Antagonistic Effects of Enrofloxacin on Carbendazim-Induced Developmental Toxicity in Zebrafish Embryos
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Zebrafish Breeding and Egg Collection
2.3. Zebrafish Embryo Exposure and Developmental Index Detection
2.4. Exposure for Transcriptomics
2.5. mRNA Library Construction
2.6. RNA-Seq Data Analyze
2.7. Real-Time Quantitative Reverse Transcription PCR
2.8. Data Analysis and Statistics
3. Results
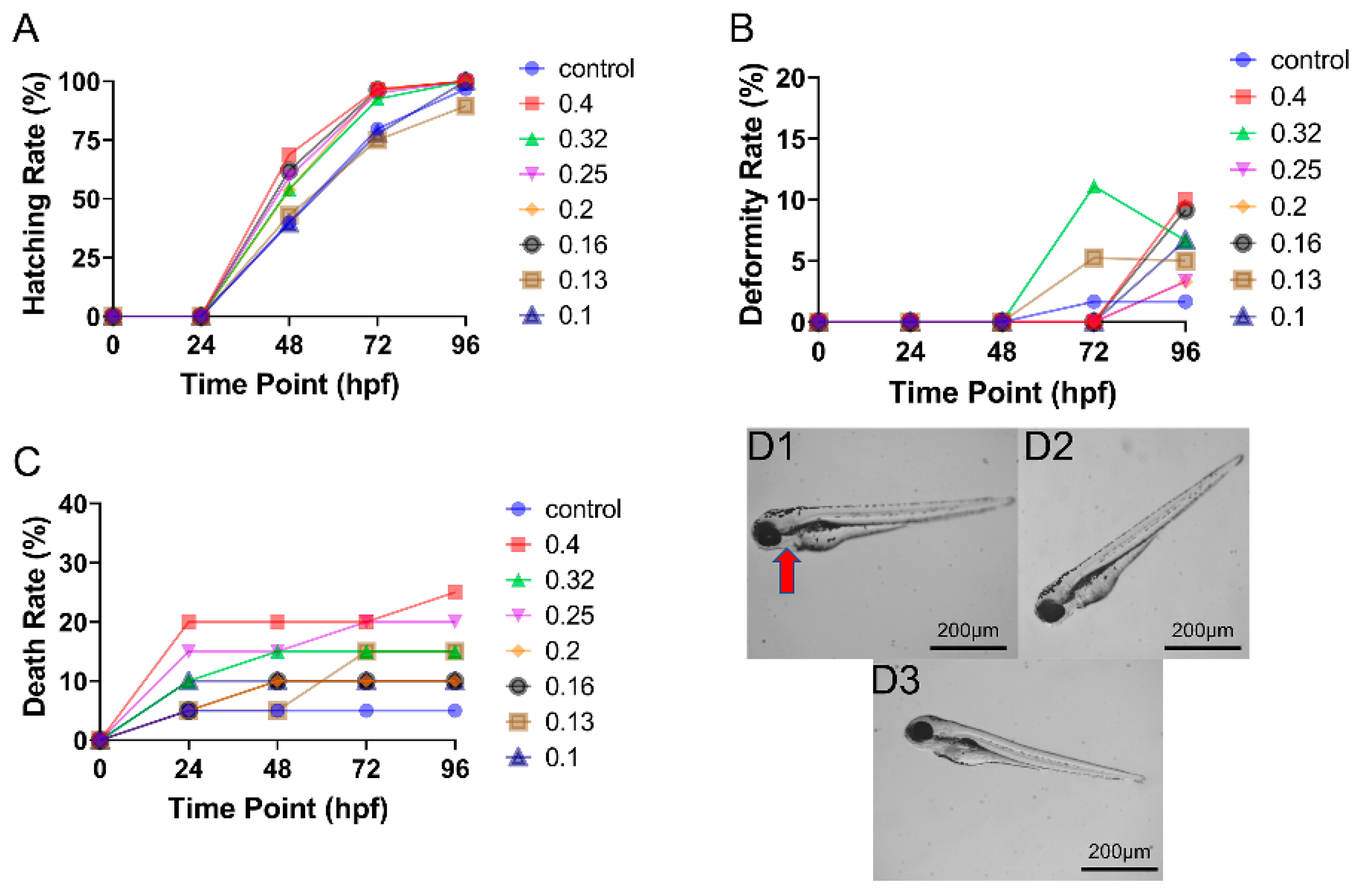
3.1. Toxicity of CAR Alone on Zebrafish Embryo Development
3.2. Toxicity of ENF Alone on Zebrafish Embryo Development
3.3. Toxicity of Mixed CAR and ENF Exposure on Zebrafish Embryo Development
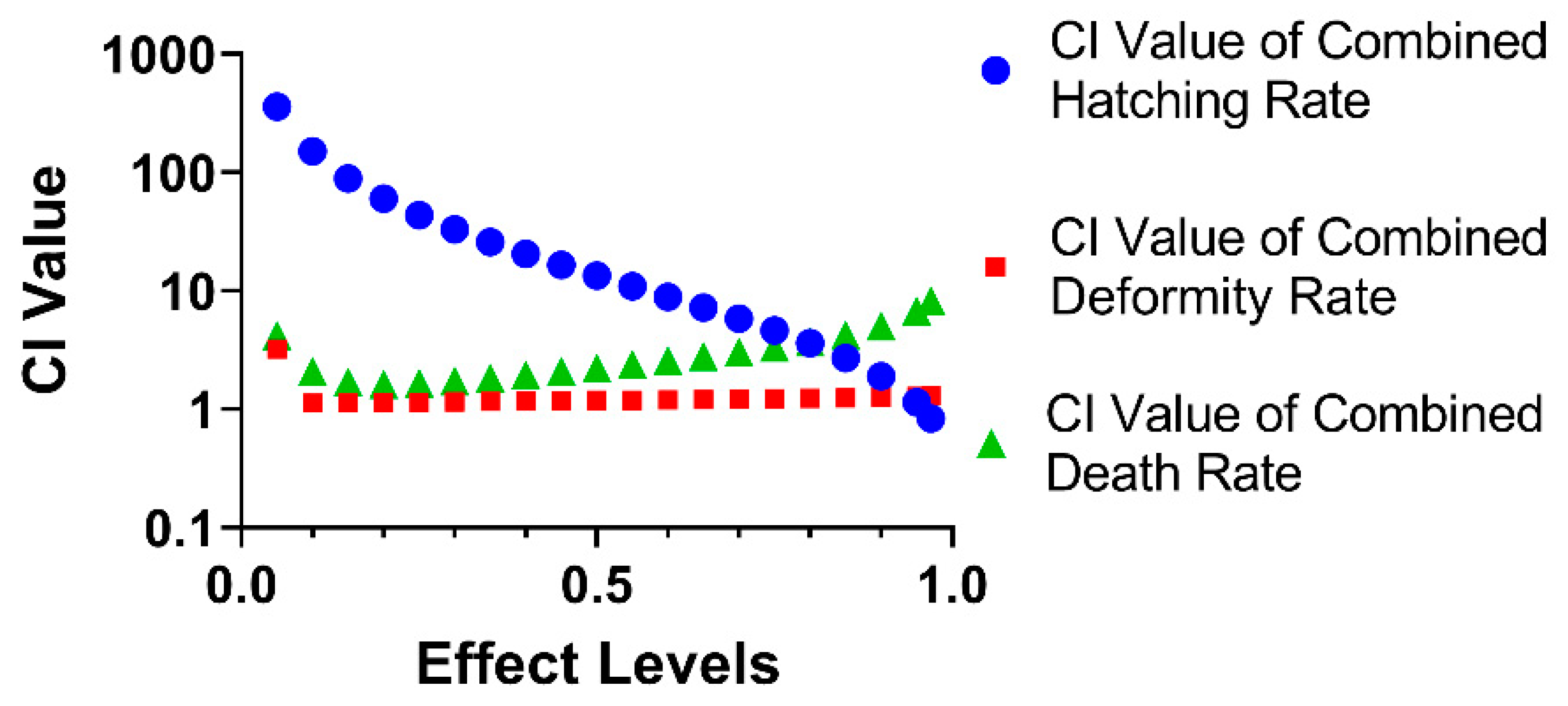
3.4. The Combined Effect of CAR and ENF on Developmental Parameters
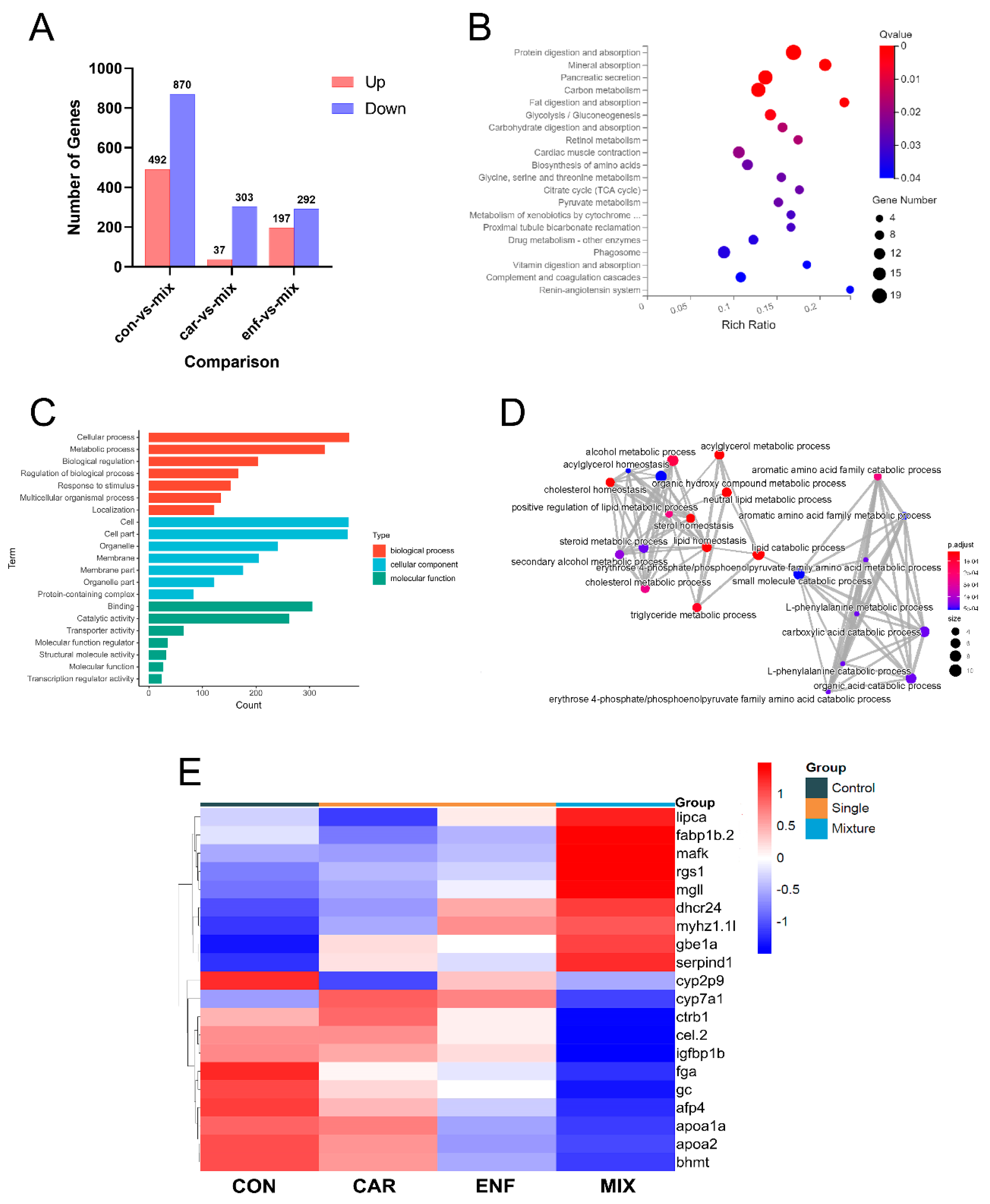
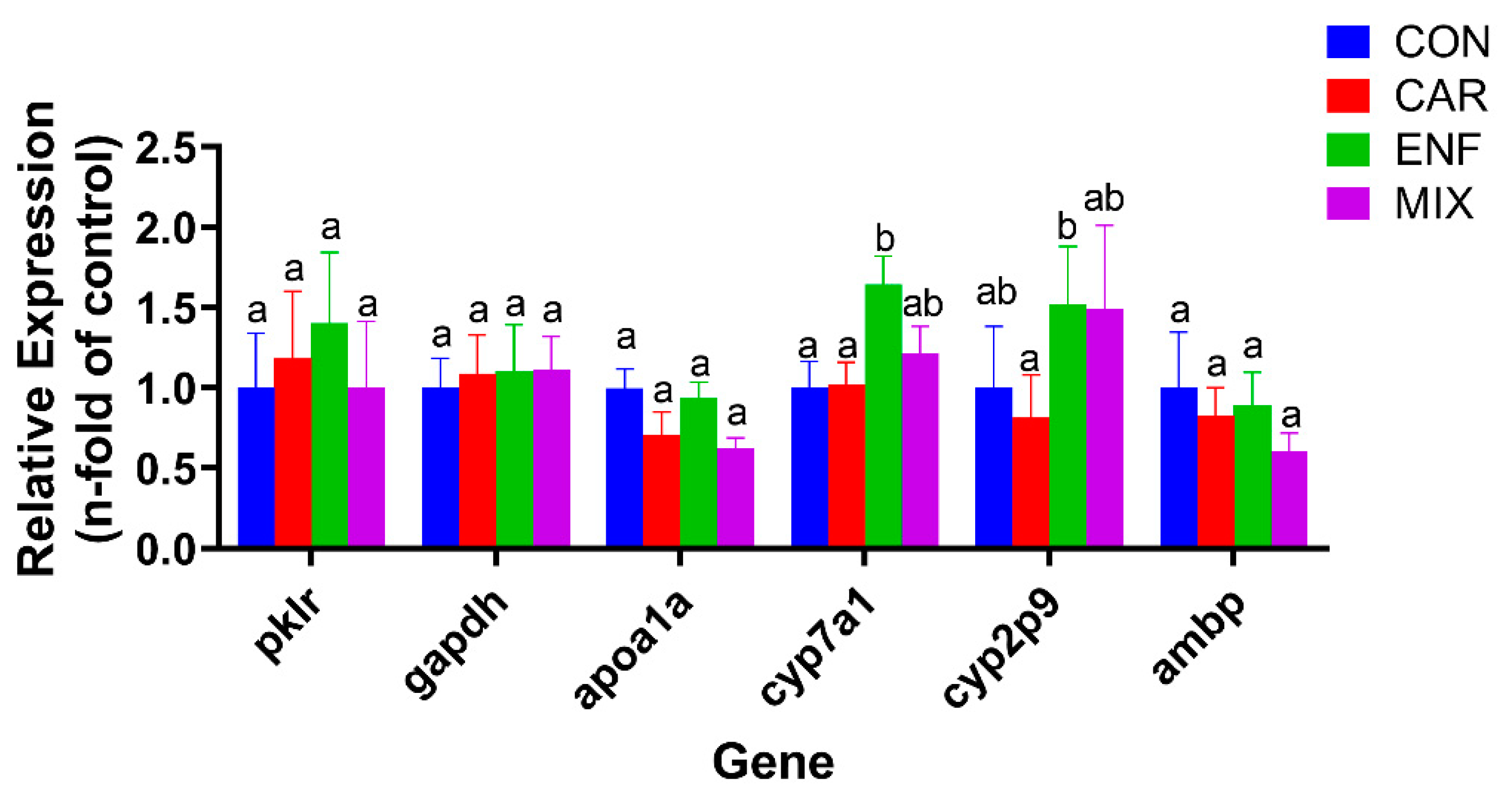
3.5. Transcriptomics and qPCR Analysis of Toxicity Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teglia, C.M.; Guinez, M.; Culzoni, M.J.; Cerutti, S. Determination of residual enrofloxacin in eggs due to long term administration to laying hens. Analysis of the consumer exposure assessment to egg derivatives. Food Chem. 2021, 351, 8. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Xu, Y.; Wang, J.; Lyu, W.; Li, R.; Tan, S.; Xiao, Y.; Tang, B.; Yang, H.; Qian, M. Multiresidue determination of antibiotics in ready-to-eat duck eggs marketed through e-commerce stores in China and subsequent assessment of dietary risks to consumers. J. Food Sci. 2021, 86, 2145–2162. [Google Scholar] [CrossRef]
- Ellerbrock, R.E.; Canisso, I.F.; Roady, P.J.; Rothrock, L.T.; Zhong, L.; Wilkins, P.; Dirikolu, L.; Lima, F.S.; Honoroto, J. Diffusion of enrofloxacin to pregnancy fluids and effects on fetal cartilage after intravenous administration to late pregnant mares. Equine Vet. J. 2019, 51, 544–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Chen, Z.; Li, Y.; Liu, Y. Effects of low concentration enrofloxacin on SPF mice intestinal microflora. Sci. Agric. Sin. 2005, 38, 1905–1910. [Google Scholar]
- Xu, X.; Chen, J.; Li, B.; Tang, L. Carbendazim residues in vegetables in China between 2014 and 2016 and a chronic carbendazim exposure risk assessment. Food Control 2018, 91, 20–25. [Google Scholar] [CrossRef]
- Songür, S.H.; Koçkaya, E.A.; Selmanoĝlu, G.; Barlas, N. Dose-dependent effects of carbendazim on rat thymus. Cell Biochem. Funct. 2005, 23, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, F.; Nakai, M.; Nasu, T. The fungicide carbendazim induces meiotic micronuclei in the spermatids of the rat testis. J. Vet. Med. Sci. 1999, 61, 573–576. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.H.; Liu, S.S.; Liu, H.L.; Liu, Z.Z. Evaluation of the combined toxicity of 15 pesticides by uniform design. Pest Manag. Sci. 2010, 66, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, A.F.; Parron, T.; Tsatsakis, A.M.; Requena, M.; Alarcon, R.; Lopez-Guarnido, O. Toxic effects of pesticide mixtures at a molecular level: Their relevance to human health. Toxicology 2013, 307, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pleiter, M.; Gonzalo, S.; Rodea-Palomares, I.; Leganes, F.; Rosal, R.; Boltes, K.; Marco, E.; Fernandez-Pinas, F. Toxicity of five antibiotics and their mixtures towards photosynthetic aquatic organisms: Implications for environmental risk assessment. Water Res. 2013, 47, 2050–2064. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, K.; Nagase, H.; Ozawa, M.; Endoh, Y.S.; Goto, K.; Hirata, K.; Miyamoto, K.; Yoshimura, H. Evaluation of antimicrobial agents for veterinary use in the ecotoxicity test using microalgae. Chemosphere 2004, 57, 1733–1738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, H.; Wang, S.; Ding, T.; Xu, N.; Yu, K.; Shen, W.; Zhao, Z.; Liu, Y.; Wu, B.; et al. Simultaneous Determination of Eight Kinds of Drug Residues in Sprouts by High Performance Liquid Chromatography—Tandem Mass Spectrometry. J. Instrum. Anal. 2015, 34, 164–170. [Google Scholar]
- Gong, Y.; Zou, X.; Zhang, L.; Wu, Y.; Dai, M.; Chen, Z.; Zhang, M. Study on detecting enrofloxacin residues in food by chemiluminescence immunoassay. Sci. Technol. Food Ind. 2014, 35, 66. [Google Scholar]
- Jagadeeswaran, P. Zebrafish: A tool to study hemostasis and thrombosis. Curr. Opin. Hematol. 2005, 12, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.J.; Teraoka, H.; Heideman, W.; Peterson, R.E. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol. Sci. 2005, 86, 6–19. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Chen, K.; Qiu, X.C.; Xu, H.; Mao, G.H.; Zhao, T.; Feng, W.W.; Okeke, E.S.; Wu, X.Y.; Yang, L.Q. The reproductive toxicity and potential mechanisms of combined exposure to dibutyl phthalate and diisobutyl phthalate in male zebrafish (Danio rerio). Chemosphere 2020, 258, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.N.; Zhang, Q.N.; Fang, J.; Ma, N.; Geng, X.; Xu, M.; Yang, H.; Jia, X.D. Hepatotoxicity study of combined exposure of DEHP and ethanol: A comprehensive analysis of transcriptomics and metabolomics. Food Chem. Toxicol. 2020, 141, 9. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Wu, Y.; Li, S.; Su, B. Effects of different concentrations of dimethylsulfoxide on development of zebrafish embryos and dopamine neurons. J. Third Mil. Med. Univ. 2019, 41, 1538–1544. [Google Scholar]
- Chen, T.H.; Wang, Y.H.; Wu, Y.H. Developmental exposures to ethanol or dimethylsulfoxide at low concentrations alter locomotor activity in larval zebrafish: Implications for behavioral toxicity bioassays. Aquat. Toxicol. 2011, 102, 162–166. [Google Scholar] [CrossRef]
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. GB 2763-2021 National Food Safety Standard—Maximum Residue Limits of Pesticides in Food; MOA: Beijing, China, 2021.
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. GB 31650-2019 National Food Safety Standard—Maximum Residue Limits for Veterinary Drugs in Foods; MOA: Beijing, China, 2019.
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.A.; Gift, J.S.; Zhao, Q.J. Introduction to benchmark dose methods and US EPA’s benchmark dose software (BMDS) version 2.1.1. Toxicol. Appl. Pharmacol. 2011, 254, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Preclinical versus clinical drug combination studies. Leuk. Lymphoma 2008, 49, 2059–2080. [Google Scholar] [CrossRef]
- Zeid, M.I.A.; Awad, M.K.; Melki, K.C.; Jawdah, Y.A.; Jammoul, A.M. Pesticides residues on Loquat: A minor crop in Lebanon. Food Control 2021, 130, 108297. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhou, B.L.; Qian, M.R.; Wang, Q.; Zhang, H. Transfer Assessment of Carbendazim Residues from Rape Flowers to Apicultural Products. J. Anal. Methods Chem. 2017, 2017, 7. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.P.; Lin, Q.; Wang, X.; Zhang, X.Z.; Xu, J.; Chen, Z.M. Residue transfer and risk assessment of carbendazim in tea. J. Sci. Food Agric. 2018, 98, 5329–5334. [Google Scholar] [CrossRef] [PubMed]
- Bailone, R.L.; Aguiar, L.K.d.; Roca, R.d.O.; Borra, R.C.; Corrêa, T.; Janke, H.; Fukushima, H.C.S. Zebrafish as an animal model for food safety research: Trends in the animal research. Food Biotechnol. 2019, 33, 283–302. [Google Scholar] [CrossRef]
- Ali, S.; van Mil, H.G.J.; Richardson, M.K. Large-Scale Assessment of the Zebrafish Embryo as a Possible Predictive Model in Toxicity Testing. PLoS ONE 2011, 6, e21076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Evaluation of Data for Acceptable Daily Intake (ADI) for Humans, Maximum Residue Levels and Supervised Trials Median Residues (STMRs); WHO: Geneva, Switzerland, 2004. [Google Scholar]
- Bartkiene, E.; Ruzauskas, M.; Bartkevics, V.; Pugajeva, I.; Zavistanaviciute, P.; Starkute, V.; Zokaityte, E.; Lele, V.; Dauksiene, A.; Grashorn, M.; et al. Study of the antibiotic residues in poultry meat in some of the EU countries and selection of the best compositions of lactic acid bacteria and essential oils against Salmonella enterica. Poult. Sci. 2020, 99, 4065–4076. [Google Scholar] [CrossRef]
- Panzenhagen, P.H.; Aguiar, W.S.; Gouvêa, R.; de Oliveira, A.M.; Barreto, F.; Pereira, V.L.; Aquino, M.H. Investigation of enrofloxacin residues in broiler tissues using ELISA and LC-MS/MS. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2016, 33, 639–643. [Google Scholar] [CrossRef]
- World Health Organization. Evaluation of Certain Veterinary Drug Residues in Food; WHO: Geneva, Switzerland, 1998. [Google Scholar]
- Lu, S.-Y.; Liao, J.-W.; Kuo, M.-L.; Hwang, J.-S.; Ueng, T.-H. Antagonistic and synergistic effects of carbendazim and flutamide exposures in utero on reproductive and developmental toxicity in rats. J. Food Drug Anal. 2006, 14, 120–132. [Google Scholar] [CrossRef]
- Dikic, D.; Mojsovic-Cuic, A.; Cupor, I.; Benkovic, V.; Horvat-Knezevic, A.; Lisicic, D.; Orsolic, N. Carbendazim combined with imazalil or cypermethrin potentiate DNA damage in hepatocytes of mice. Hum. Exp. Toxicol. 2012, 31, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.X.; Zhang, Q.; Li, J.L.; Chen, X.Y.; Lang, Q.L.; Kuang, S.P. Combined effects of erythromycin and enrofloxacin on antioxidant enzymes and photosynthesis-related gene transcription in Chlorella vulgaris. Aquat. Toxicol. 2019, 212, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.H.; Zhao, J.J.; Han, H.F.; Shen, J.Z.; Tang, S.S.; Cheng, L.L. Toxicologic effect and transcriptome analysis for short-term orally dosed enrofloxacin combined with two veterinary antimicrobials on rat liver. Ecotoxicol. Environ. Saf. 2021, 220, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, Y. Acute toxicity and joint toxicity of Cr~(6+), Mn~(7+) and Zn~(2+) on Palaemon carincauda juvenile. Mar. Environ. Sci. 2013, 32, 235–238. [Google Scholar]
- Wei, F.; Wang, D.; Li, H.; You, J. Joint toxicity of imidacloprid and azoxystrobin to Chironomus dilutus at organism, cell, and gene levels. Aquat. Toxicol. 2021, 233, 105783. [Google Scholar] [CrossRef]
- Bao, Z.; Zhao, Y.; Wu, A.; Lou, Z.; Lu, H.; Yu, Q.; Fu, Z.; Jin, Y. Sub-chronic carbendazim exposure induces hepatic glycolipid metabolism disorder accompanied by gut microbiota dysbiosis in adult zebrafish (Daino rerio). Sci. Total Environ. 2020, 739, 140081. [Google Scholar] [CrossRef] [PubMed]
- Farag, A.; Ebrahim, H.; ElMazoudy, R.; Kadous, E. Developmental Toxicity of Fungicide Carbendazim in Female Mice. Birth Defects Res. Part B Dev. Reprod. Toxicol. 2011, 92, 122–130. [Google Scholar] [CrossRef]
- Kong, B.; Wang, L.; Chiang, J.Y.L.; Zhang, Y.C.; Klaassen, C.D.; Guo, G.L. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 2012, 56, 1034–1043. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Han, Y.L.; Chai, F.N.; Xiang, H.M.; Huang, T.; Kou, S.M.; Han, B.; Gong, X.B.; Ye, X.L. The antihypercholesterolemic effect of columbamine from Rhizoma Coptidis in HFHC-diet induced hamsters through HNF-4 alpha/FTF-mediated CYP7A1 activation. Fitoterapia 2016, 115, 111–121. [Google Scholar] [CrossRef]
- Sharma, R.; Kaur, R.; Mukesh, M.; Sharma, V.L. Assessment of hepatotoxicity of first-line anti-tuberculosis drugs on Wistar rats. Naunyn-Schmiedebergs Arch. Pharmacol. 2018, 391, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Vieira, L.R.; Hissa, D.C.; de Souza, T.M.; Sa, C.A.; Evaristo, J.A.M.; Nogueira, F.C.S.; Carvalho, A.F.U.; Farias, D.F. Proteomics analysis of zebrafish larvae exposed to 3,4-dichloroaniline using the fish embryo acute toxicity test. Environ. Toxicol. 2020, 35, 849–860. [Google Scholar] [CrossRef]
- Cui, Y.; Lv, S.; Liu, J.; Nie, S.; Chen, J.; Dong, Q.; Huang, C.; Yang, D. Chronic perfluorooctanesulfonic acid exposure disrupts lipid metabolism in zebrafish. Hum. Exp. Toxicol. 2017, 36, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Yu, J.; Kong, Y.; Chen, H.; Mao, M.; Ji, C.; Shao, S.; Zhu, J.; Gu, J.; Zhao, M. Metabolomic modulations of HepG2 cells exposed to bisphenol analogues. Environ. Int. 2019, 129, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wang, M.; Liang, X.; Martyniuk, C.J.; Zha, J.; Wang, Z. Environmentally relevant concentrations of carbamazepine induce liver histopathological changes and a gender-specific response in hepatic proteome of Chinese rare minnows (Gobiocypris rarus). Environ. Pollut. 2018, 243, 480–491. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, R.; Zhang, W.; Jia, L.; Luo, S.; Liu, Y.; Jin, Y.; Li, Y.; Yuan, X.; Chen, Y. Antagonistic Effects of Enrofloxacin on Carbendazim-Induced Developmental Toxicity in Zebrafish Embryos. Toxics 2021, 9, 349. https://doi.org/10.3390/toxics9120349
Fan R, Zhang W, Jia L, Luo S, Liu Y, Jin Y, Li Y, Yuan X, Chen Y. Antagonistic Effects of Enrofloxacin on Carbendazim-Induced Developmental Toxicity in Zebrafish Embryos. Toxics. 2021; 9(12):349. https://doi.org/10.3390/toxics9120349
Chicago/Turabian StyleFan, Ruiqi, Wanjun Zhang, Li Jia, Sunlin Luo, Ying Liu, Yongpeng Jin, Yongchen Li, Xiaoyan Yuan, and Yiqiang Chen. 2021. "Antagonistic Effects of Enrofloxacin on Carbendazim-Induced Developmental Toxicity in Zebrafish Embryos" Toxics 9, no. 12: 349. https://doi.org/10.3390/toxics9120349
APA StyleFan, R., Zhang, W., Jia, L., Luo, S., Liu, Y., Jin, Y., Li, Y., Yuan, X., & Chen, Y. (2021). Antagonistic Effects of Enrofloxacin on Carbendazim-Induced Developmental Toxicity in Zebrafish Embryos. Toxics, 9(12), 349. https://doi.org/10.3390/toxics9120349





