Comparison of Artificial Neural Network and Multiple Linear Regression to Predict Cadmium Concentration in Rice: A Field Study in Guangxi, China
Abstract
1. Introduction
2. Materials and Methods
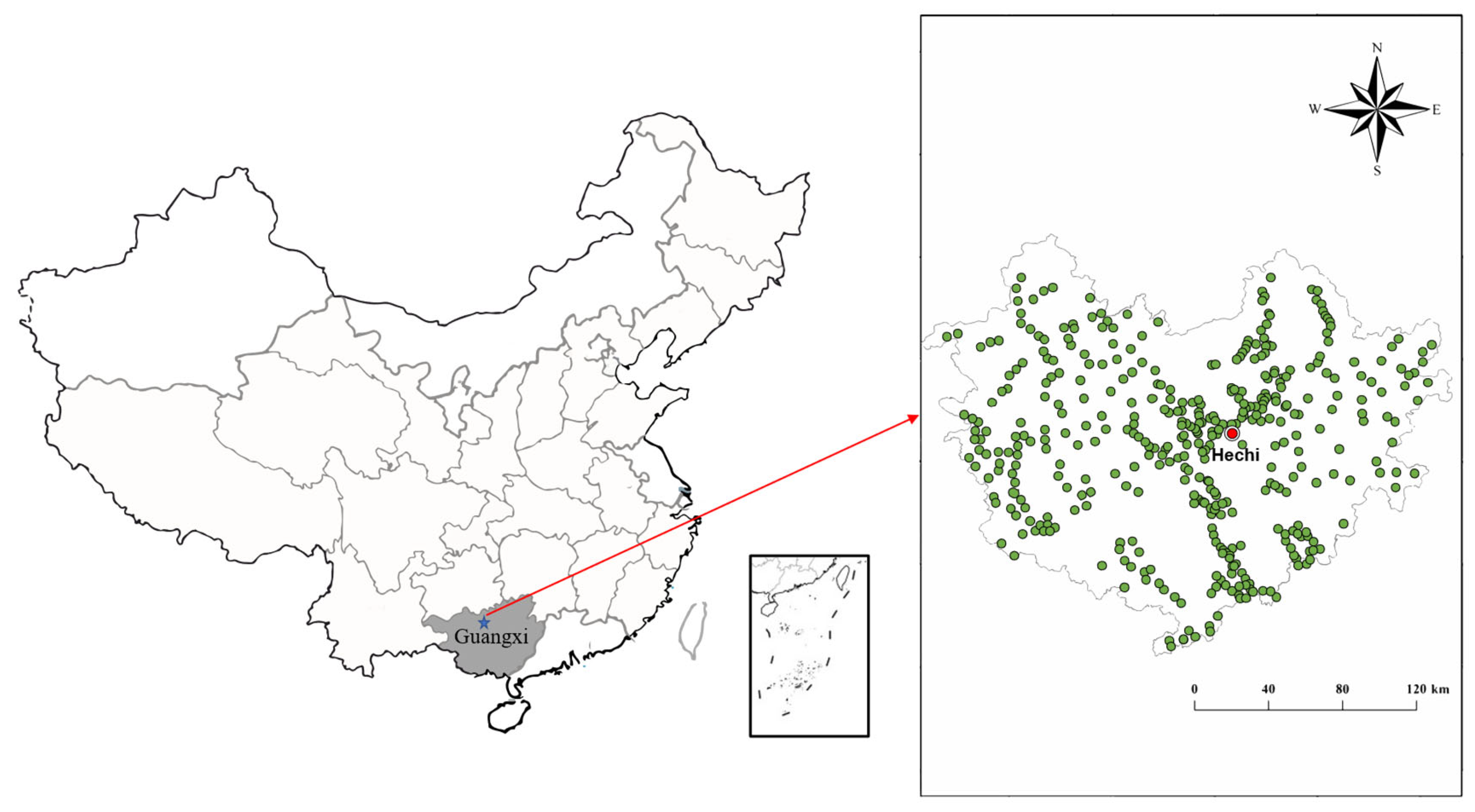
2.1. Study Area
2.2. Sampling and Pre-Treatment
2.3. Soil and Rice Sampling
2.4. Statistical Analysis
3. Development of Prediction Models
3.1. Data Pre-Processing Phase
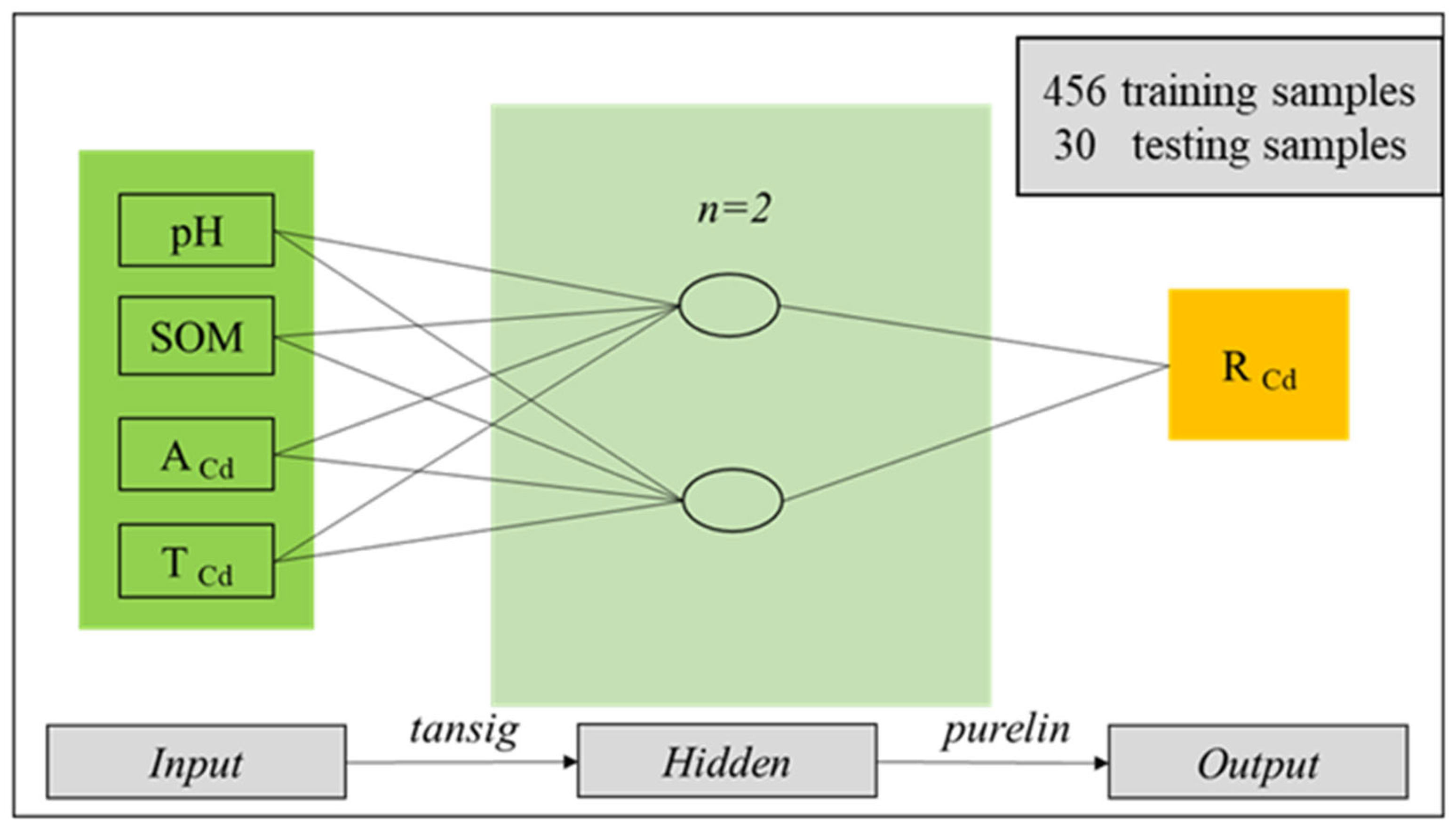
3.2. Development of BP-ANN Model
3.3. Development of MLR Model
3.4. Evaluation Criteria for Model Performance
4. Results and Discussion
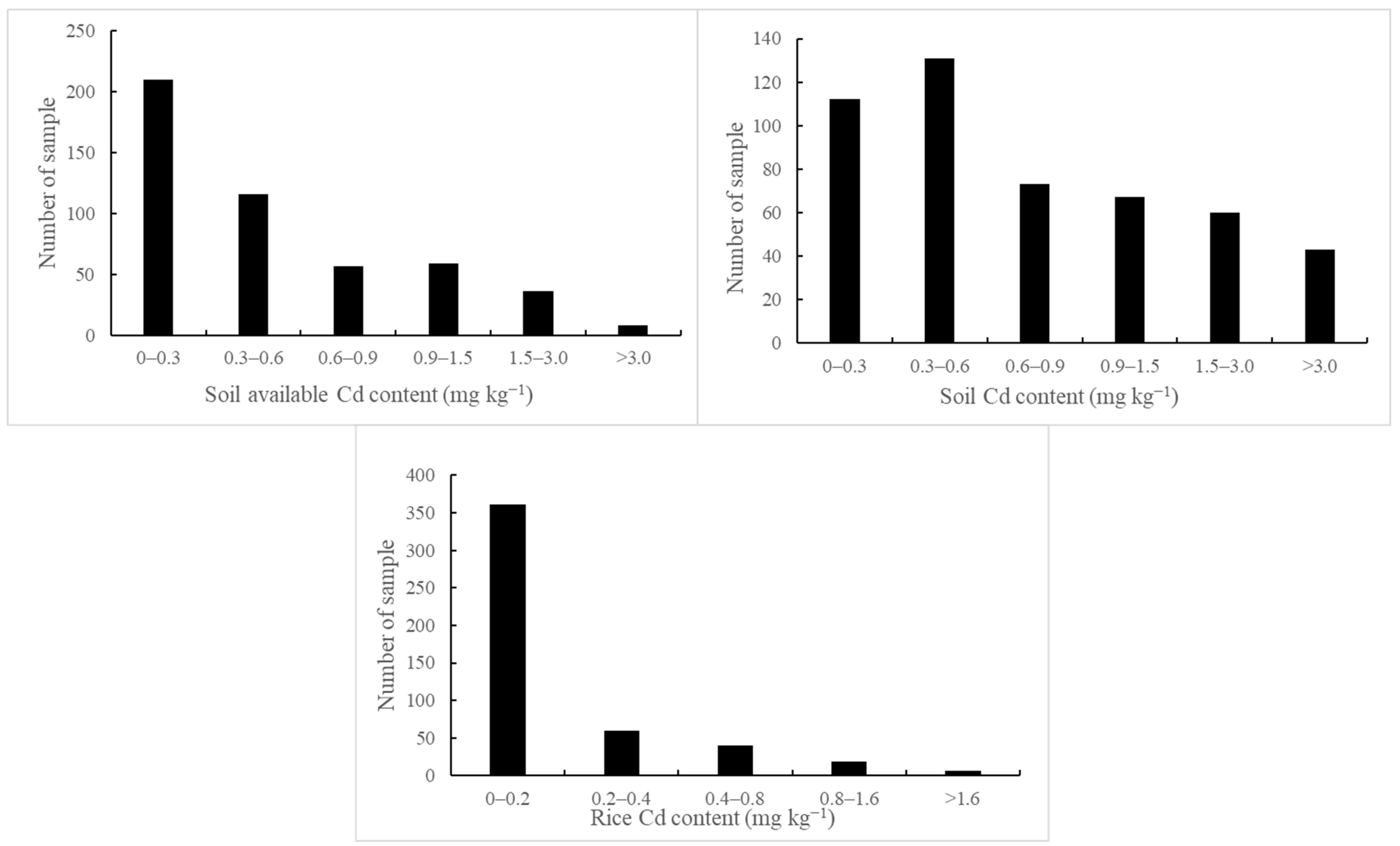
4.1. Changes in Soil Properties and Cd Concentration
4.2. Effect of Soil Properties on RCd
4.3. Development of BP-ANN Model for RCd
4.4. MLR for Predicting Cd Content in Rice
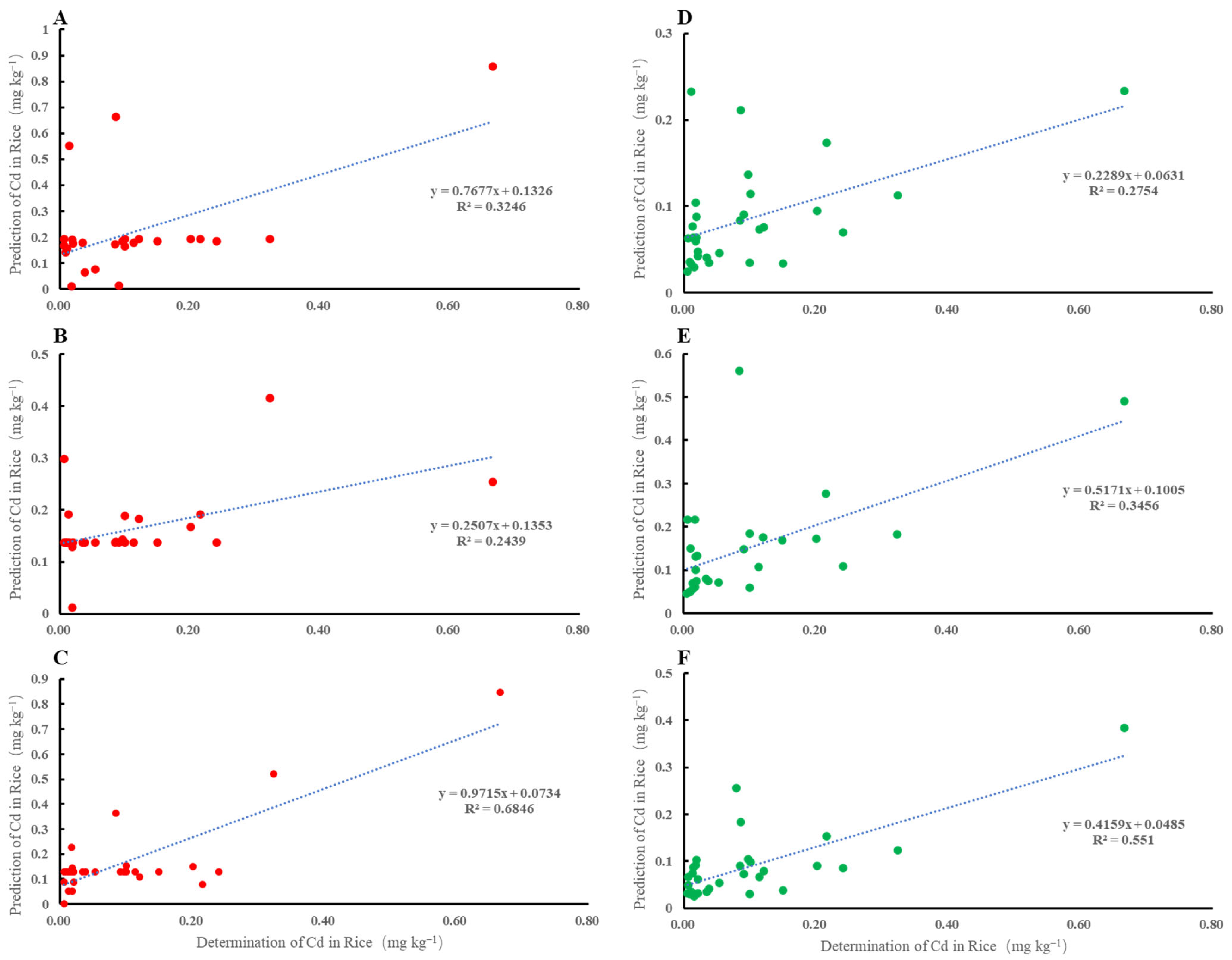
5. Comparison of Different Models
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, J.; Qin, S.; Pan, P.; Chen, D.; Tang, S.; Chen, L.; Wang, X.; Gu, M.; Tang, F.; He, J.; et al. Microbial driving mechanism of soil conditioner on reducing cadmium uptake by rice and improving soil environment. Agric. Ecosyst. Environ. 2023, 349, 108452. [Google Scholar] [CrossRef]
- Shahid, M.; Dumat, C.; Khalid, S.; Niazi, N.K.; Antunes, P.M.C. Cadmium Bioavailability, Uptake, Toxicity and Detoxification in Soil-Plant System. Rev. Environ. Contam. Toxicol. 2017, 241, 73–137. [Google Scholar] [PubMed]
- Liu, F.; Liu, X.; Ding, C.; Wu, L. The dynamic simulation of rice growth parameters under cadmium stress with the assimilation of multi-period spectral indices and crop model. Field Crops Res. 2015, 183, 225–234. [Google Scholar] [CrossRef]
- Hu, Y.; Cheng, H.; Tao, S. The Challenges and Solutions for Cadmium-contaminated Rice in China: A Critical Review. Environ. Int. 2016, 92–93, 515–532. [Google Scholar] [CrossRef]
- Li, K.; Cao, C.; Ma, Y.; Su, D.; Li, J. Identification of cadmium bioaccumulation in rice (Oryza sativa L.) by the soil-plant transfer model and species sensitivity distribution. Sci. Total Environ. 2019, 692, 1022–1028. [Google Scholar] [CrossRef]
- Zhou, T.; Wu, L.; Luo, Y.; Christie, P. Effects of organic matter fraction and compositional changes on distribution of cadmium and zinc in long-term polluted paddy soils. Environ. Pollut. 2018, 232, 514–522. [Google Scholar] [CrossRef]
- Ye, X.; Hu, H.; Li, H.; Xiong, Q.; Gao, H. Combined nitrogen fertilizer and wheat straw increases the cadmium phytoextraction efficiency of Tagetes patula. Ecotoxicol. Environ. Saf. 2019, 170, 210–217. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Li, Y.; Liang, C.; Huang, H.; Wang, S. Available heavy metals concentrations in agricultural soils: Relationship with soil properties and total heavy metals concentrations in different industries. J. Hazard. Mater. 2024, 471, 134410. [Google Scholar] [CrossRef]
- Hu, B.; Xue, J.; Zhou, Y.; Shao, S.; Fu, Z.; Li, Y.; Chen, S.; Qi, L.; Shi, Z. Modelling bioaccumulationof heavy metals in soil-crop ecosystems and identifying itscontrolling factors using machine learning. Environ. Pollut. 2020, 262, 114308. [Google Scholar] [CrossRef] [PubMed]
- Mu, T.; Zhou, T.; Li, Z.; Hu, P.; Luo, Y.; Christie, P.; Wu, L. Prediction models for rice Cd accumulation in Chinese paddy fields and the im-plications in deducing soil thresholds based on food safety standards. Environ. Pollut. 2020, 258, 113879. [Google Scholar] [CrossRef]
- Hou, Y.; Zhao, H.; Wu, K.; Li, K. Prediction of crop Cd content and zoning of safety planting based on BP neural network. Resour. Sci. 2018, 40, 2414–2424. [Google Scholar]
- Liu, H.; Tarima, S.; Borders, A.S.; Getchell, T.V.; Getchell, M.L.; Stromberg, A.J. Quadratic regression analysis for gene discovery and pattern recognition for non-cyclic short time-course microarray experiments. BMC Bioinform. 2005, 6, 106. [Google Scholar] [CrossRef]
- Fawzy, M.; Nasr, M.; Nagy, H.; Helmi, S. Artificial intelligence and regression analysis for Cd(II) ion biosorption from aqueous solution by Gossypium barbadense waste. Environ. Sci. Pollut. Res. Int. 2018, 25, 5875–5888. [Google Scholar] [CrossRef] [PubMed]
- Römkens, P.F.; Guo, H.Y.; Chu, C.L.; Liu, T.S.; Chiang, C.F.; Koopmans, G.F. Prediction of Cadmium uptake by brown rice and derivation of soil-plant transfer models to improve soil protection guidelines. Environ. Pollut. 2009, 157, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Brus, D.J.; Li, Z.; Song, J.; Koopmans, G.F.; Temminghoff, E.J.; Yin, X.; Yao, C.; Zhang, H.; Luo, Y.; Japenga, J. Predictions of spatially averaged cadmium contents in rice grains in the Fuyang Valley, P.R. China. J. Environ. Qual. 2009, 38, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Zhang, T.; Wang, X.; Zhou, F.; Yang, Y.; Yin, Y. Effects of soil type and genotype on lead concentration in rootstalk vegetables and the selection of cultivars for food safety. J. Environ. Manag. 2013, 122, 8–14. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, X.; Li, T.; Hu, S.; Ji, J.; Wang, C. Characteristics of heavy metal transfer and their influencing factors in different soil-crop systems of the industrialization region, China. Ecotoxicol. Environ. Saf. 2016, 126, 193–201. [Google Scholar] [CrossRef]
- Mojid, M.A.; Hossain, A.B.M.Z.; Ashraf, M.A. Artificial neural network model to predict transport parameters of reactive solutes from basic soil properties. Environ. Pollut. 2019, 255 Pt 2, 113355. [Google Scholar] [CrossRef]
- Liu, W.; Guo, G.; Chen, F.; Chen, Y. Meteorological pattern analysis assisted daily PM2.5 grades prediction using SVM optimized by PSO algorithm. Atmos. Pollut. Res. 2019, 10, 1482–1491. [Google Scholar] [CrossRef]
- Hou, Y.X.; Zhao, H.F.; Zhang, Z.; Wu, K.N. A novel method for predicting cadmium concentration in rice grain using genetic algorithm and back-propagation neural network based on soil properties. Environ. Sci. Pollut. Res. Int. 2018, 25, 35682–35692. [Google Scholar] [CrossRef]
- Wu, B.; Guo, S.; Zhang, L.; Li, F. Risk forewarning model for rice grain Cd pollution based on Bayes theory. Sci. Total Environ. 2018, 618, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Khazaei, J.; Naghavi, M.R.; Jahansouz, M.R.; Salimi-Khorshidi, G. Yield Estimation and Clustering of Chickpea Genotypes Using Soft Computing Techniques. Agron. J. 2008, 100, 1077–1087. [Google Scholar] [CrossRef]
- Jin, T.; Cai, S.; Jiang, D.; Liu, J. A data-driven model for real-time water quality prediction and early warning by an integration method. Environ. Sci. Pollut. Res. Int. 2019, 26, 30374–30385. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tang, L.; Xiang, M.; Zhang, C.; Ge, Y.; Chen, X. Model constructions and validations for regional cadmium coupling relationships in soilrice grain. Chin. J. Ecol. 2021, 40, 2341–2347. [Google Scholar]
- Zhang, D.; Liu, J.; Jiang, C.; Liu, A.; Xia, B. Quantitative detection of formaldehyde and ammonia gas via metal oxide-modified graphene-based sensor array combining with neural network model. Sens. Actuators B Chem. 2017, 240, 55–65. [Google Scholar] [CrossRef]
- China Geological Survey. Report on Geochemical Survey of Cultivated Land in China; China Geological Survey: Beijing, China, 2015. (In Chinese) [Google Scholar]
- Bao, S. Soil and Agricultural Chemistry Analysis; China Agricultural Press: Beijing, China, 2000. (In Chinese) [Google Scholar]
- GB/T 23739-2009; Soil Quality—Analysis of Available Lead and Cadmium Contents in Soils—Atomic Absorption Spectrometry. Ministry of Agriculture of the People’s Republic of China: Beijing, China, 2009. (In Chinese)
- Lu, H.; Qin, S.; Zhao, J.; Pan, P.; Wang, F.; Tang, S.; Chen, L.; Akhtar, K.; He, B. Silicon inhibits the upward transport of Cd in the first internode of different rice varieties in a Cd stressed farm land. J. Hazard. Mater. 2023, 458, 131860. [Google Scholar] [CrossRef]
- GBW100348; Certified Reference Material for the Chemical Composition of Rice Flour. NCS Testing Technology Co., Ltd.: Beijing, China, 2015. (In Chinese)
- Azadi, S.; Karimi-Jashni, A. Verifying the performance of artificial neural network and multiple linear regression in predicting the mean seasonal municipal solid waste generation rate: A case study of Fars province, Iran. Waste Manag. 2016, 48, 14–23. [Google Scholar] [CrossRef]
- Blagojev, N.; Kukić, D.; Vasić, V.; Šćiban, M.; Prodanović, J.; Bera, O. A new approach for modelling and optimization of Cu(II) biosorption from aqueous solutions using sugar beet shreds in a fixed-bed column. J. Hazard. Mater. 2019, 363, 366–375. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, B.; Zuo, P.; Huang, Z.; Zhang, J. Influencing factors and prediction of ambient Peroxyacetyl nitrate concentration in Beijing, China. J. Environ. Sci. 2019, 77, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Deo, R.C.; Sahin, M. Forecasting long-term global solar radiation with an ANN algorithm coupled with satellite-derived (MODIS) land surface temperature (LST) for regional locations in Queensland. Renew. Sustain. Energy Rev. 2017, 72, 828–848. [Google Scholar] [CrossRef]
- Luo, X.; Chen, R.; Kabir, M.H.; Liu, F.; Tao, Z.; Liu, L.; Kong, W. Fast Detection of Heavy Metal Content in Fritillaria thunbergii by Laser-Induced Breakdown Spectroscopy with PSO-BP and SSA-BP Analysis. Molecules 2023, 28, 3360. [Google Scholar] [CrossRef]
- An, B.; Wang, X.; Huang, X.; Kawuqiati, B. Hyperspectral Estimation of Heavy Metal Cadmium Content in Soil based on Continuous Wavelet Transform. Earth Environ. 2023, 51, 246–253. [Google Scholar]
- NY861-2004; Limits of Eight Elements in Cereals, Legume, Tubes and Its Products. Ministry of Agriculture of the People’s Republic of China: Beijing, China, 2004. (In Chinese)
- Novotná, M.; Mikeš, O.; Komprdová, K. Development and comparison of regression models for the uptake of metals into various field crops. Environ. Pollut. 2015, 207, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.L.; Zhao, F.; McGrath, S.P.; Nicholson, F.A.; Chambers, B.J. Predicting cadmium concentrations in wheat and barley grain using soil properties. J. Environ. Qual. 2004, 33, 532–541. [Google Scholar] [CrossRef]
- Zhu, B.; Liao, Q.; Zhao, X.; Gu, X.; Gu, C. A multi-surface model to predict Cd phytoavailability to wheat (Triticum aestivum L.). Sci. Total Environ. 2018, 630, 1374–1380. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, J.; Zhou, T.; Li, Z.; Jiang, J.; Zhu, D.; Hou, J.; Wang, Z.; Luo, Y.; Christie, P. Estimating cadmium availability to the hyperaccumulator Sedum plumbizincicola in a wide range of soil types using a piecewise function. Sci. Total Environ. 2018, 637–638, 1342–1350. [Google Scholar] [CrossRef]
- Wan, Y.; Huang, Q.; Wang, Q.; Yu, Y.; Su, D.; Qiao, Y.; Li, H. Accumulation and bioavailability of heavy metals in an acid soil and their uptake by paddy rice under continuous application of chicken and swine manure. J. Hazard. Mater. 2020, 384, 121293. [Google Scholar] [CrossRef]
- Xu, W.; Li, Y.; He, J.; Ma, Q.; Zhang, X.; Chen, G.; Wang, H.; Zhang, H. Cd uptake in rice cultivars treated with organic acids and EDTA. J. Environ. Sci. 2010, 22, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Fu, W.; Ye, Z.; Zhang, C. Contamination and spatial variation of heavy metals in the soil-rice system in Nanxun County, Southeastern China. Int. J. Environ. Res. Public Health 2015, 12, 1577–1594. [Google Scholar] [CrossRef] [PubMed]
- Prakash Maran, J.; Sivakumar, V.; Thirugnanasambandham, K.; Sridhar, R. Artificial neural network and response surface methodology modeling in mass transfer parameters predictions during osmotic dehydration of Carcia papaya L. Int. J. Food Eng. 2013, 52, 507–516. [Google Scholar]
- Olawoyin, R. Application of backpropagation artificial neural network prediction model for the PAH bioremediation of polluted soil. Chemosphere 2016, 161, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzi, A.; Sarmadian, F. Comparison of artificial neural network and multivariate regression methods in prediction of soil cation exchange capacity. Int. J. Environ. Chem. Ecol. Geo. GeoEng 2010, 4, 644–649. [Google Scholar]
- Shi, J.; Xu, J.; Yao, Y.; Xu, B. Concept learning through deep reinforcement learning with memory-augmented neural networks. Neural Netw. 2019, 110, 47–54. [Google Scholar] [CrossRef] [PubMed]

| Parameters | Mean | Minimum | Maximum |
|---|---|---|---|
| pH | 6.14 | 4.56 | 8.00 |
| SOM (g kg−1) | 37.29 | 4.07 | 75.43 |
| ACd (mg kg−1) | 0.61 | 0.002 | 4.86 |
| TCd (mg kg−1) | 1.13 | 0.093 | 8.76 |
| RCd (mg kg−1) | 0.200 | 0.001 | 4.431 |
| Indications | pH | SOM | ACd | TCd | RCd |
|---|---|---|---|---|---|
| pH | 1 | 0.123 ** | 0.112 * | 0.232 ** | 0.216 ** |
| SOM | 1 | 0.256 ** | 0.286 ** | −0.078 | |
| ACd | 1 | 0.827 ** | 0.220 ** | ||
| TCd | 1 | 0.124 ** | |||
| RCd | 1 |
| Model | Input | Output |
|---|---|---|
| Model I | pH, SOM, TCd | RCd |
| Model II | pH, SOM, ACd | RCd |
| Model III | pH, TCd, ACd | RCd |
| Implied Layer of Neurons | Training Set and Calibration Set Ratio | RPD | ||
|---|---|---|---|---|
| Model I | Model II | Model III | ||
| 2 | 1:1 | 0.992 | 1.488 | 2.368 |
| 2:1 | 2.165 | 2.488 | 2.036 | |
| 3:1 | 1.892 | 2.409 | 2.015 | |
| 3 | 1:1 | 1.172 | 1.504 | 2.225 |
| 2:1 | 1.683 | 2.315 | 2.422 | |
| 3:1 | 1.464 | 1.328 | 2.282 | |
| 4 | 1:1 | 1.037 | 1.389 | 2.221 |
| 2:1 | 1.165 | 1.246 | 1.944 | |
| 3:1 | 1.492 | 2.386 | 2.172 | |
| 5 | 1:1 | 0.846 | 1.703 | 2.278 |
| 2:1 | 1.040 | 1.578 | 2.185 | |
| 3:1 | 1.132 | 1.153 | 2.047 | |
| 6 | 1:1 | 1.024 | 0.981 | 2.296 |
| 2:1 | 0.901 | 1.312 | 2.081 | |
| 3:1 | 0.910 | 1.749 | 2.157 | |
| Model | Training and Calibration Set Ratio | MLR | RPD |
|---|---|---|---|
| Model I | 1:1 | log RCd = 0.517 − 0.23 pH + 0.483 logTCd | 2.032 |
| 2:1 | log RCd = 0.973 − 0.263 pH − 0.007 SOM + 0.615 logTCd | 2.226 | |
| 3:1 | log RCd = 1.0115 − 0.265 pH − 0.008 SOM + 0.596 logTCd | 2.224 | |
| Model II | 1:1 | logRCd = 0.385 − 0.204 pH + 0.391 logACd | 2.314 |
| 2:1 | log RCd = 0.851 − 0.226 pH − 0.007 SOM + 0.557 logACd | 2.342 | |
| 3:1 | log RCd = 0.814 − 0.221 pH − 0.008 SOM + 0.502 logACd | 2.232 | |
| Model III | 1:1 | log RCd = 0.517 − 0.236 pH + 0.48 logTCd | 2.032 |
| 2:1 | log RCd = 0.645 − 0.258 pH + 0.533 logTCd | 2.172 | |
| 3:1 | log RCd = 0.682 − 0.262 pH + 0.509 logTCd | 2.164 |
| Model | Model Type | RPD | RMSE |
|---|---|---|---|
| Mode I | BP-ANN | 2.670 | 0.119 |
| MLR | 2.573 | 0.135 | |
| Mode II | BP-ANN | 2.853 | 0.102 |
| MLR | 2.581 | 0.123 | |
| Mode III | BP-ANN | 2.778 | 0.104 |
| MLR | 2.398 | 0.049 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Zheng, F.; Yu, B.; Qin, G.; Meng, S.; Qiu, Y.; He, B. Comparison of Artificial Neural Network and Multiple Linear Regression to Predict Cadmium Concentration in Rice: A Field Study in Guangxi, China. Toxics 2025, 13, 645. https://doi.org/10.3390/toxics13080645
Zhao J, Zheng F, Yu B, Qin G, Meng S, Qiu Y, He B. Comparison of Artificial Neural Network and Multiple Linear Regression to Predict Cadmium Concentration in Rice: A Field Study in Guangxi, China. Toxics. 2025; 13(8):645. https://doi.org/10.3390/toxics13080645
Chicago/Turabian StyleZhao, Junyang, Fuhai Zheng, Baoshan Yu, Guanchun Qin, Shunpiao Meng, Yuhang Qiu, and Bing He. 2025. "Comparison of Artificial Neural Network and Multiple Linear Regression to Predict Cadmium Concentration in Rice: A Field Study in Guangxi, China" Toxics 13, no. 8: 645. https://doi.org/10.3390/toxics13080645
APA StyleZhao, J., Zheng, F., Yu, B., Qin, G., Meng, S., Qiu, Y., & He, B. (2025). Comparison of Artificial Neural Network and Multiple Linear Regression to Predict Cadmium Concentration in Rice: A Field Study in Guangxi, China. Toxics, 13(8), 645. https://doi.org/10.3390/toxics13080645






