Comparative Analysis of Chemical Distribution Models for Quantitative In Vitro to In Vivo Extrapolation
Abstract
1. Introduction
2. Materials and Methods
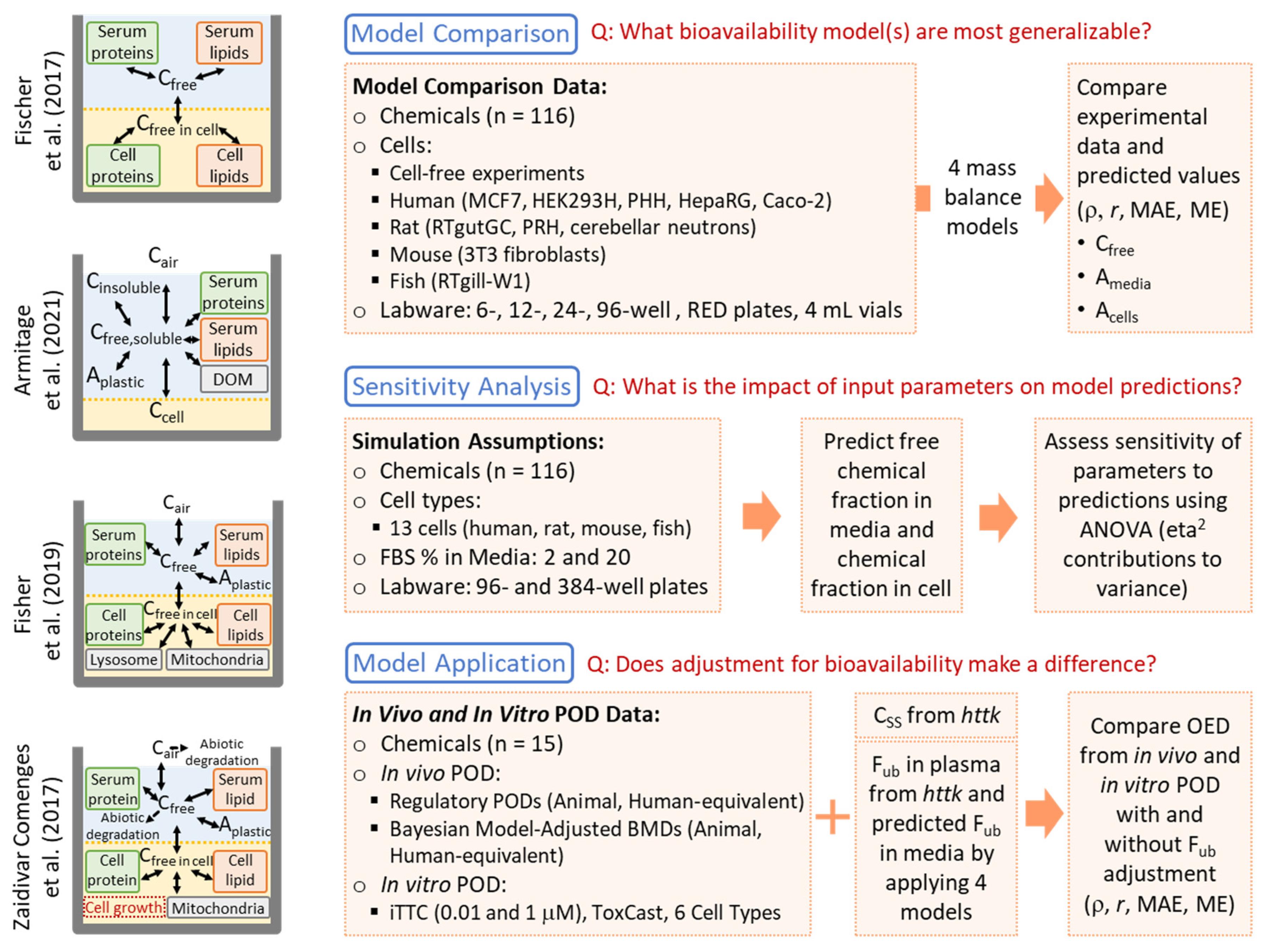
2.1. Model Comparison and Performance Evaluation
2.1.1. Models Selected for Analysis
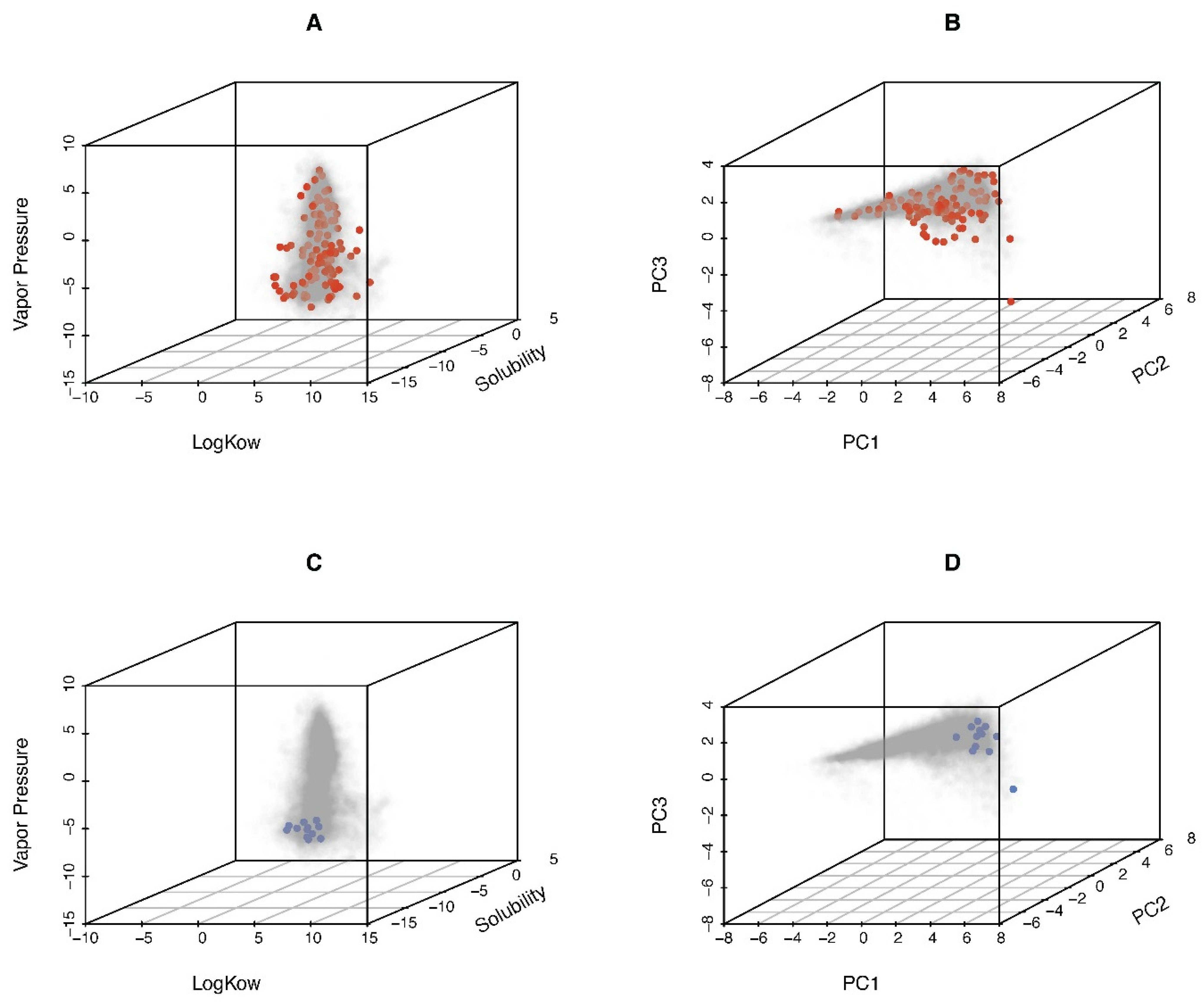
2.1.2. Data Used for Comparisons
- (1)
- Ratio of free to nominal concentration
- (2)
- Chemical amount in media and in cells
2.1.3. Model Parametrization
- (1)
- Chemical-related parameters
- (2)
- Cell-, media-, and labware-related parameters
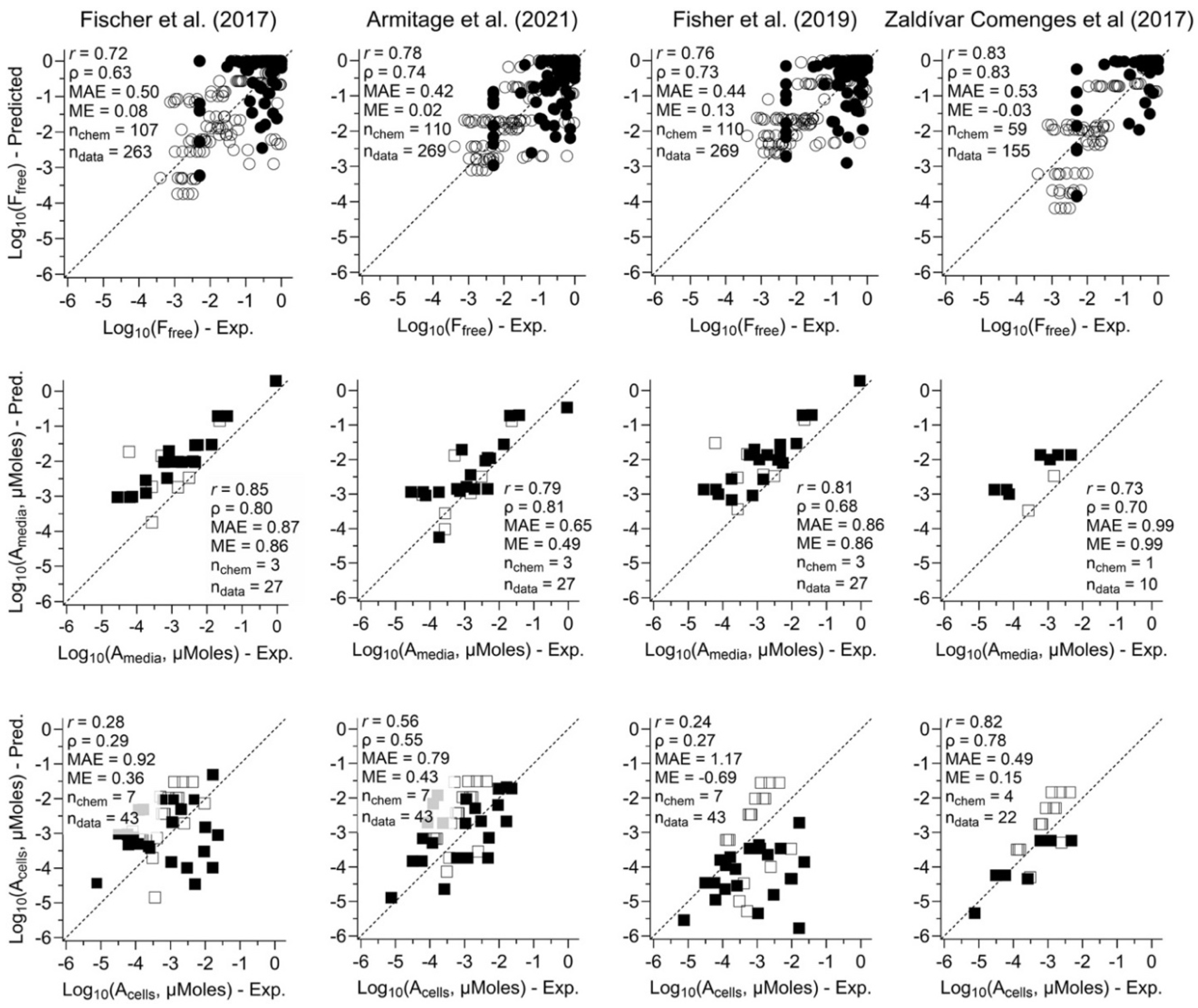
2.1.4. Model Assumptions, Execution, and Evaluation
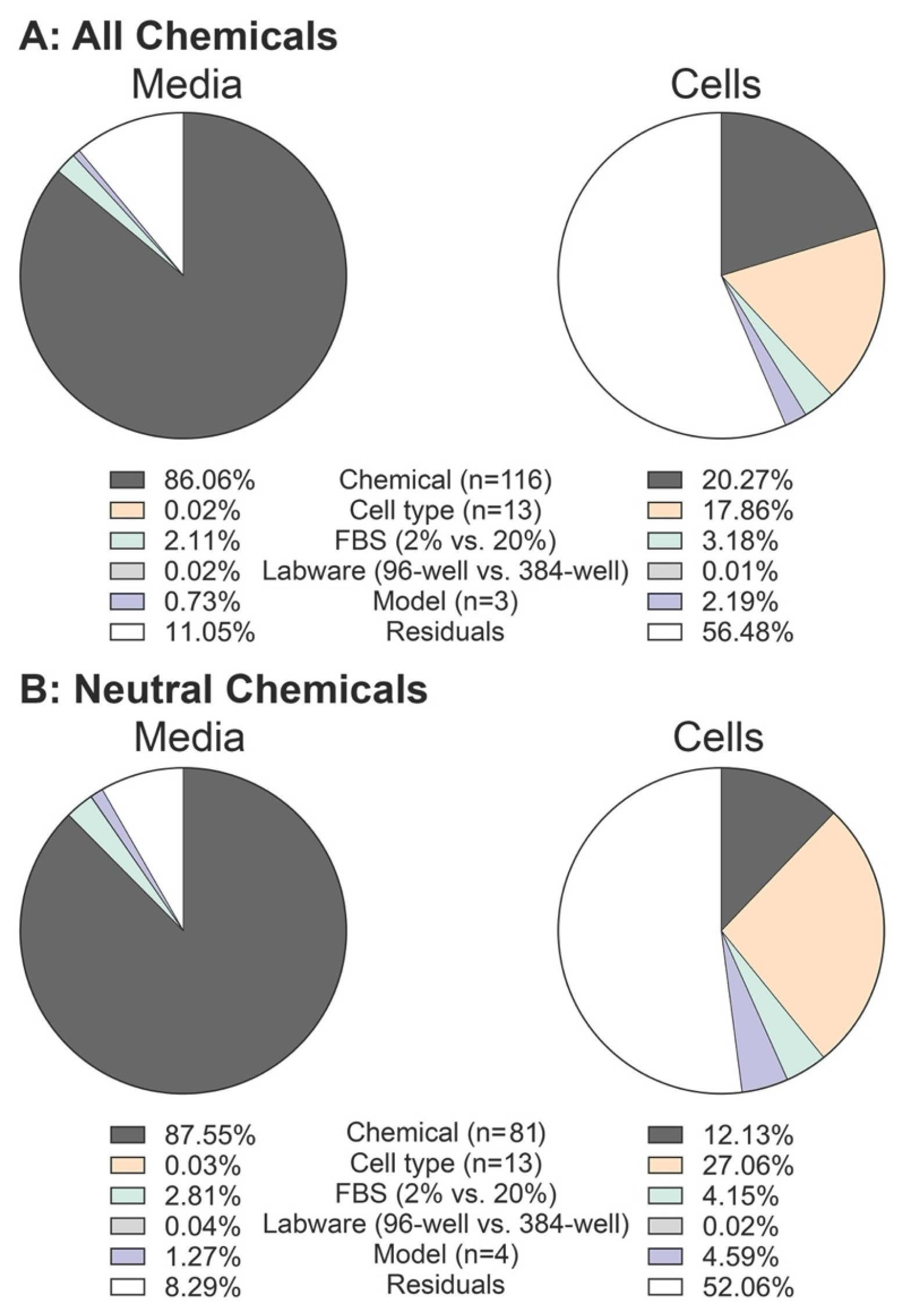
2.2. Sensitivity Analysis
2.3. Chemical Distribution Model Application: Assessing In Vitro–In Vivo Concordance of PODs
3. Results
3.1. Comparing Performance of Four In Vitro Mass Balance Models
3.2. Determining the Impact of Input Parameters on In Vitro Bioavailability Predictions
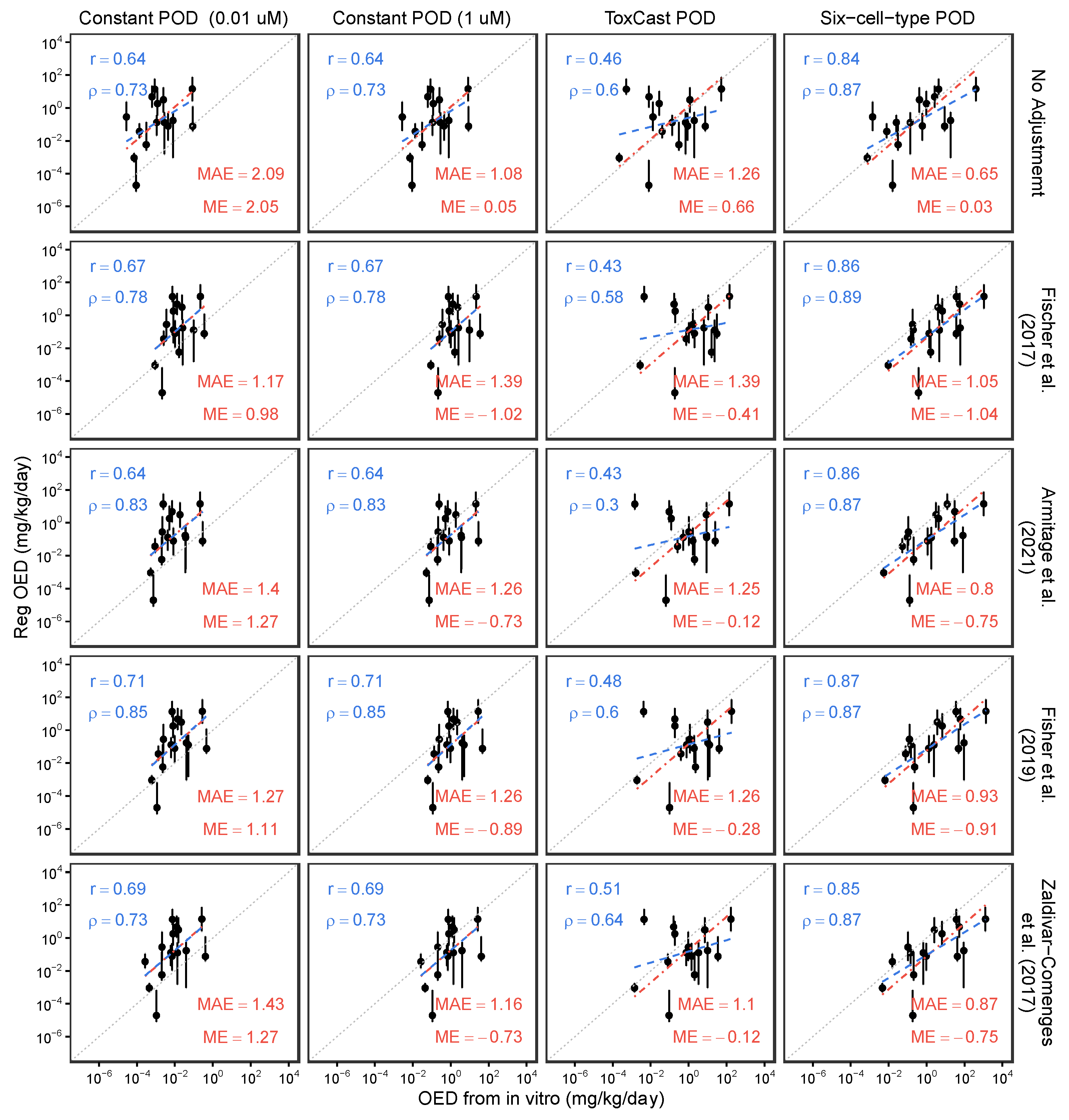
3.3. Model Application: Effects of In Vitro Bioavailability Adjustment on In Vitro–In Vivo Concordance of POD Estimations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| QIVIVE | Quantitative In Vitro to In Vivo Extrapolation |
| Tox21 | Toxicity Testing in the 21st Century |
| HTS | High-Throughput Screening |
| BED | Biologically Effective Dose |
| PBK | Physiologically Based Kinetics |
| iPSCs | Induced Pluripotent Stem Cells |
| POD | Point of Departure |
| IOC | Ionizable Organic Chemical |
| PFAS | Per- and Polyfluoroalkyl Substances |
| PBDEs | Polybrominated Diphenyl Ethers |
| PRH | Primary Rat Hepatocyte |
| PHH | Primary Human Hepatocyte |
| RED | Rapid Equilibrium Dialysis |
| FBS | Fetal Bovine Serum |
| CERAPP | Collaborative Estrogen Receptor Activity Prediction Project |
| PCA | Principal Component Analysis |
| MAE | Mean Absolute Error |
| ME | Mean Error |
| MW | Molecular Weight |
| MP | Melting Point |
| OED | Oral Equivalent Dose |
| iTTC | Internal Threshold of Toxicological Concern |
References
- Dix, D.J.; Houck, K.A.; Martin, M.T.; Richard, A.M.; Setzer, R.W.; Kavlock, R.J. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 2007, 95, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Krimsky, S. The unsteady state and inertia of chemical regulation under the US Toxic Substances Control Act. PLoS Biol. 2017, 15, e2002404. [Google Scholar] [CrossRef]
- National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy; National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Richard, A.M.; Huang, R.; Waidyanatha, S.; Shinn, P.; Collins, B.J.; Thillainadarajah, I.; Grulke, C.M.; Williams, A.J.; Lougee, R.R.; Judson, R.S.; et al. The Tox21 10K Compound Library: Collaborative Chemistry Advancing Toxicology. Chem. Res. Toxicol. 2021, 34, 189–216. [Google Scholar] [CrossRef] [PubMed]
- Judson, R.; Richard, A.; Dix, D.J.; Houck, K.; Martin, M.; Kavlock, R.; Dellarco, V.; Henry, T.; Holderman, T.; Sayre, P.; et al. The toxicity data landscape for environmental chemicals. Environ. Health Perspect 2009, 117, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Paul Friedman, K.; Gagne, M.; Loo, L.H.; Karamertzanis, P.; Netzeva, T.; Sobanski, T.; Franzosa, J.A.; Richard, A.M.; Lougee, R.R.; Gissi, A.; et al. Utility of In Vitro Bioactivity as a Lower Bound Estimate of In Vivo Adverse Effect Levels and in Risk-Based Prioritization. Toxicol. Sci. 2020, 173, 202–225. [Google Scholar] [CrossRef]
- Thomas, R.S.; Bahadori, T.; Buckley, T.J.; Cowden, J.; Deisenroth, C.; Dionisio, K.L.; Frithsen, J.B.; Grulke, C.M.; Gwinn, M.R.; Harrill, J.A.; et al. The Next Generation Blueprint of Computational Toxicology at the U.S. Environmental Protection Agency. Toxicol. Sci. 2019, 169, 317–332. [Google Scholar] [CrossRef]
- Chang, X.; Tan, Y.-M.; Allen, D.G.; Bell, S.; Brown, P.C.; Browning, L.; Ceger, P.; Gearhart, J.; Hakkinen, P.J.; Kabadi, S.V. IVIVE: Facilitating the use of in vitro toxicity data in risk assessment and decision making. Toxics 2022, 10, 232. [Google Scholar] [CrossRef]
- Wetmore, B.A.; Wambaugh, J.F.; Allen, B.; Ferguson, S.S.; Sochaski, M.A.; Setzer, R.W.; Houck, K.A.; Strope, C.L.; Cantwell, K.; Judson, R.S. Incorporating high-throughput exposure predictions with dosimetry-adjusted in vitro bioactivity to inform chemical toxicity testing. Toxicol. Sci. 2015, 148, 121–136. [Google Scholar] [CrossRef]
- Wetmore, B.A.; Wambaugh, J.F.; Ferguson, S.S.; Sochaski, M.A.; Rotroff, D.M.; Freeman, K.; Clewell, H.J., 3rd; Dix, D.J.; Andersen, M.E.; Houck, K.A.; et al. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol. Sci. 2012, 125, 157–174. [Google Scholar] [CrossRef]
- Derendorf, H.; Schmidt, S. Rowland and Tozer’s Clinical Pharmacokinetics and Pharmacodynamics: Concepts and Applications, 5th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2019. [Google Scholar]
- Gülden, M.; Seibert, H. Influence of protein binding and lipophilicity on the distribution of chemical compounds in in vitro systems. Toxicol. Vitr. 1997, 11, 479–483. [Google Scholar] [CrossRef]
- Proença, S.; Escher, B.I.; Fischer, F.C.; Fisher, C.; Grégoire, S.; Hewitt, N.J.; Nicol, B.; Paini, A.; Kramer, N.I. Effective exposure of chemicals in in vitro cell systems: A review of chemical distribution models. Toxicol. Vitr. 2021, 73, 105133. [Google Scholar] [CrossRef] [PubMed]
- Gülden, M.; Mörchel, S.; Seibert, H. Factors influencing nominal effective concentrations of chemical compounds in vitro: Cell concentration. Toxicol. Vitr. 2001, 15, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Heringa, M.B.; Schreurs, R.H.; Busser, F.; Van Der Saag, P.T.; Van Der Burg, B.; Hermens, J.L. Toward more useful in vitro toxicity data with measured free concentrations. Environ. Sci. Technol. 2004, 38, 6263–6270. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.; Simeon, S.; Jamei, M.; Gardner, I.; Bois, Y. VIVD: Virtual in vitro distribution model for the mechanistic prediction of intracellular concentrations of chemicals in in vitro toxicity assays. Toxicol. Vitr. 2019, 58, 42–50. [Google Scholar] [CrossRef]
- Mielke, H.; Di Consiglio, E.; Kreutz, R.; Partosch, F.; Testai, E.; Gundert-Remy, U. The importance of protein binding for the in vitro–in vivo extrapolation (IVIVE)—Example of ibuprofen, a highly protein-bound substance. Arch. Toxicol. 2017, 91, 1663–1670. [Google Scholar] [CrossRef]
- Honda, G.S.; Pearce, R.G.; Pham, L.L.; Setzer, R.; Wetmore, B.A.; Sipes, N.S.; Gilbert, J.; Franz, B.; Thomas, R.S.; Wambaugh, J.F. Using the concordance of in vitro and in vivo data to evaluate extrapolation assumptions. PLoS ONE 2019, 14, e0217564. [Google Scholar] [CrossRef]
- Armitage, J.M.; Sangion, A.; Parmar, R.; Looky, A.B.; Arnot, J.A. Update and evaluation of a high-throughput in vitro mass balance distribution model: IV-MBM EQP v2. 0. Toxics 2021, 9, 315. [Google Scholar] [CrossRef]
- Kramer, N.I. Measuring, Modeling, and Increasing the Free Concentration of Test Chemicals in Cell Assays; Utrecht University: Utrecht, The Netherlands, 2010. [Google Scholar]
- Fischer, F.C.; Cirpka, O.A.; Goss, K.-U.; Henneberger, L.; Escher, B.I. Application of experimental polystyrene partition constants and diffusion coefficients to predict the sorption of neutral organic chemicals to multiwell plates in in vivo and in vitro bioassays. Environ. Sci. Technol. 2018, 52, 13511–13522. [Google Scholar] [CrossRef]
- Fischer, F.C.; Henneberger, L.; König, M.; Bittermann, K.; Linden, L.; Goss, K.-U.; Escher, B.I. Modeling exposure in the Tox21 in vitro bioassays. Chem. Res. Toxicol. 2017, 30, 1197–1208. [Google Scholar] [CrossRef]
- Zaldivar-Comenges, J.; Joossens, E.; Benito, J.S.; Worth, A.; Paini, A. Theoretical and mathematical foundation of the virtual cell based assay—A review. Toxicol. Vitr. 2017, 45, 209–221. [Google Scholar] [CrossRef]
- Huchthausen, J.; Mühlenbrink, M.; König, M.; Escher, B.I.; Henneberger, L. Experimental exposure assessment of ionizable organic chemicals in in vitro cell-based bioassays. Chem. Res. Toxicol. 2020, 33, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Tanneberger, K.; Knöbel, M.; Busser, F.J.; Sinnige, T.L.; Hermens, J.L.; Schirmer, K. Predicting fish acute toxicity using a fish gill cell line-based toxicity assay. Environ. Sci. Technol. 2013, 47, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Schug, H.; Maner, J.; Hülskamp, M.; Begnaud, F.; Debonneville, C.; Berthaud, F.; Gimeno, S.; Schirmer, K. Extending the concept of predicting fish acute toxicity in vitro to the intestinal cell line RTgutGC. Altex 2020, 37, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Nicol, B.; Vandenbossche-Goddard, E.; Thorpe, C.; Newman, R.; Patel, H.; Yates, D. A workflow to practically apply true dose considerations to in vitro testing for next generation risk assessment. Toxicology 2024, 505, 153826. [Google Scholar] [CrossRef]
- Valdiviezo, A.; Luo, Y.-S.; Chen, Z.; Chiu, W.A.; Rusyn, I. Quantitative in vitro-to-in vivo extrapolation for mixtures: A case study of superfund priority list pesticides. Toxicol. Sci. 2021, 183, 60–69. [Google Scholar] [CrossRef]
- Blanchette, A.D.; Grimm, F.A.; Dalaijamts, C.; Hsieh, N.H.; Ferguson, K.; Luo, Y.S.; Rusyn, I.; Chiu, W.A. Thorough QT/QTc in a dish: An in vitro human model that accurately predicts clinical concentration-QTc relationships. Clin. Pharmacol. Ther. 2019, 105, 1175–1186. [Google Scholar] [CrossRef]
- Bellwon, P.; Truisi, G.; Bois, F.Y.; Wilmes, A.; Schmidt, T.; Savary, C.; Parmentier, C.; Hewitt, P.; Schmal, O.; Josse, R. Kinetics and dynamics of cyclosporine A in three hepatic cell culture systems. Toxicol. Vitr. 2015, 30, 62–78. [Google Scholar] [CrossRef]
- Broeders, J.J.; Blaauboer, B.J.; Hermens, J.L. In vitro biokinetics of chlorpromazine and the influence of different dose metrics on effect concentrations for cytotoxicity in Balb/c 3T3, Caco-2 and HepaRG cell cultures. Toxicol. Vitr. 2013, 27, 1057–1064. [Google Scholar] [CrossRef]
- Broeders, J.J.; Parmentier, C.; Truisi, G.L.; Jossé, R.; Alexandre, E.; Savary, C.C.; Hewitt, P.G.; Mueller, S.O.; Guillouzo, A.; Richert, L. Biokinetics of chlorpromazine in primary rat and human hepatocytes and human HepaRG cells after repeated exposure. Toxicol. Vitr. 2015, 30, 52–61. [Google Scholar] [CrossRef]
- Kodavanti, P.R.S.; Ward, T.R.; Ludewig, G.; Robertson, L.W.; Birnbaum, L.S. Polybrominated diphenyl ether (PBDE) effects in rat neuronal cultures: 14C-PBDE accumulation, biological effects, and structure-activity relationships. Toxicol. Sci. 2005, 88, 181–192. [Google Scholar] [CrossRef]
- Pomponio, G.; Zurich, M.-G.; Schultz, L.; Weiss, D.G.; Romanelli, L.; Gramowski-Voss, A.; Di Consiglio, E.; Testai, E. Amiodarone biokinetics, the formation of its major oxidative metabolite and neurotoxicity after acute and repeated exposure of brain cell cultures. Toxicol. Vitr. 2015, 30, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Truisi, G.L.; Di Consiglio, E.; Parmentier, C.; Savary, C.C.; Pomponio, G.; Bois, F.; Lauer, B.; Jossé, R.; Hewitt, P.G.; Mueller, S.O. Understanding the biokinetics of ibuprofen after single and repeated treatments in rat and human in vitro liver cell systems. Toxicol. Lett. 2015, 233, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, K.; Abdelaziz, A.; Rybacka, A.; Roncaglioni, A.; Tropsha, A.; Varnek, A.; Zakharov, A.; Worth, A.; Richard, A.M.; Grulke, C.M. CERAPP: Collaborative estrogen receptor activity prediction project. Environ. Health Perspect. 2016, 124, 1023–1033. [Google Scholar] [CrossRef]
- Pan, X.; Wang, H.; Li, C.; Zhang, J.Z.; Ji, C. MolGpka: A web server for small molecule p K a prediction using a graph-convolutional neural network. J. Chem. Inf. Model. 2021, 61, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Goss, K.-U. Serum albumin binding of structurally diverse neutral organic compounds: Data and models. Chem. Res. Toxicol. 2011, 24, 2293–2301. [Google Scholar] [CrossRef]
- Endo, S.; Escher, B.I.; Goss, K.-U. Capacities of membrane lipids to accumulate neutral organic chemicals. Environ. Sci. Technol. 2011, 45, 5912–5921. [Google Scholar] [CrossRef]
- Bois, F.Y. GNU MCSim: Bayesian statistical inference for SBML-coded systems biology models. Bioinformatics 2009, 25, 1453–1454. [Google Scholar] [CrossRef]
- Lu, E.-H.; Ford, L.C.; Chen, Z.; Burnett, S.D.; Rusyn, I.; Chiu, W.A. Evaluating scientific confidence in the concordance of in vitro and in vivo protective points of departure. Regul. Toxicol. Pharmacol. 2024, 148, 105596. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Wright, F.A.; Chiu, W.A.; Rusyn, I. Rapid hazard characterization of environmental chemicals using a compendium of human cell lines from different organs. Altex 2020, 37, 623. [Google Scholar] [CrossRef]
- Ford, L.C.; Jang, S.; Chen, Z.; Zhou, Y.-H.; Gallins, P.J.; Wright, F.A.; Chiu, W.A.; Rusyn, I. A population-based human in vitro approach to quantify inter-individual variability in responses to chemical mixtures. Toxics 2022, 10, 441. [Google Scholar] [CrossRef]
- Arnot, J.A.; Toose, L.; Armitage, J.M.; Sangion, A.; Looky, A.; Brown, T.N.; Li, L.; Becker, R.A. Developing an internal threshold of toxicological concern (iTTC). J. Expo. Sci. Environ. Epidemiol. 2022, 32, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, K.L.; Carr, G.; Rose, J.L.; Selman, B.G. An interim internal Threshold of Toxicologic Concern (iTTC) for chemicals in consumer products, with support from an automated assessment of ToxCast™ dose response data. Regul. Toxicol. Pharmacol. 2020, 114, 104656. [Google Scholar] [CrossRef] [PubMed]
- Dimitrijevic, D.; Fabian, E.; Nicol, B.; Funk-Weyer, D.; Landsiedel, R. Toward realistic dosimetry in vitro: Determining effective concentrations of test substances in cell culture and their prediction by an in silico mass balance model. Chem. Res. Toxicol. 2022, 35, 1962–1973. [Google Scholar] [CrossRef] [PubMed]
- Bloch, S.; Arnot, J.A.; Kramer, N.I.; Armitage, J.M.; Verner, M.-A. Dynamic Mass Balance Modeling for Chemical Distribution Over Time in In Vitro Systems With Repeated Dosing. Front. Toxicol. 2022, 4, 911128. [Google Scholar] [CrossRef]
- Cirit, M.; Stokes, C.L. Maximizing the impact of microphysiological systems with in vitro–in vivo translation. Lab A Chip 2018, 18, 1831–1837. [Google Scholar] [CrossRef]
- Ramadan, Q.; Fardous, R.S.; Hazaymeh, R.; Alshmmari, S.; Zourob, M. Pharmacokinetics-on-a-chip: In vitro microphysiological models for emulating of drugs ADME. Adv. Biol. 2021, 5, 2100775. [Google Scholar] [CrossRef]
- Schurdak, M.; Vernetti, L.; Bergenthal, L.; Wolter, Q.K.; Shun, T.Y.; Karcher, S.; Taylor, D.L.; Gough, A. Applications of the microphysiology systems database for experimental ADME-Tox and disease models. Lab A Chip 2020, 20, 1472–1492. [Google Scholar] [CrossRef]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab A Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef]
- Sung, J.H.; Koo, J.; Shuler, M.L. Mimicking the human physiology with microphysiological systems (MPS). BioChip J. 2019, 13, 115–126. [Google Scholar] [CrossRef]
- Van Meer, B.; de Vries, H.; Firth, K.; van Weerd, J.; Tertoolen, L.; Karperien, H.; Jonkheijm, P.; Denning, C.; IJzerman, A.; Mummery, C. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem. Biophys. Res. Commun. 2017, 482, 323–328. [Google Scholar] [CrossRef]
- Farhang Doost, N.; Srivastava, S.K. A Comprehensive Review of Organ-on-a-Chip Technology and Its Applications. Biosensors 2024, 14, 225. [Google Scholar] [CrossRef] [PubMed]
- Leedale, J.A.; Kyffin, J.A.; Harding, A.L.; Colley, H.E.; Murdoch, C.; Sharma, P.; Williams, D.P.; Webb, S.D.; Bearon, R.N. Multiscale modelling of drug transport and metabolism in liver spheroids. Interface Focus 2020, 10, 20190041. [Google Scholar] [CrossRef] [PubMed]
- Aly, N.A.; Luo, Y.-S.; Liu, Y.; Casillas, G.; McDonald, T.J.; Kaihatu, J.M.; Jun, M.; Ellis, N.; Gossett, S.; Dodds, J.N.; et al. Temporal and spatial analysis of per and polyfluoroalkyl substances in surface waters of Houston ship channel following a large-scale industrial fire incident. Environ. Pollut. 2020, 265, 115009. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.R.; Janiak, T.; Frei, T. 52-Week Oral Toxicity (Feeding) Study with Azinphos-Methyl (E1582) in the Dog; Mobay Corporation, Ed.; U.S. Environmental Protection Agency: Washington, DC, USA, 1990.
- BCL. Unpublished Subchronic Toxicity Study: Naphthalene (C52904), Fischer 344 Rats; Battelle’s Columbus Laboratories: Columbus, OH, USA, 1980; Prepared by Battelle Laboratories under NTP Subcontract No. 76-34-106002. [Google Scholar]
- BRRC. Developmental Toxicity Evaluation of o-, m- or p-Cresol Administered by Gavage to Rabbits and Rats with Cover Letter Dated 07/06/88. Final Project Report 51-508. TSCA Section 4 Submission. U.S. EPA Doc. No. 40-8860253. Fiche No. OTS0517695; Bushy Run Research Center: Export, PA, USA, 1988. [Google Scholar]
- Côté, M.; Chu, I.; Villeneuve, D.C.; Secours, V.E.; Valli, V.E. Trichlorobenzenes: Results of a Thirteen Week Feeding Study in the Rat. Drug Chem. Toxicol. 1988, 11, 11–28. [Google Scholar] [CrossRef]
- Fitzhugh, O.; Nelson, A.; Quaife, M. Chronic oral toxicity of aldrin and dieldrin in rats and dogs. Food Cosmet. Toxicol. 1964, 2, 551–562. [Google Scholar] [CrossRef]
- Hoechst. Endosulfan -Substance Technical (Code HOE 02671 OI ZD97 0003): Combined Chronic Toxicity/Carcinogenicity Study: 104-Week Feeding in Rats; Huntington Research Centre: Cambridgeshire, UK, 1989; Project no. HST 289/881067.: Conducted for Hoechst Aktiengesellschaft, Frankfurt, Germany. [Google Scholar]
- Littlefield, N.A.; Nelson, C.J.; Frith, C.H. Benzidine dihydrochloride: Toxicological assessment in mice during chronic exposures. J. Toxicol. Environ. Heal. 1983, 12, 671–685. [Google Scholar] [CrossRef]
- McCollister, S.B.; Kociba, R.J.; Humiston, C.G.; McCollister, D.D. Studies on the acute and long-term oral toxicity fo chlorpyrifos (0,0-diethyl-0(3,5,6-trichloro-2-pyridyl) phosphorothioate). Food Cosmet. Toxicol. 1974, 12, 45–61. [Google Scholar] [CrossRef]
- Mecler, F. Fifty-Two Week Repeated Dose Chronic Oral Study of Pentachlorophenol Administered via Capsule to Dogs. Study Conducted by TSI Mason Laboratories, Worcester, MA.; TSI Report #ML-PTF-J31-95-94. Submitted to the Pentachlorophenol Task Force, c/o SRA International, Inc., Washington, DC. MRID 439827-01. Unpublished Report; U.S. Environmental Protection Agency: Washington, DC, USA, 1996.
- Meera, P.; Rao, P.; Shanker, R.; Tripathi, O. Immunomodulatory Effects of ?-HCH (Lindane) in Mice. Immunopharmacol. Immunotoxicol. 1992, 14, 261–282. [Google Scholar] [CrossRef]
- Smialowicz, R.J.; Williams, W.C.; Copeland, C.B.; Harris, M.W.; Overstreet, D.; Davis, B.J.; Chapin, R.E. The Effects of Perinatal/Juvenile Heptachlor Exposure on Adult Immune and Reproductive System Function in Rats. Toxicol. Sci. 2001, 61, 164–175. [Google Scholar] [CrossRef][Green Version]
- Tegeris, A. Dicofol (Kelthane Technical Miticide): One Year Dietary Toxicity Study in Beagle Dogs: Final Report: Report No. 86014; Tegeris Laboratories, Inc.: Laurel, MD, USA, 1988; 2094p. [Google Scholar]
- U.S.EPA. 13-Week Mouse Oral Subchronic Toxicity Study; Toxicity Research Laboratories, Ltd., Muskegon, MI for the Office of Solid Waste: Washington, DC, USA, 1988. [Google Scholar]
- USEPA. Recommendations for and Documentation of Biological Values For Use in Risk Assessment; U.S. Environmental Protection Agency: Washington, DC, USA, 1988.
- U.S.EPA. Mouse Oral Subchronic Study with Acenaphthene. Study Conducted by Hazelton Laboratories, Inc., for the Office of Solid Waste; U.S. Environmental Protection Agency: Washington, DC, USA, 1989.
- Walker, A.; Stevenson, D.; Robinson, J.; Thorpe, E.; Roberts, M. The toxicology and pharmacodynamics of dieldrin (HEOD): Two-year oral exposures of rats and dogs. Toxicol. Appl. Pharmacol. 1969, 15, 345–373. [Google Scholar] [CrossRef]
| Model Reference | Applicable In Vitro System | Applicable Chemicals | Model Type | Partitioning Included a | Other Factors/ Processes | |||
|---|---|---|---|---|---|---|---|---|
| Media | Cell | Lab-Ware | Head-Space | |||||
| Fischer et al. [22] | Generic | Neutral/ionized; Non-volatile | Equilibrium partitioning model | √ | √ (protein and lipid) | |||
| Armitage et al. [19] | Generic; culture media with serum and monolayer cell | Neutral/ionized; Non-volatile/volatile | Equilibrium partitioning model | √ | √ | √ | √ | Solubility |
| Fisher et al. [16] | Generic | Neutral/ionized; Non-volatile/volatile | Time-dependent model | √ | √ (protein, lipid, lysosome and mitochondria) | √ | √ | Metabolism |
| Zaldivar-Comenges et al. [23] | Generic | Neutral; Non-volatile/volatile | Time-dependent model | √ | √ (protein, lipid and mitochondria) | √ | √ | Evaporation; abiotic degradation; cell growth |
| Parameters (Abbreviation) [Unit] | Model | |||
|---|---|---|---|---|
| Fischer et al. [22] | Armitage et al. [19] | Fisher et al. [16] | Zaldivar-Comenges et al. [23] | |
| Molecular weight (MW) | √ | √ | √ | |
| Melting point (MP) [°C] | √ | √ | ||
| Octanol–water partition coefficient (log KOW) | √ a | √ | √ | √ |
| Air–water partition coefficient (log KAW) | √ | √ | ||
| Solubility (CSAT,W) [mg/L] | √ | |||
| Salting-out constant (Ksalt) | √ b | |||
| pKa | √ a | √ | √ | |
| IOC type | √ | √ | ||
| Henry’s constant at 37 °C (H37) [Pa × m3/mol] | √ | |||
| Molecular volume (Vb) | √ | |||
| Distribution ratio at pH 7.4 between bovine serum albumin (BSA) and water (log DBSA/w) | √ a | |||
| Distribution ratio at pH 7.4 between phospho-lipid liposomes (lip) and water (log Dlip/w) | √ a | |||
| Dataset | Chemical (s) a | Cell Type (s) b | Endpoint Measurement |
|---|---|---|---|
| Ratio of free to nominal concentration | |||
| Huchthausen et al. [24] | Neutral and IOC (n = 12) | MCF7, HEK293H | Measured freely dissolved IC10 conc., then obtain ratio |
| Tanneberger et al. [25] | Neutral and IOC (n = 27) | Fish RTgill-W1 | Ratio of conc. in medium at the end and beginning of experiments (C24h/C0h) |
| Schug et al. [26] | Neutral organics (n = 9) | Fish RTgutGC | Ratio of conc. in medium at the end and beginning of experiments (C24h/C0h) |
| Nicol et al. [27] | Neutral and IOC (n = 30) | No cell | Ratio of conc. in buffer and in total matrix. These measurements were performed using a Rapid Equilibrium Dialysis (RED) plate |
| Valdiviezo et al. [28] | Pesticides (n = 20) | No cell | Ratio of response ratios measured from free media and initial exposure media |
| Blanchette et al. [29] | Neutral and IOC (n = 30) | No cell | Ratio of response ratios measured from free media and initial exposure media |
| This study | PFAS (n = 14) | No cell | Ratio of response ratios measured from free media and initial exposure media |
| Amount of chemicals in media and/or cells | |||
| Bellwon et al. [30] | Cyclosporine A (n = 1) | PRH, PHH, HepaRG | Measured chemical amount in cell lysate and media at multiple timepoint (≤24 h) |
| Broeders et al. [31] | Chlorpromazine (n = 1) | Balb/c 3T3, Caco-2, HepaRG | Measured chemical distribution % in cells and medium at 48 h (Balb/c 3T3 and Caco-2 cells) or 72 h (HepaRG cells) |
| Broeders et al. [32] | Chlorpromazine (n = 1) | PRH, PHH, HepaRG | Measured chemical amount in cell lysate and media at multiple timepoint (≤24 h) |
| Kodavanti et al. [33] | PBDEs (n = 3) | Rat cerebellar granule cell | Measured chemical amount in cell lysate at multiple timepoint (≤24 h) |
| Pomponio et al. [34] | Amiodarone (n = 1) | PHH, HepaRG | Measured chemical amount in cell lysate at multiple timepoint (≤24 h) |
| Truisi et al. [35] | Ibuprofen (n = 1) | PRH, PHH, HepaRG | Measured chemical amount in cell lysate and media at multiple timepoint (≤24 h) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-C.; Ford, L.C.; Rusyn, I.; Chiu, W.A. Comparative Analysis of Chemical Distribution Models for Quantitative In Vitro to In Vivo Extrapolation. Toxics 2025, 13, 439. https://doi.org/10.3390/toxics13060439
Lin H-C, Ford LC, Rusyn I, Chiu WA. Comparative Analysis of Chemical Distribution Models for Quantitative In Vitro to In Vivo Extrapolation. Toxics. 2025; 13(6):439. https://doi.org/10.3390/toxics13060439
Chicago/Turabian StyleLin, Hsing-Chieh, Lucie C. Ford, Ivan Rusyn, and Weihsueh A. Chiu. 2025. "Comparative Analysis of Chemical Distribution Models for Quantitative In Vitro to In Vivo Extrapolation" Toxics 13, no. 6: 439. https://doi.org/10.3390/toxics13060439
APA StyleLin, H.-C., Ford, L. C., Rusyn, I., & Chiu, W. A. (2025). Comparative Analysis of Chemical Distribution Models for Quantitative In Vitro to In Vivo Extrapolation. Toxics, 13(6), 439. https://doi.org/10.3390/toxics13060439






