Gene-Expression Analysis of Human Fibroblasts Affected by 3D-Printed Carboxylated Nanocellulose Constructs
Abstract
1. Introduction
2. Materials and Methods
2.1. Production and Characterization of CNF Grades
2.2. Viscosity of the CNFs
2.3. Three-Dimensional-Printing Trials and Characterization
2.4. Nano-Mechanical Assessment of 3D-Printed Constructs
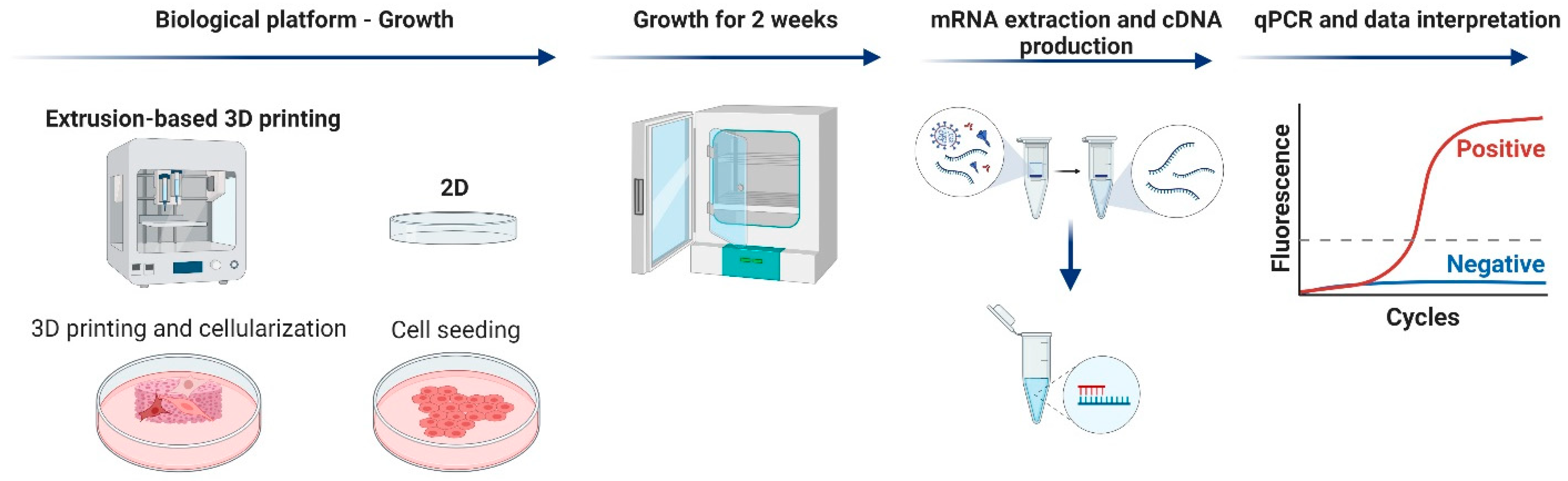
2.5. Gene-Expression Analysis
2.5.1. Cell Culture
2.5.2. RNA Extraction and qPCR
2.6. Statistical Analysis
3. Results and Discussion
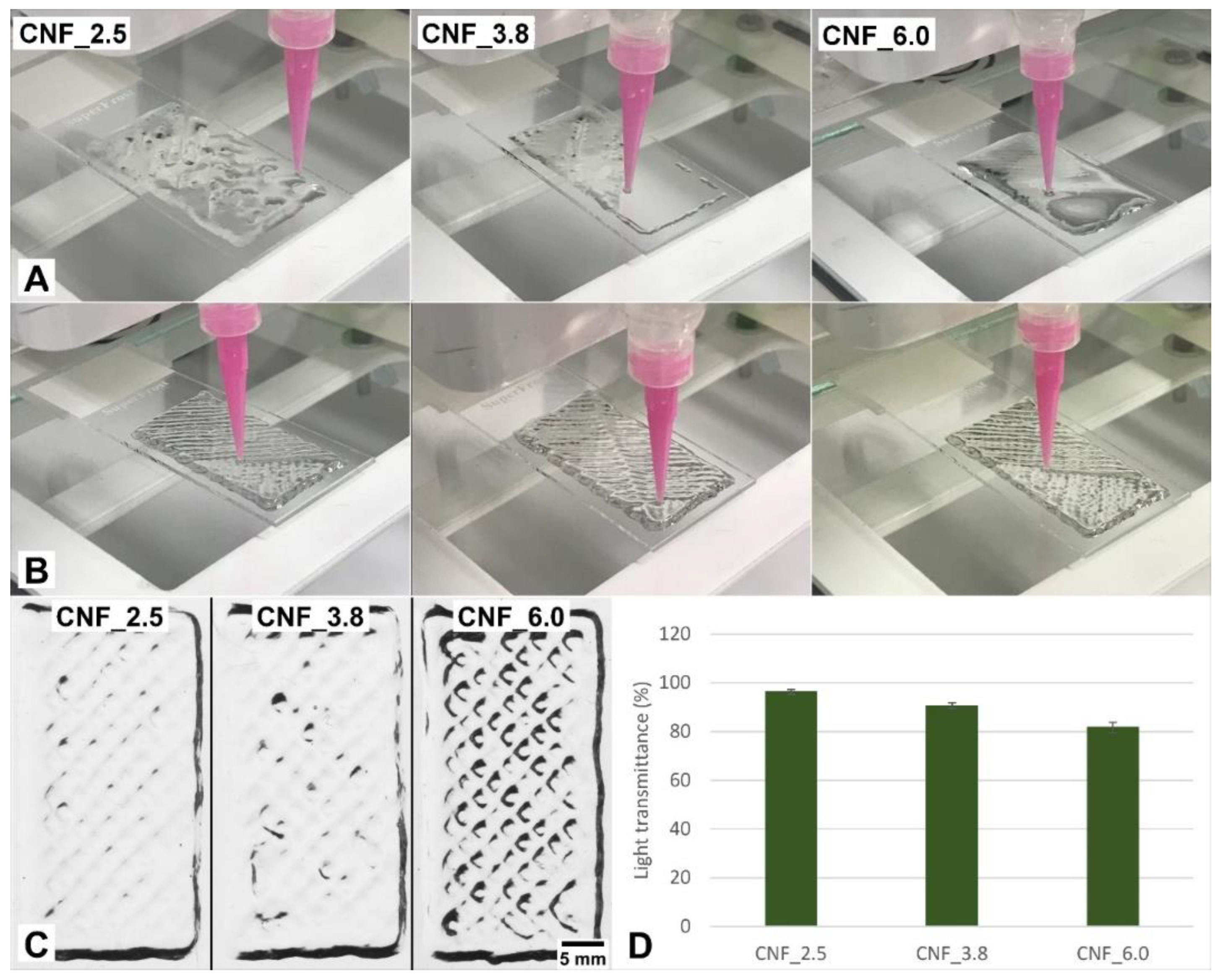
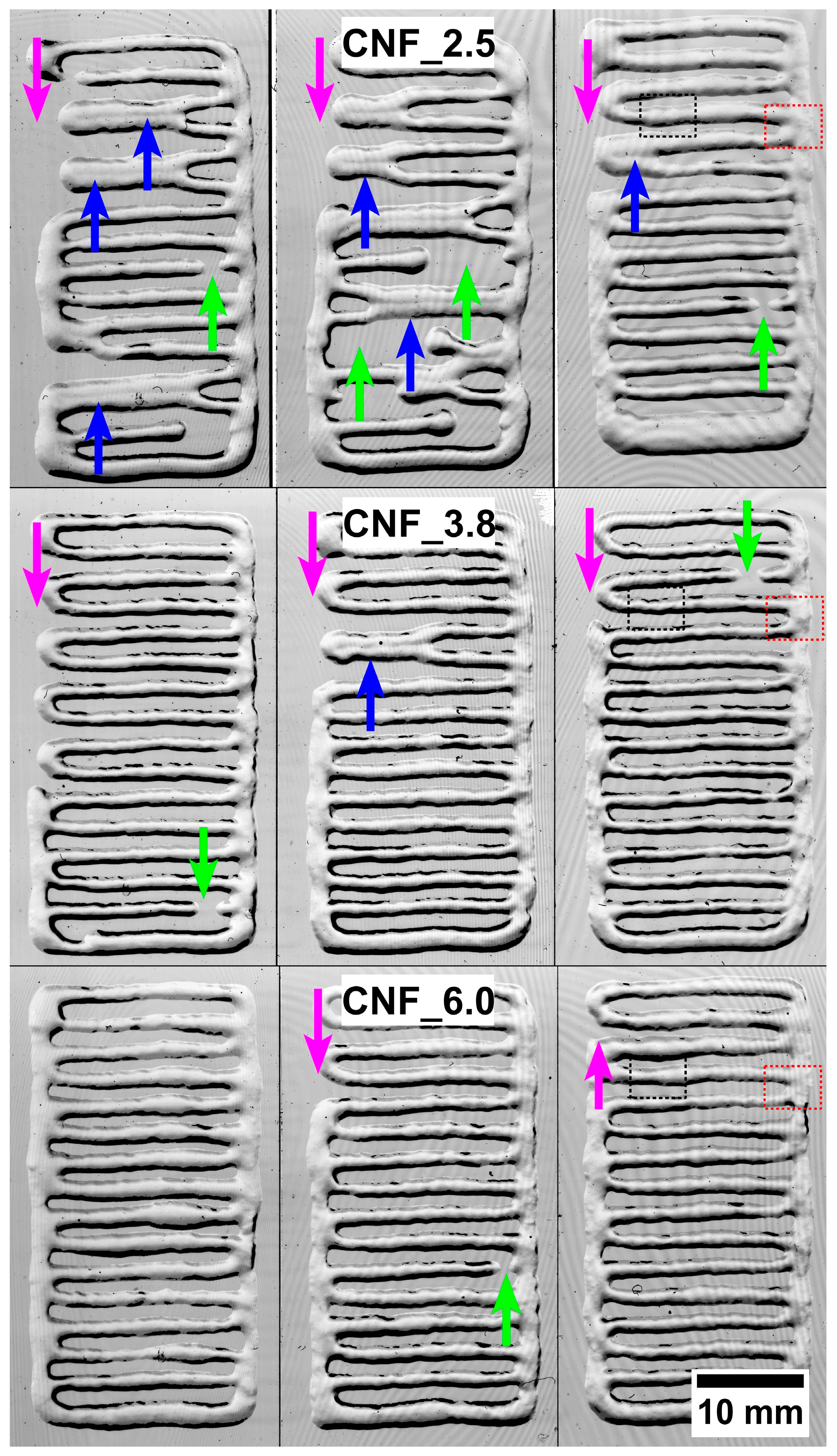
3.1. Three-Dimensional Printing
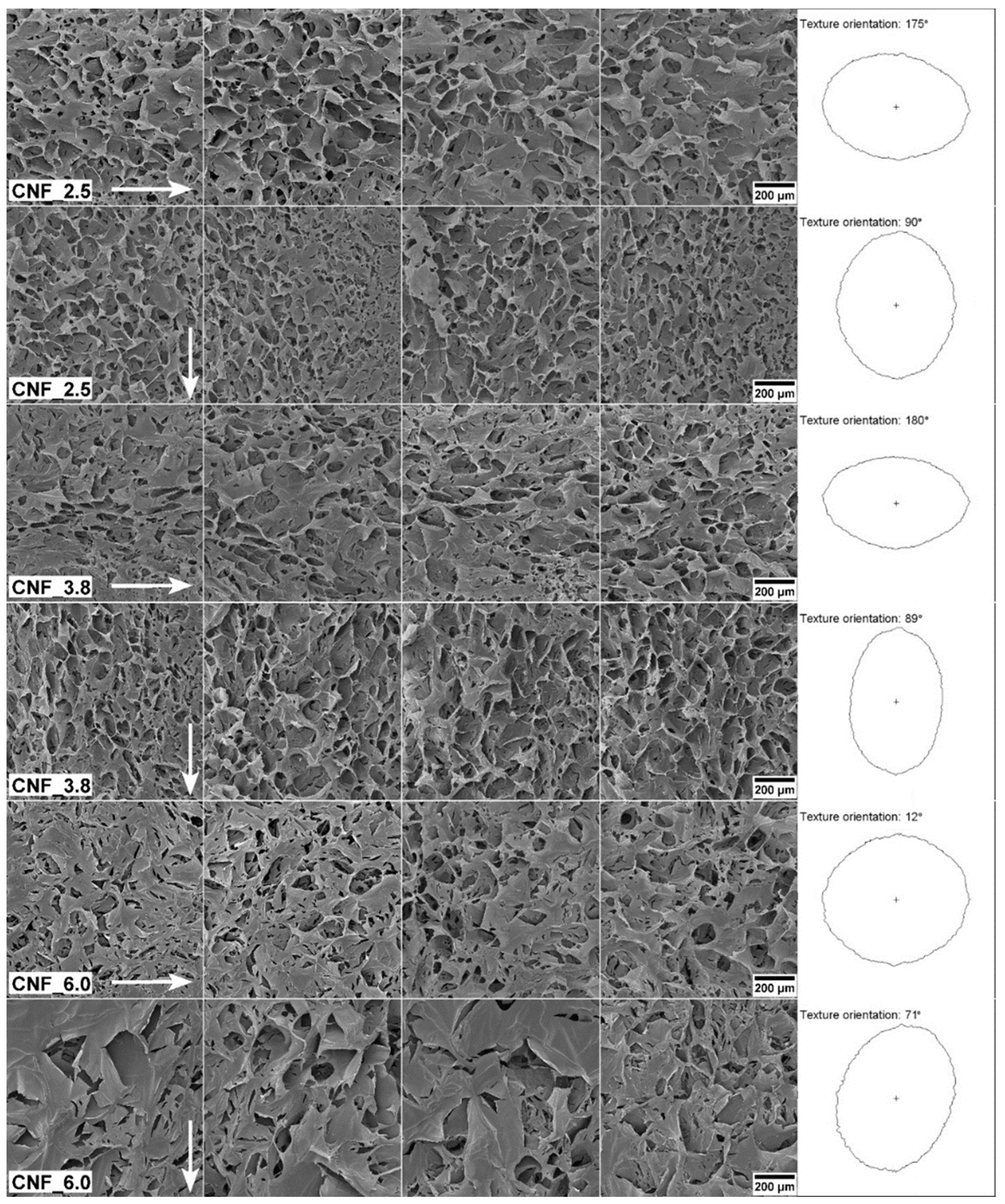
3.2. Structural and Mechanical Analysis
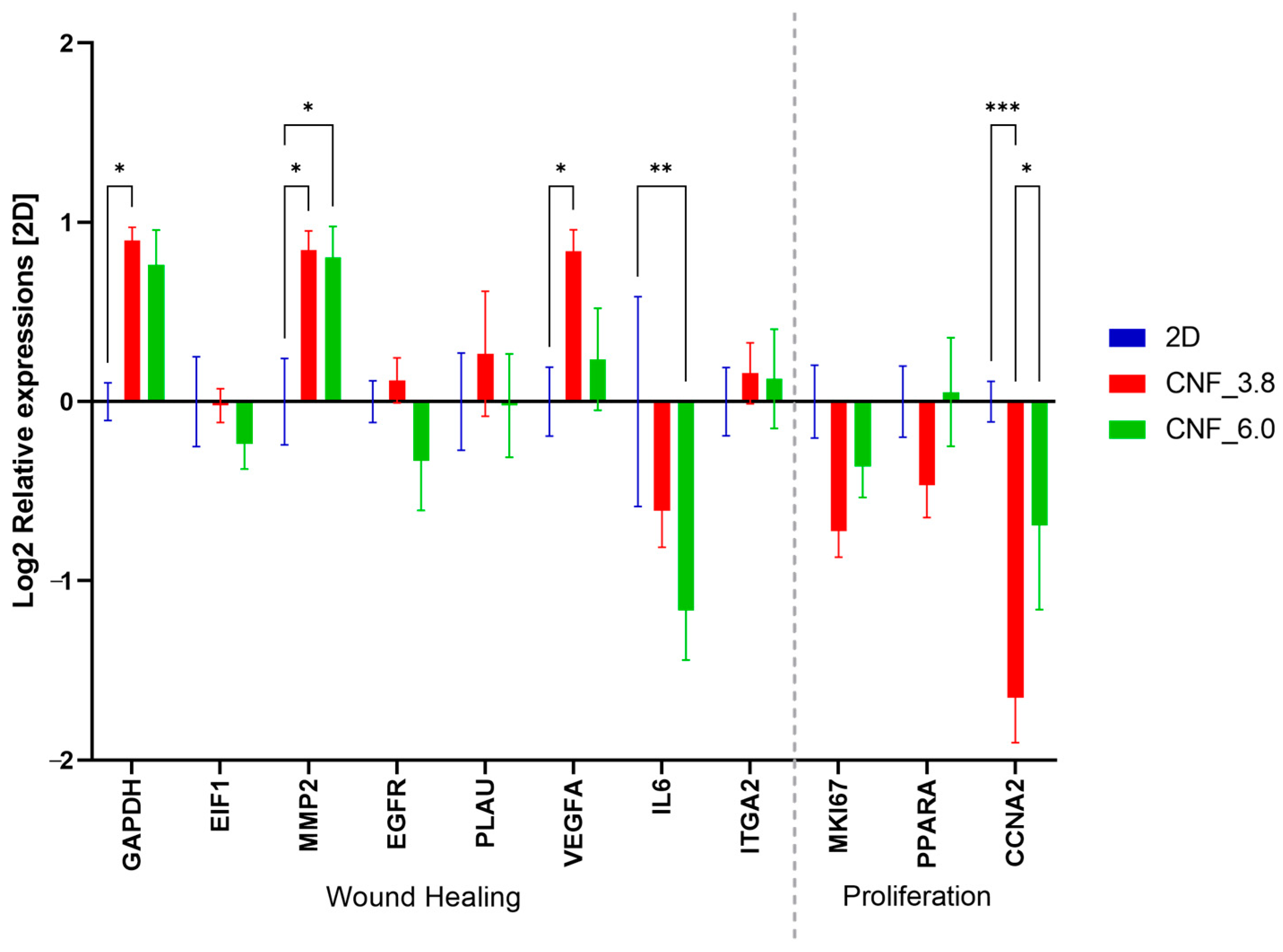
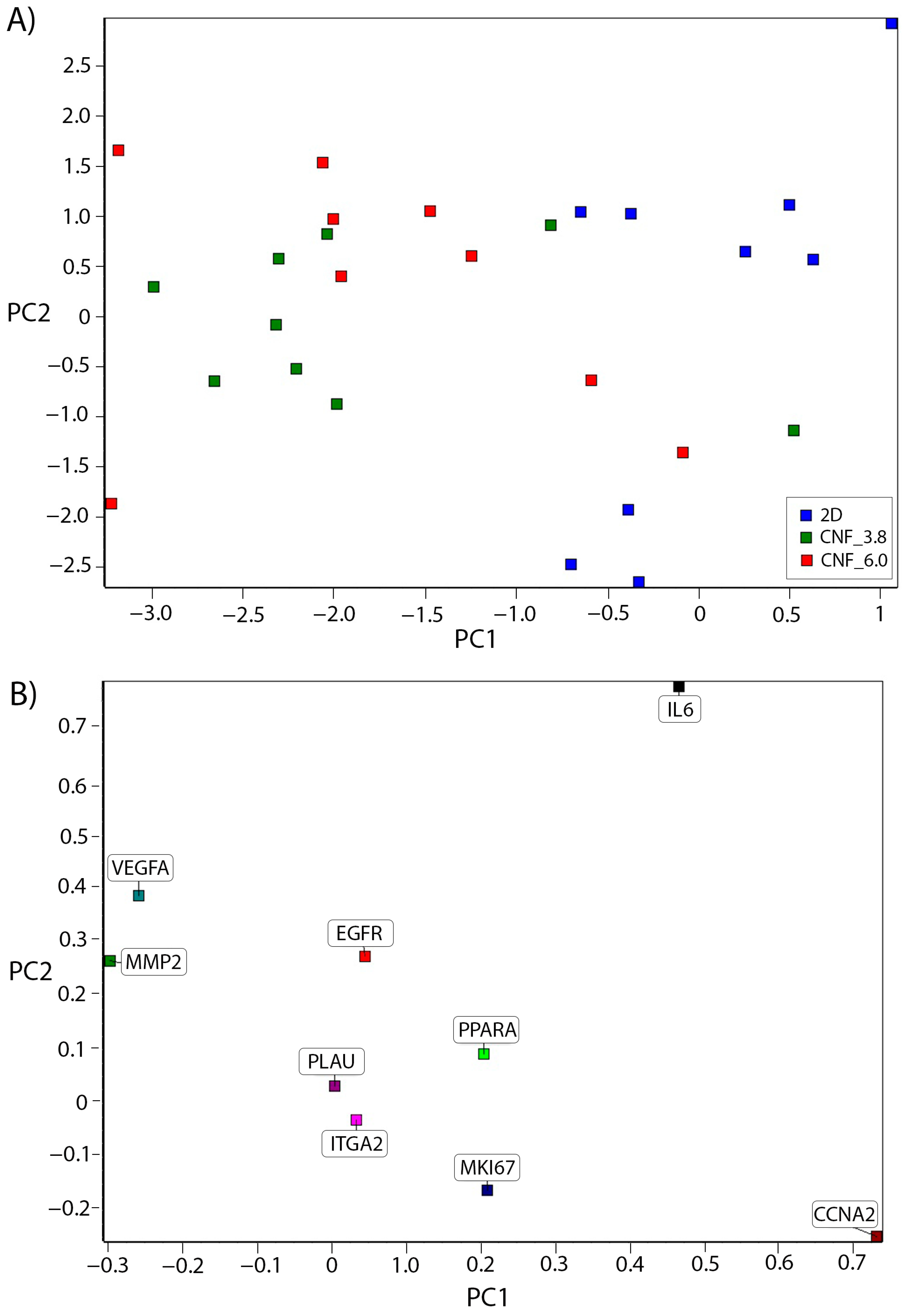
3.3. Cell Culture and Gene-Expression Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alexandrescu, L.; Syverud, K.; Gatti, A.; Chinga-Carrasco, G. Cytotoxicity tests of cellulose nanofibril-based structures. Cellulose 2013, 20, 1765–1775. [Google Scholar] [CrossRef]
- Rashad, A.; Mustafa, K.; Heggset, E.B.; Syverud, K. Cytocompatibility of Wood-Derived Cellulose Nanofibril Hydrogels with Different Surface Chemistry. Biomacromolecules 2017, 18, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Rashad, A.; Mohamed-Ahmed, S.; Ojansivu, M.; Berstad, K.; Yassin, M.A.; Kivijarv, T.; Heggset, E.B.; Syverud, K.; Mustafa, K. Coating 3D Printed Polycaprolactone Scaffolds with Nanocellulose Promotes Growth and Differentiation of Mesenchymal Stem Cells. Biomacromolecules 2018, 19, 4307–4319. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Q.; Sun, J.Z.; Yao, Q.; Ji, C.C.; Liu, J.; Zhu, Q.Q. 3D printing with cellulose materials. Cellulose 2018, 25, 4275–4301. [Google Scholar] [CrossRef]
- Bacakova, L.; Pajorova, J.; Bacakova, M.; Skogberg, A.; Kallio, P.; Kolarova, K.; Svorcik, V. Versatile Application of Nanocellulose: From Industry to Skin Tissue Engineering and Wound Healing. Nanomaterials 2019, 9, 164. [Google Scholar] [CrossRef]
- Jack, A.A.; Nordli, H.R.; Powell, L.C.; Powell, K.A.; Kishnani, H.; Johnsen, P.O.; Pukstad, B.; Thomas, D.W.; Chinga-Carrasco, G.; Hill, K.E. The interaction of wood nanocellulose dressings and the wound pathogen P. aeruginosa. Carbohydr. Polym. 2017, 157, 1955–1962. [Google Scholar] [CrossRef]
- Nordli, H.R.; Chinga-Carrasco, G.; Rokstad, A.M.; Pukstad, B. Producing ultrapure wood cellulose nanofibrils and evaluating the cytotoxicity using human skin cells. Carbohydr. Polym. 2016, 150, 65–73. [Google Scholar] [CrossRef]
- Nordli, H.R.; Pukstad, B.; Chinga-Carrasco, G.; Rokstad, A.M. Ultrapure Wood Nanocellulose—Assessments of Coagulation and Initial Inflammation Potential. ACS Appl. Bio. Mater. 2019, 2, 1107–1118. [Google Scholar] [CrossRef]
- Rees, A.; Powell, L.C.; Chinga-Carrasco, G.; Gethin, D.T.; Syverud, K.; Hill, K.E.; Thomas, D.W. 3D Bioprinting of Carboxymethylated-Periodate Oxidized Nanocellulose Constructs for Wound Dressing Applications. Biomed Res. Int. 2015, 2015, 925757. [Google Scholar] [CrossRef]
- Dai, L.; Cheng, T.; Duan, C.; Zhao, W.; Zhang, W.P.; Zou, X.J.; Aspler, J.; Ni, Y.H. 3D printing using plant-derived cellulose and its derivatives: A review. Carbohydr. Polym. 2019, 203, 71–86. [Google Scholar] [CrossRef]
- Leppiniemi, J.; Lahtinen, P.; Paajanen, A.; Mahlberg, R.; Metsa-Kortelainen, S.; Pinornaa, T.; Pajari, H.; Vikholm-Lundin, I.; Pursula, P.; Hytonen, V.P. 3D-Printable Bioactivated Nanocellulose-Alginate Hydrogels. ACS Appl. Mater. Interfaces 2017, 9, 21959–21970. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, G.; Kokkinis, D.; Libanori, R.; Hausmann, M.K.; Gladman, A.S.; Neels, A.; Tingaut, P.; Zimmermann, T.; Lewis, J.A.; Studart, A.R. Cellulose Nanocrystal Inks for 3D Printing of Textured Cellular Architectures. Adv. Funct. Mater. 2017, 27, 1604619. [Google Scholar] [CrossRef]
- Xu, W.Y.; Wang, X.J.; Sandler, N.; Willfor, S.; Xu, C.L. Three-Dimensional Printing of Wood-Derived Biopolymers: A Review Focused on Biomedical Applications. ACS Sustain. Chem. Eng. 2018, 6, 5663–5680. [Google Scholar] [CrossRef] [PubMed]
- Heggset, E.B.; Strand, B.L.; Sundby, K.W.; Simon, S.; Chinga-Carrasco, G.; Syverud, K. Viscoelastic properties of nanocellulose based inks for 3D printing and mechanical properties of CNF/alginate biocomposite gels. Cellulose 2019, 26, 581–595. [Google Scholar] [CrossRef]
- Chinga-Carrasco, G. Potential and Limitations of Nanocelluloses as Components in Biocomposite Inks for Three-Dimensional Bioprinting and for Biomedical Devices. Biomacromolecules 2018, 19, 701–711. [Google Scholar] [CrossRef]
- Piras, C.C.; Fernandez-Prieto, S.; De Borggraeve, W.M. Nanocellulosic materials as bioinks for 3D bioprinting. Biomater. Sci. 2017, 5, 1988–1992. [Google Scholar] [CrossRef]
- Zhang, X.; Morits, M.; Jonkergouw, C.; Ora, A.; Valle-Delgado, J.J.; Farooq, M.; Ajdary, R.; Huan, S.; Linder, M.; Rojas, O.; et al. Three-Dimensional Printed Cell Culture Model Based on Spherical Colloidal Lignin Particles and Cellulose Nanofibril-Alginate Hydrogel. Biomacromolecules 2020, 21, 1875–1885. [Google Scholar] [CrossRef]
- Jeong, J.H.; Liang, Y.; Jang, M.; Cha, C.; Chu, C.; Lee, H.; Jung, W.; Kim, J.W.; Boppart, S.A.; Kong, H. Stiffness-Modulated Water Retention and Neovascularization of Dermal Fibroblast-Encapsulating Collagen Gel. Tissue Eng. Part A 2013, 19, 1275–1284. [Google Scholar] [CrossRef]
- Yi, B.; Xu, Q.; Liu, W. An overview of substrate stiffness guided cellular response and its applications in tissue regeneration. Bioact. Mater. 2022, 15, 82–102. [Google Scholar] [CrossRef] [PubMed]
- Selby, A.; Maldonado-Codina, C.; Derby, B. Influence of specimen thickness on the nanoindentation of hydrogels: Measuring the mechanical properties of soft contact lenses. J. Mech. Behav. Biomed. Mater. 2014, 35, 144–156. [Google Scholar] [CrossRef]
- Xia, T.; Liu, W.; Yang, L. A review of gradient stiffness hydrogels used in tissue engineering and regenerative medicine. J. Biomed. Mater. Res. Part A 2017, 105, 1799–1812. [Google Scholar] [CrossRef] [PubMed]
- Richbourg, N.R.; Rausch, M.K.; Peppas, N.A. Cross-evaluation of stiffness measurement methods for hydrogels. Polymer 2022, 258, 125316. [Google Scholar] [CrossRef]
- Li, X.; Bhushan, B. A review of nanoindentation continuous stiffness measurement technique and its applications. Mater. Charact. 2002, 48, 11–36. [Google Scholar] [CrossRef]
- Zhang, P.; Li, S.X.; Zhang, Z.F. General relationship between strength and hardness. Mater. Sci. Eng. A 2011, 529, 62–73. [Google Scholar] [CrossRef]
- Balak, D.M. Fumaric acid esters in the management of psoriasis. Psoriasis 2015, 5, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Tourrière, H.; Chebli, K.; Tazi, J. mRNA degradation machines in eukaryotic cells. Biochimie 2002, 84, 821–837. [Google Scholar] [CrossRef] [PubMed]
- Tabriz, A.G.; Douroumis, D. Recent advances in 3D printing for wound healing: A systematic review. J. Drug Deliv. Sci. Technol. 2022, 74, 103564. [Google Scholar] [CrossRef]
- Sinno, H.; Prakash, S. Complements and the wound healing cascade: An updated review. Plast. Surg. Int. 2013, 2013, 146764. [Google Scholar] [CrossRef]
- Saito, T.; Isogai, A. TEMPO-mediated oxidation of native cellulose. The effect of oxidation conditions on chemical and crystal structures of the water-insoluble fractions. Biomacromolecules 2004, 5, 1983–1989. [Google Scholar] [CrossRef]
- Chinga-Carrasco, G.J.; Johansson, J.; Heggset, E.B.; Leirset, I.; Björn, C.; Agrenius, K.; Stevanic, J.S.; Håkansson, J. Characterisation and antibacterial properties of autoclaved carboxylated wood nanocellulose. Biomacromolecules 2021, 22, 2779–2789. [Google Scholar] [CrossRef]
- Chinga, G.; Johnsen, P.O.; Dougherty, R.; Berli, E.L.; Walter, J. Quantification of the 3D microstructure of SC surfaces. J. Microsc. 2007, 227, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Moberg, T.; Sahlin, K.; Yao, K.; Geng, S.Y.; Westman, G.; Zhou, Q.; Oksman, K.; Rigdahl, M. Rheological properties of nanocellulose suspensions: Effects of fibril/particle dimensions and surface characteristics. Cellulose 2017, 24, 2499–2510. [Google Scholar] [CrossRef]
- Naderi, A.; Lindstrom, T.; Sundstrom, J. Carboxymethylated nanofibrillated cellulose: Rheological studies. Cellulose 2014, 21, 1561–1571. [Google Scholar] [CrossRef]
- Iotti, M.; Gregersen, Ø.W.; Moe, S.; Lenes, M. Rheological Studies of Microfibrillar Cellulose Water Dispersions. J. Polym. Environ. 2011, 19, 137–145. [Google Scholar] [CrossRef]
- Schmitt, J.; Calabrese, V.; da Silva, M.A.; Lindhoud, S.; Alfredsson, V.; Scott, J.L.; Edler, K.J. TEMPO-oxidised cellulose nanofibrils; probing the mechanisms of gelation via small angle X-ray scattering. Phys. Chem. Chem. Phys. 2018, 20, 16012–16020. [Google Scholar] [CrossRef]
- Fukuzumi, H.; Saito, T.; Okita, Y.; Isogai, A. Thermal stabilization of TEMPO-oxidized cellulose. Polym. Degrad. Stabil. 2010, 95, 1502–1508. [Google Scholar] [CrossRef]
- Lasseuguette, E.; Roux, D.; Nishiyama, Y. Rheological properties of microfibrillar suspension of TEMPO-oxidized pulp. Cellulose 2008, 15, 425–433. [Google Scholar] [CrossRef]
- Fukuzumi, H.; Saito, T.; Isogai, A. Influence of TEMPO-oxidized cellulose nanofibril length on film properties. Carbohyd. Polym. 2013, 93, 172–177. [Google Scholar] [CrossRef]
- Hausmann, M.K.; Ruhs, P.A.; Siqueira, G.; Lauger, J.; Libanori, R.; Zimmermann, T.; Studart, A.R. Dynamics of Cellulose Nanocrystal Alignment during 3D Printing. ACS Nano 2018, 12, 6926–6937. [Google Scholar] [CrossRef]
- Gadalamaria, F.; Parsi, F. Measurement of Fiber Orientation in Short-Fiber Composites Using Digital Image-Processing. Polym. Compos. 1993, 14, 126–131. [Google Scholar] [CrossRef]
- Yoshigi, M.; Clark, E.B.; Yost, H.J. Quantification of stretch-induced cytoskeletal remodeling in vascular endothelial cells by image processing. Cytom. Part A 2003, 55A, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Ottesen, V.; Kumar, V.; Toivakka, M.; Chinga-Carrasco, G.; Syverud, K.; Gregersen, O.W. Viability and properties of roll-to-roll coating of cellulose nanofibrils on recycled paperboard. Nord Pulp Pap. Res. J. 2017, 32, 179–188. [Google Scholar] [CrossRef]
- Wu, S.H.; Peng, H.; Li, X.H.; Streubel, P.N.; Liu, Y.; Duan, B. Effect of scaffold morphology and cell co-culture on tenogenic differentiation of HADMSC on centrifugal melt electrospun poly (L-lactic acid) fibrous meshes. Biofabrication 2017, 9, 044106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liao, K.; Li, C.; Lai, A.C.K.; Foo, J.J.; Chan, V. Progress in Integrative Biomaterial Systems to Approach Three-Dimensional Cell Mechanotransduction. Bioengineering 2017, 4, 72. [Google Scholar] [CrossRef] [PubMed]
- Handorf, A.M.; Zhou, Y.; Halanski, M.A.; Li, W.J. Tissue stiffness dictates development, homeostasis, and disease progression. Organogenesis 2015, 11, 1–15. [Google Scholar] [CrossRef]
- Jones, D.S.; Woolfson, A.D.; Brown, A.F. Textural, viscoelastic and mucoadhesive properties of pharmaceutical gels composed of cellulose polymers. Int. J. Pharm. 1997, 151, 223–233. [Google Scholar] [CrossRef]
- Sezer, A.D.; Cevher, E.; Hatipoglu, F.; Ogurtan, Z.; Bas, A.L.; Akbuga, J. Preparation of fucoidan-chitosan hydrogel and its application as burn healing accelerator on rabbits. Biol. Pharm. Bull. 2008, 31, 2326–2333. [Google Scholar] [CrossRef]
- Rosendahl, J.; Svanström, A.; Berglin, M.; Petronis, S.; Bogestål, Y.; Stenlund, P.; Standoft, S.; Ståhlberg, A.; Landberg, G.; Chinga-Carrasco, G.; et al. 3D Printed Nanocellulose Scaffolds as a Cancer Cell Culture Model System. Bioengineering 2021, 8, 97. [Google Scholar] [CrossRef]
- Sun, F.; Nordli, H.R.; Pukstad, B.; Kristofer Gamstedt, E.; Chinga-Carrasco, G. Mechanical characteristics of nanocellulose-PEG bionanocomposite wound dressings in wet conditions. J. Mech. Behav. Biomed. Mater. 2017, 69, 377–384. [Google Scholar] [CrossRef]
- Syverud, K.; Pettersen, S.R.; Draget, K.; Chinga-Carrasco, G. Controlling the elastic modulus of cellulose nanofibril hydrogels—Scaffolds with potential in tissue engineering. Cellulose 2015, 22, 473–481. [Google Scholar] [CrossRef]
- Panteli, P.A.; Patrickios, C.S.; Constantinou, M.; Constantinides, G. Multiple Network Hydrogels: A Study of Their Nanoindentation Hardness. Macromol. Symp. 2019, 385, 1800201. [Google Scholar] [CrossRef]
- Buffinton, C.M.; Tong, K.J.; Blaho, R.A.; Buffinton, E.M.; Ebenstein, D.M. Comparison of mechanical testing methods for biomaterials: Pipette aspiration, nanoindentation, and macroscale testing. J. Mech. Behav. Biomed. Mater. 2015, 51, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Landberg, G.; Fitzpatrick, P.; Isakson, P.; Jonasson, E.; Karlsson, J.; Larsson, E.; Svanström, A.; Rafnsdottir, S.; Persson, E.; Gustafsson, A.; et al. Patient-derived scaffolds uncover breast cancer promoting properties of the microenvironment. Biomaterials 2020, 235, 119705. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.T.; Salerno, S.; Ranji, P.; Håkansson, J.; Bogestål, Y.; Wettergren, Y.; Ståhlberg, A.; Bexe Lindskog, E.; Landberg, G. Patient-derived scaffolds as a model of colorectal cancer. Cancer Med. 2021, 10, 867–882. [Google Scholar] [CrossRef]
- Svanström, A.; Rosendahl, J.; Salerno, S.; Leiva, M.C.; Gregersson, P.; Berglin, M.; Bogestål, Y.; Lausmaa, J.; Oko, A.; Chinga-Carrasco, G.; et al. Optimized alginate-based 3D printed scaffolds as a model of patient derived breast cancer microenvironments in drug discovery. Biomed. Mater. 2021, 16, 045046. [Google Scholar] [CrossRef] [PubMed]
- Tracy, L.E.; Minasian, R.A.; Caterson, E.J. Extracellular Matrix and Dermal Fibroblast Function in the Healing Wound. Adv. Wound Care 2016, 5, 119–136. [Google Scholar] [CrossRef]
- Bainbridge, P. Wound healing and the role of fibroblasts. J. Wound Care 2013, 22, 407–408, 410–412. [Google Scholar] [CrossRef] [PubMed]
- Pratsinis, H.; Giannouli, C.C.; Zervolea, I.; Psarras, S.; Stathakos, D.; Kletsas, D. Differential proliferative response of fetal and adult human skin fibroblasts to transforming growth factor-beta. Wound Repair Regen. 2004, 12, 374–383. [Google Scholar] [CrossRef]
- Chen, Y.S.; Lee, S.M.; Lin, Y.J.; Chiang, S.H.; Lin, C.C. Effects of Danshensu and Salvianolic Acid B from Salvia miltiorrhiza Bunge (Lamiaceae) on cell proliferation and collagen and melanin production. Molecules 2014, 19, 2029–2041. [Google Scholar] [CrossRef]
- Chiang, J.H.; Tsai, F.J.; Lin, T.H.; Yang, J.S.; Chiu, Y.J. Tremella fuciformis Inhibits Melanogenesis in B16F10 Cells and Promotes Migration of Human Fibroblasts and Keratinocytes. In Vivo 2022, 36, 713–722. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Cheng, M.C.; Lee, C.C.; Chiou, T.Y.; Tsai, T.Y. Effect of ethanol extract from Lactobacillus plantarum TWK10-fermented soymilk on wound healing in streptozotocin-induced diabetic rat. AMB Express 2019, 9, 163. [Google Scholar] [CrossRef]
- Lin, H.I.; Chu, S.J.; Perng, W.C.; Wu, C.P.; Lin, Z.Y.; Huang, K.L. Hyperbaric oxygen attenuates cell growth in skin fibroblasts cultured in a high-glucose medium. Wound Repair Regen. 2008, 16, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.B.; Chen, C.C.; Chen, L.C.; Chen, H.H. The bioactive composite film prepared from bacterial cellulose and modified by hydrolyzed gelatin peptide. J. Biomater. Appl. 2015, 29, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Montoya, A.; Daza, A.; Muñoz, D.; Ríos, K.; Taylor, V.; Cedeño, D.; Vélez, I.D.; Echeverri, F.; Robledo, S.M. Development of a novel formulation with hypericin to treat cutaneous leishmaniasis based on photodynamic therapy in in vitro and in vivo studies. Antimicrob. Agents Chemother. 2015, 59, 5804–5813. [Google Scholar] [CrossRef]
- Yoon, D.; Yoon, D.; Sim, H.; Hwang, I.; Lee, J.S.; Chun, W. Accelerated Wound Healing by Fibroblasts Differentiated from Human Embryonic Stem Cell-Derived Mesenchymal Stem Cells in a Pressure Ulcer Animal Model. Stem. Cells Int. 2018, 2018, 4789568. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.M.; Raposo, N.R.; Brayner, R.; Teixeira, E.M.; Oliveira, V.; Quintão, C.C.; Camargo, L.S.; Mattoso, L.H.; Brandão, H.M. Cytotoxicity and expression of genes involved in the cellular stress response and apoptosis in mammalian fibroblast exposed to cotton cellulose nanofibers. Nanotechnology 2013, 24, 075103. [Google Scholar] [CrossRef] [PubMed]
- Cuylen, S.; Blaukopf, C.; Politi, A.Z.; Müller-Reichert, T.; Neumann, B.; Poser, I.; Ellenberg, J.; Hyman, A.A.; Gerlich, D.W. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 2016, 535, 308–312. [Google Scholar] [CrossRef]
- Ali, M.U.; Ur Rahman, M.S.; Jia, Z.; Jiang, C. Eukaryotic translation initiation factors and cancer. Tumour Biol. 2017, 39, 1010428317709805. [Google Scholar] [CrossRef]
- Epidermal Growth Factor and Epidermal Growth Factor Receptor: The Yin and Yang in the Treatment of Cutaneous Wounds and Cancer. Adv. Wound Care 2013, 2, 24–29. [CrossRef]
- Smith, S.A.; Mutch, N.J.; Baskar, D.; Rohloff, P.; Docampo, R.; Morrissey, J.H. Polyphosphate modulates blood coagulation and fibrinolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 903–908. [Google Scholar] [CrossRef]
- Turabelidze, A.; Guo, S.; DiPietro, L.A. Importance of housekeeping gene selection for accurate reverse transcription-quantitative polymerase chain reaction in a wound healing model. Wound Repair Regen. 2010, 18, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.A.; Cardinale, G.F. Young’s modulus and Poisson’s ratio of CVD diamond. Diam. Relat. Mater. 1993, 2, 918–923. [Google Scholar] [CrossRef]


| Analyses | CNF_2.5 | CNF_3.8 | CNF_6.0 |
|---|---|---|---|
| pH (0.6 wt%) | 6.81 ± 0.02 | 6.83 ± 0.02 | 6.89 ± 0.01 |
| Carboxylic-acid content (μmol/g) | 1036 ± 41 | 1285 ± 42 | 1593 ± 10 |
| Viscosity at 10 RPM (mPas) | 13,855 ± 17 | 18,157 ± 25 | 18,208 ± 35 |
| Mean object size (nm) | 708 | 509 | 498 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosendahl, J.; Zarna, C.; Håkansson, J.; Chinga-Carrasco, G. Gene-Expression Analysis of Human Fibroblasts Affected by 3D-Printed Carboxylated Nanocellulose Constructs. Bioengineering 2023, 10, 121. https://doi.org/10.3390/bioengineering10010121
Rosendahl J, Zarna C, Håkansson J, Chinga-Carrasco G. Gene-Expression Analysis of Human Fibroblasts Affected by 3D-Printed Carboxylated Nanocellulose Constructs. Bioengineering. 2023; 10(1):121. https://doi.org/10.3390/bioengineering10010121
Chicago/Turabian StyleRosendahl, Jennifer, Chiara Zarna, Joakim Håkansson, and Gary Chinga-Carrasco. 2023. "Gene-Expression Analysis of Human Fibroblasts Affected by 3D-Printed Carboxylated Nanocellulose Constructs" Bioengineering 10, no. 1: 121. https://doi.org/10.3390/bioengineering10010121
APA StyleRosendahl, J., Zarna, C., Håkansson, J., & Chinga-Carrasco, G. (2023). Gene-Expression Analysis of Human Fibroblasts Affected by 3D-Printed Carboxylated Nanocellulose Constructs. Bioengineering, 10(1), 121. https://doi.org/10.3390/bioengineering10010121







