.; Ke, Y.; McMenamin, M.G.; Gerwin, B.I.; Harris, C.C. SV40-induced immortalization and ras-transformation of human bronchial epithelial cells. Int. J. Cancer 1995, 61, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.Q.; Yang, Z.M.; Qu, Y. SV40 and cell immortalization. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi = Zhongguo Xiufu Chongjian Waike Zazhi = Chin. J. Reparative Reconstr. Surg. 2000, 14, 170–174. [Google Scholar]
- Giusti, I.; Cervelli, C.; D’Ascenzo, S.; Di Francesco, M.; Ligas, C.; D’Alessandro, E.; Papola, F.; Dolo, V. The human ovarian cancer cell line CABA I: A peculiar genetic evolution. Int. J. Mol. Med. 2016, 37, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Zhao, H.; Jiang, E.; Shang, Z. 3D Co-culture of Cancer-Associated Fibroblast with Oral Cancer Organoids. J. Dent. Res. 2021, 100, 201–208. [Google Scholar] [CrossRef]
- Kretschmer, I.; Freudenberger, T.; Twarock, S.; Yamaguchi, Y.; Grandoch, M.; Fischer, J. Esophageal Squamous Cell Carcinoma Cells Modulate Chemokine Expression and Hyaluronan Synthesis in Fibroblasts. J. Biol. Chem. 2016, 291, 4091–4106. [Google Scholar] [CrossRef]
- Pikuła, M.; Langa, P.; Kosikowska, P.; Trzonkowski, P. Stem cells and growth factors in wound healing. Postep. Hig. I Med. Dosw. (Online) 2015, 69, 874–885. [Google Scholar] [CrossRef]
- Fu, R.; Ma, X.; Bian, Z.; Ma, J. Digital separation of diaminobenzidine-stained tissues via an automatic color-filtering for immunohistochemical quantification. Biomed. Opt. Express. 2015, 6, 544–558. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brennan, J.; Lu, M.L.; Kang, Y. A New Model of Esophageal Cancers by Using a Detergent-Free Decellularized Matrix in a Perfusion Bioreactor. Bioengineering 2023, 10, 96. https://doi.org/10.3390/bioengineering10010096
Brennan J, Lu ML, Kang Y. A New Model of Esophageal Cancers by Using a Detergent-Free Decellularized Matrix in a Perfusion Bioreactor. Bioengineering. 2023; 10(1):96. https://doi.org/10.3390/bioengineering10010096
Chicago/Turabian StyleBrennan, Jordan, Michael L. Lu, and Yunqing Kang. 2023. "A New Model of Esophageal Cancers by Using a Detergent-Free Decellularized Matrix in a Perfusion Bioreactor" Bioengineering 10, no. 1: 96. https://doi.org/10.3390/bioengineering10010096
APA StyleBrennan, J., Lu, M. L., & Kang, Y. (2023). A New Model of Esophageal Cancers by Using a Detergent-Free Decellularized Matrix in a Perfusion Bioreactor. Bioengineering, 10(1), 96. https://doi.org/10.3390/bioengineering10010096