A Deep Learning Approach for Microplastic Segmentation in Microscopic Images
Abstract
1. Introduction
2. Methodology
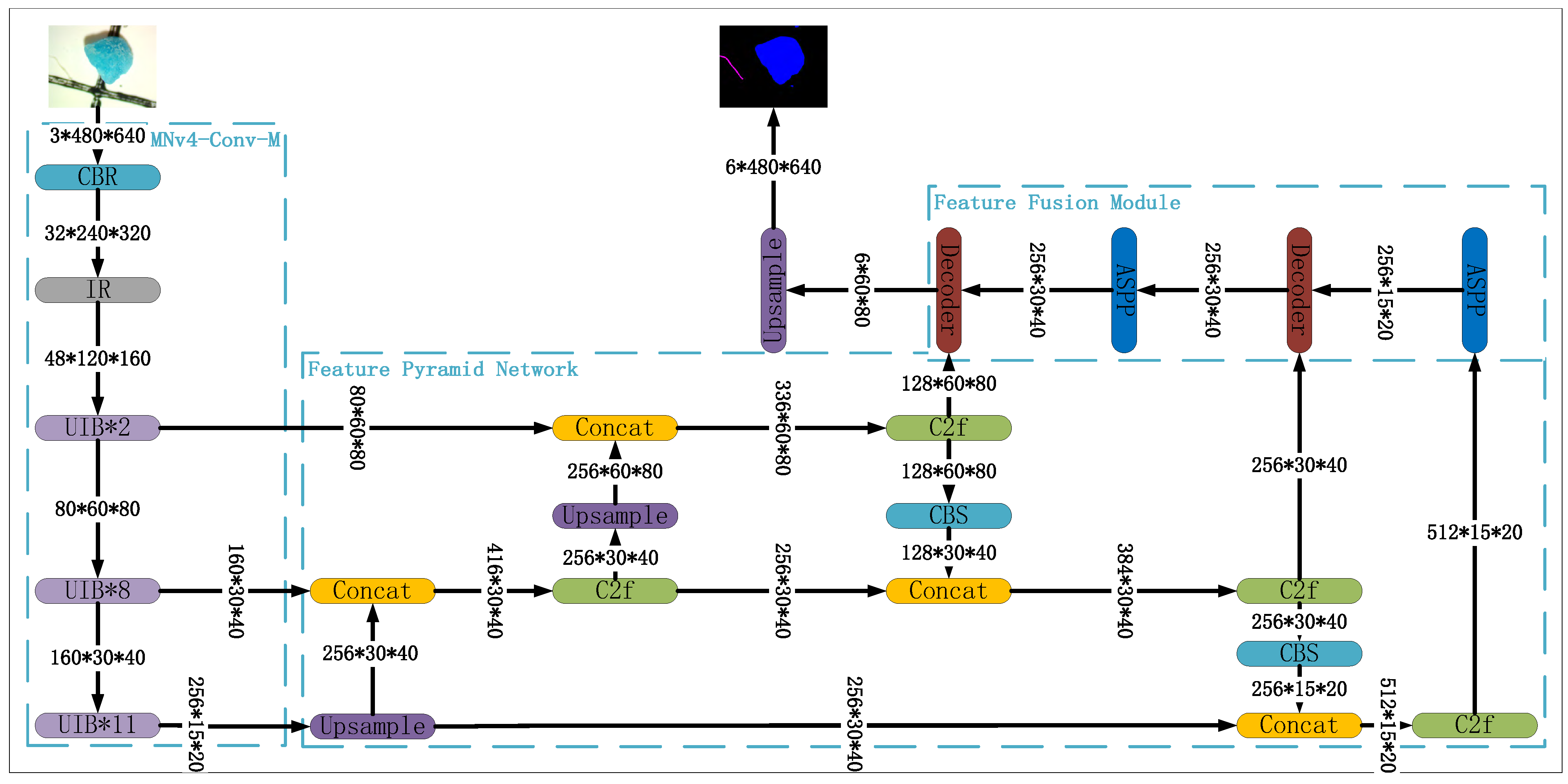
2.1. Deep Learning Model
2.1.1. BackBone Network
- CBR: Stands for Conv2d + Batch Normalization + ReLU, a widely used combination in deep learning models introduced in [34] that has proven highly effective in various image-related tasks.
- IR: Stands for Inverted Residual, introduced in [35]. The IR module applies the residual connection to high-dimensional feature maps. This inverted residual structure helps maintain the model’s performance even in relatively shallow network architectures.
- UIB: Stands for Universal Inverted Bottleneck, introduced in [33]. It builds on key components of MobileNetV4, specifically separable depthwise (DW) convolution and pointwise (PW) expansion and projection. UIB extends the Inverted Bottleneck (IB) block introduced in [35] and has become a standard building block for efficient neural network architectures.
2.1.2. Feature Pyramid Network
- Upsample: upsamples the input tensor using the bilinear upsampling method;
- Concat: concatenates tensors along the channel dimension;
- C2f: Introduced in [37], this module uses two convolutional layers and a fusion layer. It takes feature maps from different spatial scales as input, then applies a series of convolution and concatenation operations to fuse these multi-scale feature maps, allowing the model to capture and integrate information from different spatial resolutions;
- CBS: Stands for Conv2d + Batch Normalization + SiLU. It is similar to the standard CBR module, but replaces the ReLU activation function with SiLU for potentially improved performance.
2.1.3. Feature Fusion Module
- ASPP: Introduced in [39], atrous spatial pyramid pooling employs parallel atrous convolutions with varying dilation rates, enabling the model to capture features across multiple scales. This approach improves the model’s ability to handle objects of different sizes.
- Decoder: We directly incorporate the decoder from DeepLabV3+ [38], which combines the outputs from the ASPP module and FPN. This decoder upscales the feature maps, resulting in a richer and more detailed feature representation, ultimately contributing to more accurate segmentation.
2.1.4. Transfer Learning
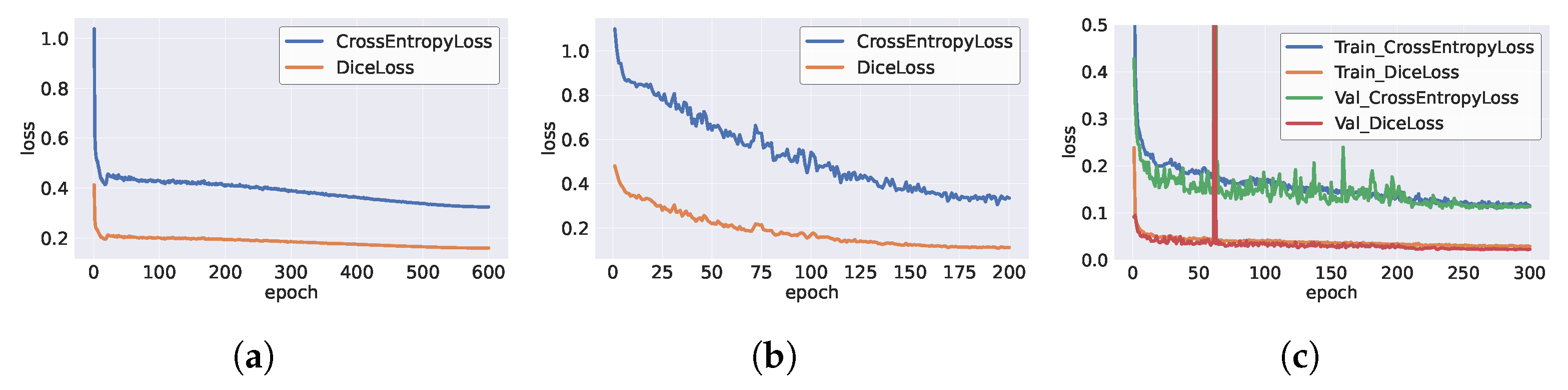
2.1.5. Loss Function
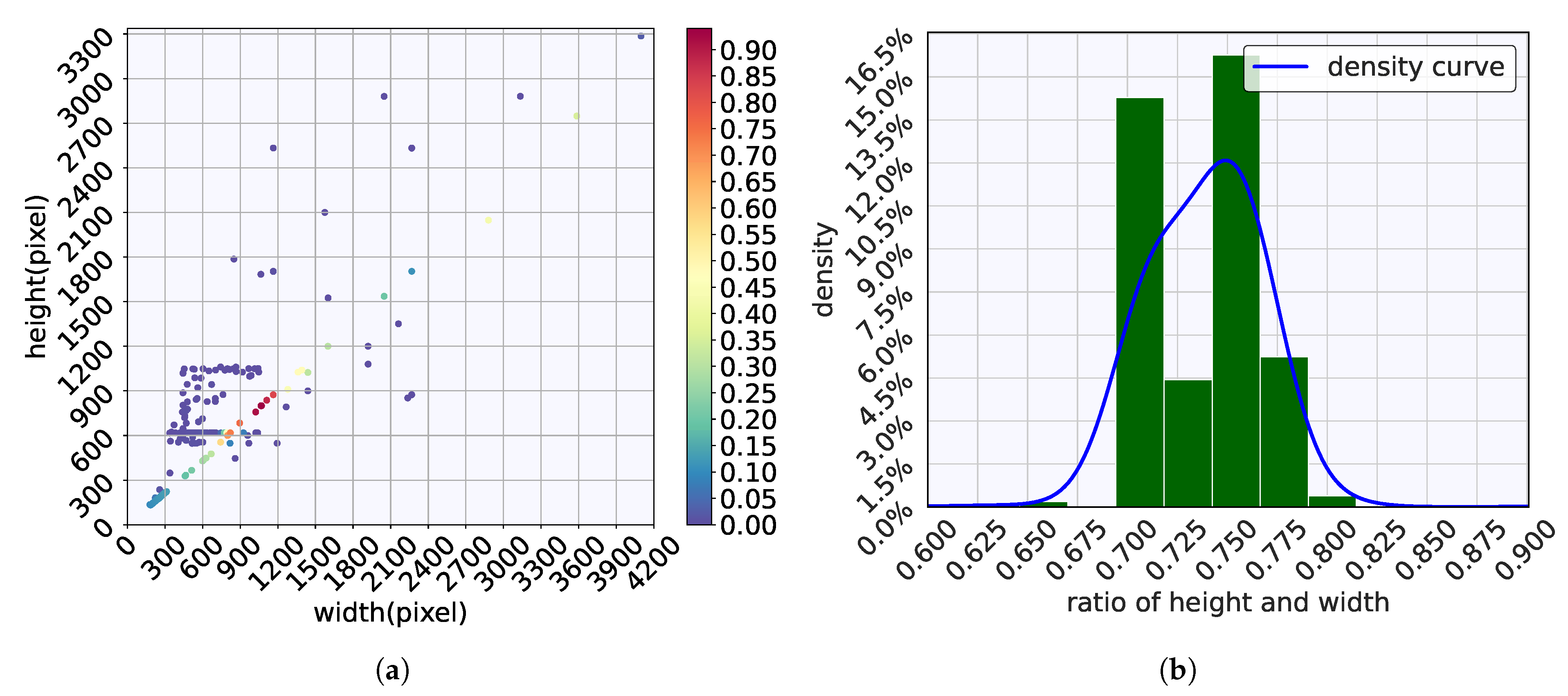
2.2. Datasets
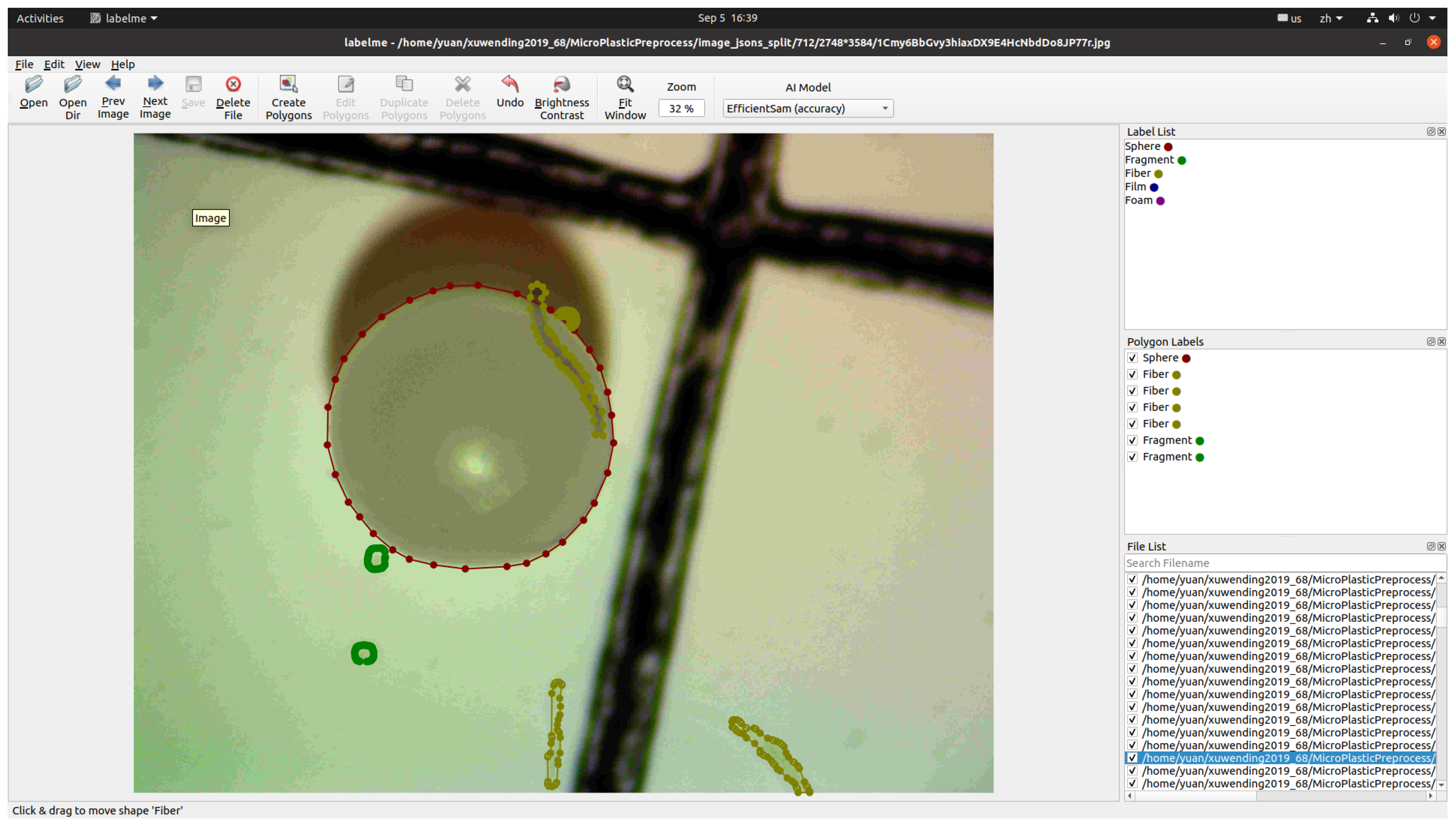
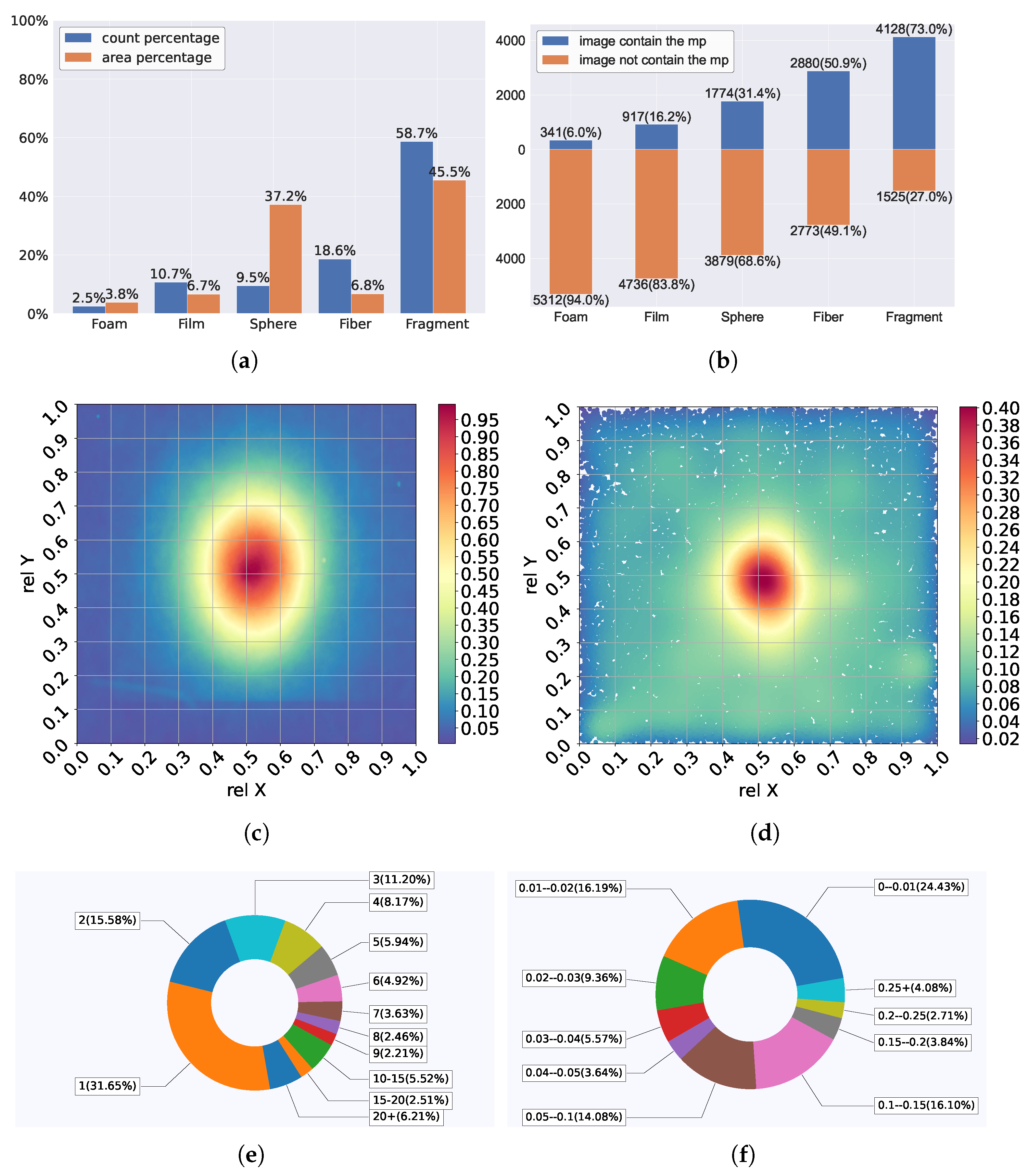
2.2.1. Microplastic Dataset
2.2.2. Cityscapes Dataset
2.3. Data Preprocessing and Augmentation
2.4. Evaluation Metrics
- mIoU: This metric measures the average overlap between the predicted segmentation mask and the ground truth mask. A higher mIoU value indicates better agreement between the predicted and actual segmentation.
- F1: This metric is calculated as the harmonic mean of precision and recall. It provides a balanced measure by considering both false positives and false negatives.
- params: This metric quantifies the number of learnable parameters in a model. While a higher parameter count can improve the model’s capacity to learn complex patterns, it also increases the risk of overfitting and can lead to longer inference times.
- FLOPs: This metric represents the total number of floating-point operations required for a model’s computation. A higher FLOPs value indicates greater computation resource usage.
- Inference time: This metric refers to the average time taken to process a single input and generate an output. A good segmentation model typically achieves an inference time of only a few milliseconds.
3. Experiments
3.1. Experiment Setup
3.2. Comparison Study
- FCN [45]: The Fully Convolutional Network (FCN) is a pioneering model that introduced the concept of fully convolutional architectures for dense pixel-wise predictions, eliminating the need for fully connected layers.
- BiSeNetV1 [46]: BiSeNetV1 achieves real-time performance by utilizing a dual-path architecture—a spatial path for capturing detailed spatial information and a context path for rich semantic context.
- BiSeNetV2 [47]: This is an enhanced version of BiSeNetV1, featuring a bilateral structure, adaptive aggregation module, and spatial enhancement modules to further improve efficiency and accuracy.
- DeepLabV3+ [38]: Known for its high accuracy, DeepLabV3+ uses an encoder–decoder architecture with atrous convolution and a spatial pyramid pooling module to capture multi-scale contextual information. It offers three backbone options: ResNet, MobileNet, and Xception.
- HRNet [27]: HRNet maintains high-resolution representations throughout the network by employing parallel multi-resolution subnetworks, enabling the capture of both rich spatial details and contextual information.
- U-Net [22]: U-Net utilizes an encoder-decoder architecture with skip connections, which help to preserve spatial information during the segmentation process.
- Nested U-Net [48]: An extension of U-Net, Nested U-Net incorporates a nested and multi-scale architecture to capture hierarchical features, leading to improved segmentation accuracy.
- Swin U-Net [49]: Swin U-Net combines the Swin Transformer with the U-Net architecture, effectively modeling both local and global spatial relationships for enhanced segmentation performance.
3.3. Experiment Results
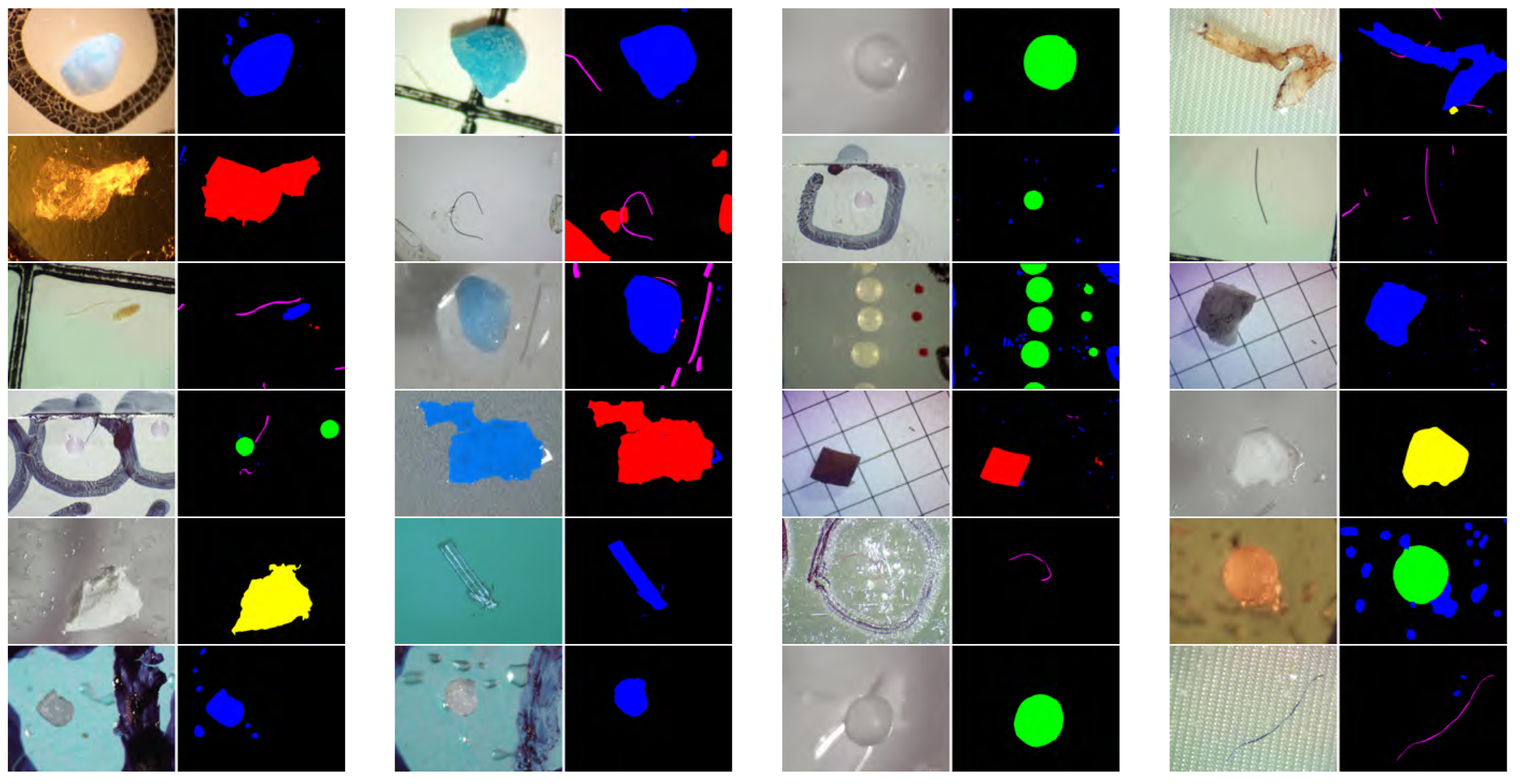
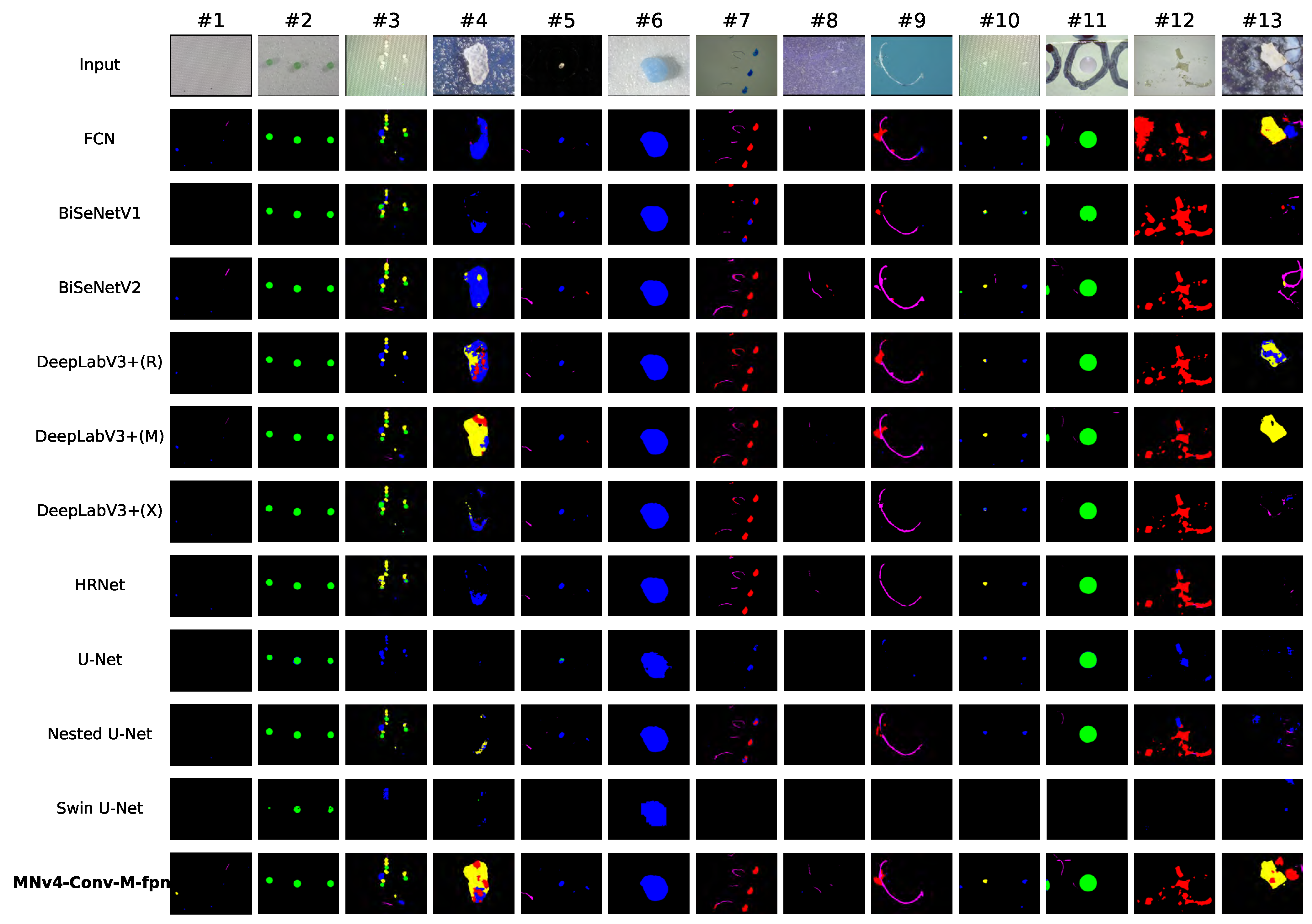
3.3.1. Visual Comparison
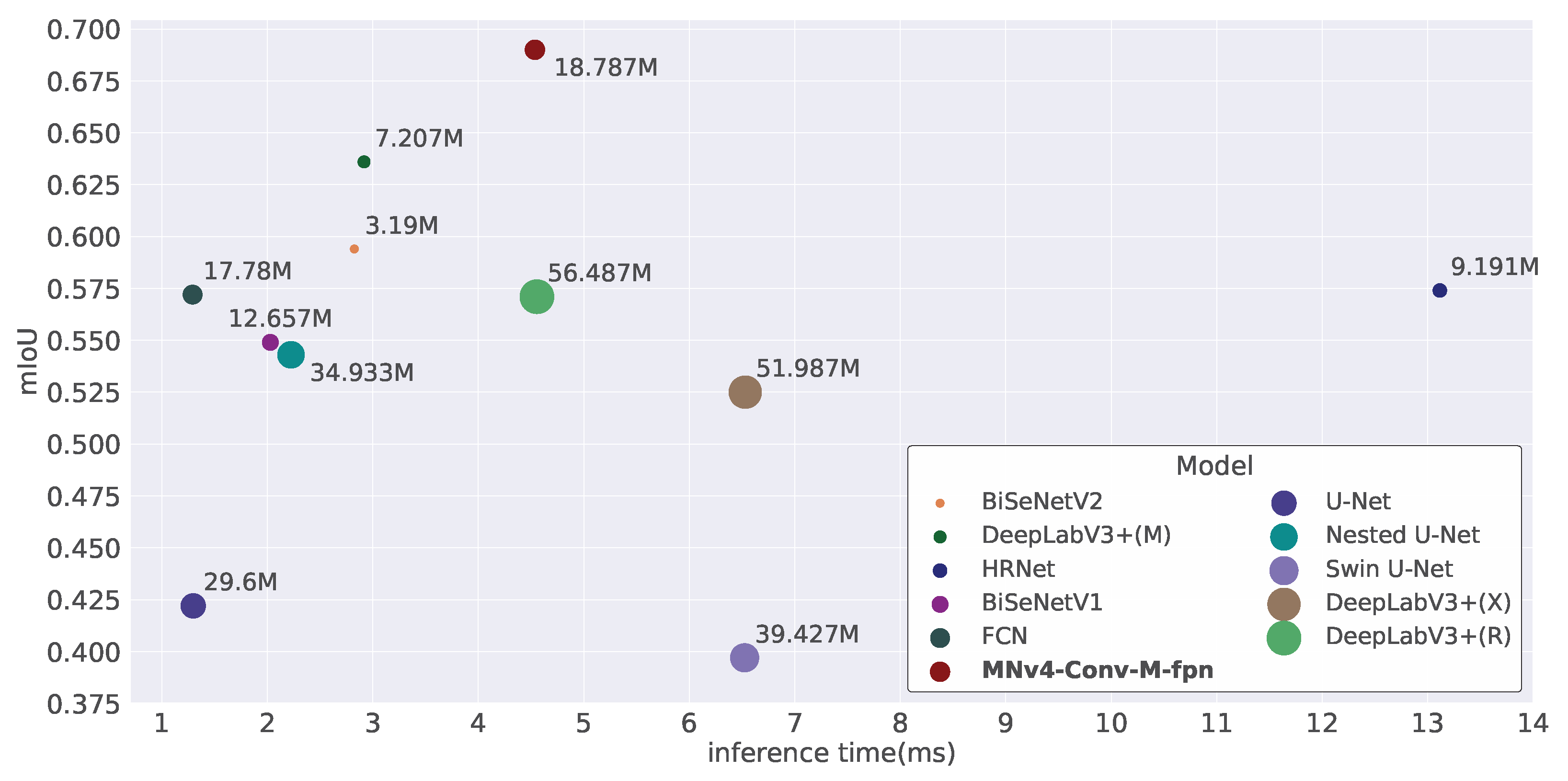
3.3.2. Evaluation Results
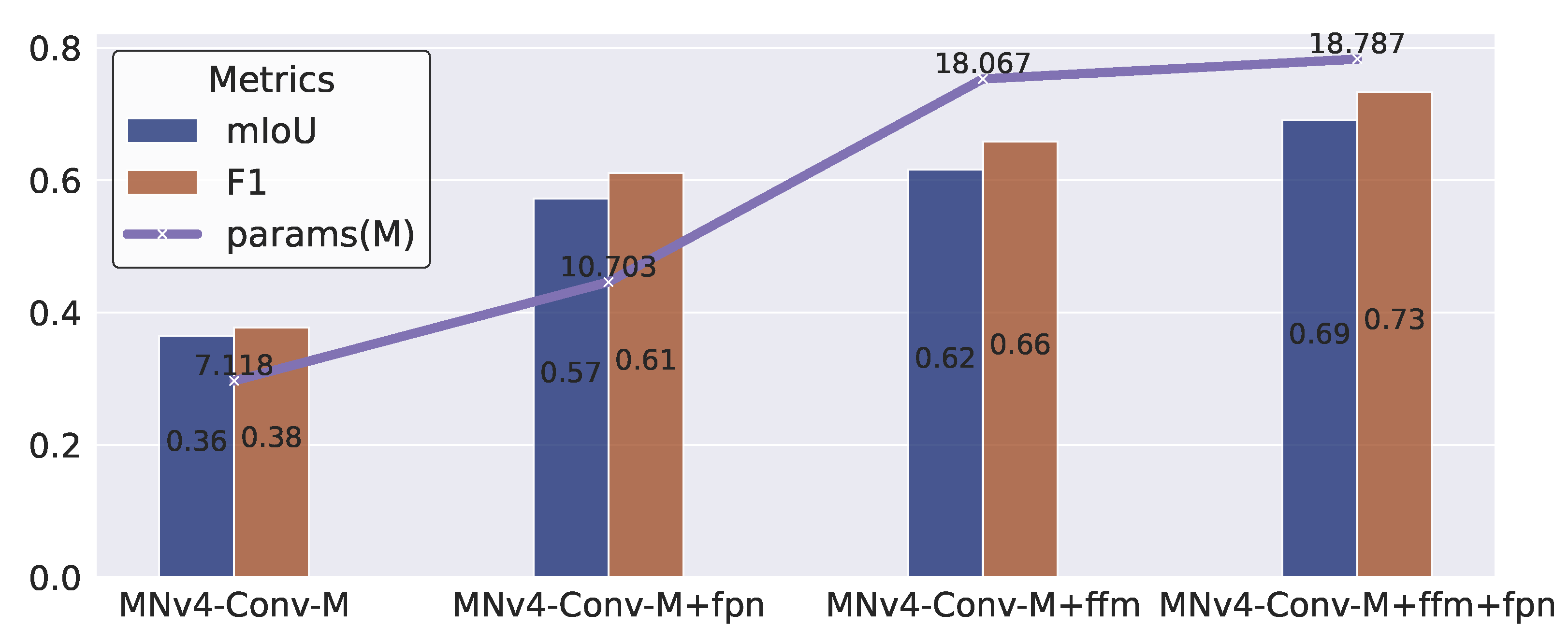
3.4. Ablation Study
4. Discussion
4.1. The Scientific and Technical Advancement
4.2. Environmental Deployment and Policy Impact
4.3. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warguła, J. Not only foils and packaging. Part 1: About the applications of polymers in ophthalmology, dentistry and medical equipment. Prospect. Pharm. Sci. 2024, 22, 135–145. [Google Scholar] [CrossRef]
- Kwon, B.G. Behavior of invisible microplastics in aquatic environments. Mar. Pollut. Bull. 2026, 222, 118739. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.M.; Martín, M.N.D.; Pérez, E.M.R.; Amey, J.; Borges, J.H.; Nuez, E.F.; Machín, F.; Moreno, D.V. Vertical distribution and composition of microplastics and marine litter in the open ocean surrounding the Canary Islands (0–1200 m depth). Mar. Pollut. Bull. 2026, 222, 118836. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez, J.R.; Metian, M.; Swarzenski, P.W.; Bank, M.S.; Bjorøy, Ø.; Cajas, J.; Bucheli, R.; González-Muñoz, R.; Lynch, J.; Piguave, E.; et al. Marine microplastics on the rise in the Eastern Tropical Pacific: Abundance doubles in 11 years and a ten-fold increase is projected by 2100. Mar. Pollut. Bull. 2025, 221, 118437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Choi, C. A transport mechanism for deep-sea microplastics: Hydroplaning of clay-laden sediment gravity flows. Mar. Pollut. Bull. 2025, 218, 118191. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, L.; Wei, N.; Bai, M.; Liu, K.; Wang, X.; Jiang, C.; Zong, C.; Zhang, F.; Li, C.; et al. Transport mechanism of microplastics from a still water system to a dynamic estuarine system: A case study in Macao SAR. Mar. Pollut. Bull. 2025, 218, 118223. [Google Scholar] [CrossRef]
- Zeng, H.; Liu, D.; Liu, S.; Li, Z.; Ding, Y.; Wang, T. Occurrence, characteristics and ecological risk assessment of microplastics in the surface water of the Central South China Sea. Mar. Pollut. Bull. 2026, 222, 118760. [Google Scholar] [CrossRef]
- Aytan, Ü.; Şentürk Koca, Y.; Pasli, S.; Güven, O.; Ceylan, Y.; Basaran, B. Microplastics in commercial fish and their habitats in the important fishing ground of the Black Sea: Characteristic, concentration, and risk assessment. Mar. Pollut. Bull. 2025, 221, 118434. [Google Scholar] [CrossRef]
- Arienzo, M.; Donadio, C. Microplastic-Pharmaceuticals Interaction in Water Systems. J. Mar. Sci. Eng. 2023, 11, 1437. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Li, Q.; Li, B.; Ma, H.; Wang, S.; Ying, Q.; Ran, S.; Leonteva, E.O.; Shilin, M.B.; et al. Microplastic exposure reshapes the virome and virus–bacteria networks with implications for immune regulation in Mytilus coruscus. Mar. Pollut. Bull. 2026, 222, 118785. [Google Scholar] [CrossRef]
- Remanan, C.; Sahadevan, P.; Anju, K.; Ankitha, C.; Lal, M.; Sanjeevan, V. Field and laboratory-based evidence of microplastic ingestion by the Asian green mussel, Perna viridis from the northern Malabar coast of India. Mar. Pollut. Bull. 2025, 220, 118468. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.; Feng, W.; Li, X.; Kalaipandian, S.; Ngien, S.K.; Yu, X. Exploring the complex interactions between microplastics and marine contaminants. Mar. Pollut. Bull. 2026, 222, 118697. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.; Singha, T.; Willans, M.; Hackett, M.; Pham, D.S. A Domain-Adaptive Deep Learning Approach for Microplastic Classification. Microplastics 2025, 4, 69. [Google Scholar] [CrossRef]
- Wootton, N.; Gillanders, B.M.; Leterme, S.; Noble, W.; Wilson, S.P.; Blewitt, M.; Swearer, S.E.; Reis-Santos, P. Research priorities on microplastics in marine and coastal environments: An Australian perspective to advance global action. Mar. Pollut. Bull. 2024, 205, 116660. [Google Scholar] [CrossRef]
- Hartmann, N.B.; Huffer, T.; Thompson, R.C.; Hassellov, M.; Verschoor, A.; Daugaard, A.E.; Rist, S.; Karlsson, T.; Brennholt, N.; Cole, M.; et al. Are we speaking the same language? Recommendations for a definition and categorization framework for plastic debris. Environ. Sci. Technol. 2019, 53, 1039–1047. [Google Scholar] [CrossRef]
- Villegas-Camacho, O.; Francisco-Valencia, I.; Alejo-Eleuterio, R.; Granda-Gutiérrez, E.E.; Martínez-Gallegos, S.; Villanueva-Vásquez, D. FTIR-Based Microplastic Classification: A Comprehensive Study on Normalization and ML Techniques. Recycling 2025, 10, 46. [Google Scholar] [CrossRef]
- Huang, H.; Cai, H.; Qureshi, J.U.; Mehdi, S.R.; Song, H.; Liu, C.; Di, Y.; Shi, H.; Yao, W.; Sun, Z. Proceeding the categorization of microplastics through deep learning-based image segmentation. Sci. Total Environ. 2023, 896, 165308. [Google Scholar] [CrossRef]
- Odhiambo, J.M.; Mvurya, M. Deep Learning Algorithm for Identifying Microplastics in Open Sewer Systems: A Systematic Review. Int. J. Eng. Sci. 2022, 11, 11–18. [Google Scholar]
- Fajardo-Urbina, J.M.; Liu, Y.; Georgievska, S.; Gräwe, U.; Clercx, H.J.; Gerkema, T.; Duran-Matute, M. Efficient deep learning surrogate method for predicting the transport of particle patches in coastal environments. Mar. Pollut. Bull. 2024, 209, 117251. [Google Scholar] [CrossRef]
- Zhao, F.; Huang, B.; Wang, J.; Shao, X.; Wu, Q.; Xi, D.; Liu, Y.; Chen, Y.; Zhang, G.; Ren, Z.; et al. Seafloor debris detection using underwater images and deep learning-driven image restoration: A case study from Koh Tao, Thailand. Mar. Pollut. Bull. 2025, 214, 117710. [Google Scholar] [CrossRef]
- Baek, J.Y.; de Guzman, M.K.; Park, H.m.; Park, S.; Shin, B.; Velickovic, T.C.; Van Messem, A.; De Neve, W. Developing a Segmentation Model for Microscopic Images of Microplastics Isolated from Clams. In Proceedings of the International Conference on Pattern Recognition, Virtual Event, 10–15 January 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 86–97. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015. [Google Scholar] [CrossRef]
- Lee, G.; Jhang, K. Neural network analysis for microplastic segmentation. Sensors 2021, 21, 7030. [Google Scholar] [CrossRef]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Z. Efficient and accurate microplastics identification and segmentation in urban waters using convolutional neural networks. Sci. Total Environ. 2024, 911, 168696. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, K.; Cheng, T.; Jiang, B.; Deng, C.; Zhao, Y.; Liu, D.; Mu, Y.; Tan, M.; Wang, X.; et al. Deep High-Resolution Representation Learning for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 3349–3364. [Google Scholar] [CrossRef]
- Jeong, J.; Lee, G.; Jeong, J.; Kim, J.; Kim, J.; Jhang, K. Microplastic Binary Segmentation with Resolution Fusion and Large Convolution Kernels. J. Comput. Sci. Eng 2024, 18, 29–35. [Google Scholar] [CrossRef]
- Royer, S.J.; Wolter, H.; Delorme, A.E.; Lebreton, L.; Poirion, O.B. Computer vision segmentation model—Deep learning for categorizing microplastic debris. Front. Environ. Sci. 2024, 12, 1386292. [Google Scholar] [CrossRef]
- Lee, K.S.; Chen, H.L.; Ng, Y.S.; Maul, T.; Gibbins, C.; Ting, K.N.; Amer, M.; Camara, M. U-Net skip-connection architectures for the automated counting of microplastics. Neural Comput. Appl. 2022, 34, 7283–7297. [Google Scholar] [CrossRef]
- Park, H.m.; Park, S.; de Guzman, M.K.; Baek, J.Y.; Cirkovic Velickovic, T.; Van Messem, A.; De Neve, W. MP-Net: Deep learning-based segmentation for fluorescence microscopy images of microplastics isolated from clams. PLoS ONE 2022, 17, e0269449. [Google Scholar] [CrossRef]
- Shi, B.; Patel, M.; Yu, D.; Yan, J.; Li, Z.; Petriw, D.; Pruyn, T.; Smyth, K.; Passeport, E.; Miller, R.D.; et al. Automatic quantification and classification of microplastics in scanning electron micrographs via deep learning. Sci. Total Environ. 2022, 825, 153903. [Google Scholar] [CrossRef]
- Qin, D.; Leichner, C.; Delakis, M.; Fornoni, M.; Luo, S.; Yang, F.; Wang, W.; Banbury, C.; Ye, C.; Akin, B.; et al. MobileNetV4-Universal Models for the Mobile Ecosystem. arXiv 2024, arXiv:2404.10518. [Google Scholar]
- Karen, S. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2018. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Sun, Y.; Chen, G.; Zhou, T.; Zhang, Y.; Liu, N. Context-aware Cross-level Fusion Network for Camouflaged Object Detection. arXiv 2021. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial Pyramid Pooling in Deep Convolutional Networks for Visual Recognition. In Computer Vision—ECCV 2014; Springer International Publishing: Berlin/Heidelberg, Germany, 2014; pp. 346–361. [Google Scholar] [CrossRef]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A comprehensive survey on transfer learning. Proc. IEEE 2020, 109, 43–76. [Google Scholar] [CrossRef]
- Cordts, M.; Omran, M.; Ramos, S.; Rehfeld, T.; Enzweiler, M.; Benenson, R.; Franke, U.; Roth, S.; Schiele, B. The Cityscapes Dataset for Semantic Urban Scene Understanding. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Mao, A.; Mohri, M.; Zhong, Y. Cross-entropy loss functions: Theoretical analysis and applications. In Proceedings of the International Conference on Machine Learning, PMLR, Honolulu, HI, USA, 23–29 July 2023; pp. 23803–23828. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Luo, L.; Xiong, Y.; Liu, Y.; Sun, X. Adaptive Gradient Methods with Dynamic Bound of Learning Rate. arXiv 2019. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. arXiv 2015. [Google Scholar] [CrossRef]
- Yu, C.; Wang, J.; Peng, C.; Gao, C.; Yu, G.; Sang, N. BiSeNet: Bilateral Segmentation Network for Real-time Semantic Segmentation. arXiv 2018. [Google Scholar] [CrossRef]
- Yu, C.; Gao, C.; Wang, J.; Yu, G.; Shen, C.; Sang, N. BiSeNet V2: Bilateral Network with Guided Aggregation for Real-time Semantic Segmentation. arXiv 2020. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. arXiv 2018. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. arXiv 2021. [Google Scholar] [CrossRef]
- Lusher, A.L.; Bråte, I.L.N.; Munno, K.; Hurley, R.R.; Welden, N.A. Is it or isn’t it: The importance of visual classification in microplastic characterization. Appl. Spectrosc. 2020, 74, 1139–1153. [Google Scholar] [CrossRef]
- Zhu, Y.; Yeung, C.H.; Lam, E.Y. Microplastic pollution monitoring with holographic classification and deep learning. J. Phys. Photonics 2021, 3, 024013. [Google Scholar] [CrossRef]
- Teng, T.W.; Sheikh, U.U. Deep learning based microplastic classification. In Proceedings of the AIP Conference Proceedings, Male’ City, Maldives, 15–17 November 2023; AIP Publishing: Melville, NY, USA, 2024; Volume 3245. [Google Scholar]
- Ravi, N.; Gabeur, V.; Hu, Y.T.; Hu, R.; Ryali, C.; Ma, T.; Khedr, H.; Rädle, R.; Rolland, C.; Gustafson, L.; et al. Sam 2: Segment anything in images and videos. arXiv 2024, arXiv:2408.00714. [Google Scholar]
| Research Work | Number of Categories | Model |
|---|---|---|
| Ji Yeon Baek et al. [21] | 2 | U-Net |
| Gwanghee Lee et al. [23] | 2 | U-Net, MultiResU-Net |
| Ka Shing Lee et al. [30] | 2 | U-Net |
| Ho-min Park et al. [31] | 2 | U-Net |
| Bin Shi et al. [32] | 2 | MultiResU-Net |
| Hui Huang et al. [17] | 2 | ResNet + FPN |
| Jiongji Xu et al. [26] | 2 | U-Net, U-Net2Plus, U-Net3Plus |
| Sarah-Jeanne Royer et al. [29] | 5 | ResNet |
| Jaeheon Jeong et al. [28] | 2 | HRNet |
| Our Work | 6 | MNv4+FPN+FFM |
| Category | Description | Count | Color |
|---|---|---|---|
| Sphere | Round, smooth particles; clear or colored | 3220 | green |
| Fragment | Irregular, jagged pieces; dull | 19,871 | blue |
| Fiber | Thread-like, colorful structures | 6312 | magenta |
| Film | Thin, flexible, transparent sheets | 3626 | red |
| Foam | Light, spongy texture; usually white | 851 | yellow |
| Method | Description |
|---|---|
| Rotation | Rotate the input image by angle with a given probability. |
| ColorJitter | Change the brightness, contrast, saturation and hue of an image. |
| GaussianBlur | Blurs image with a given Gaussian blur kernel. |
| VerticalFlip | Flip the input image vertically with a given probability. |
| HorizontalFlip | Flip the input image horizontally with a given probability. |
| ElasticTransform | Randomly offset pixels with given and . |
| Component | Configuration |
|---|---|
| CPU | Intel(R) Core(TM) i9-14900K |
| GPU | GeForce RTX4090 (24 GB) |
| Memory | 64 G |
| Hard Disk | 2 T |
| Operating System | Ubuntu 20.04.6 LTS |
| CUDA | 12.4 |
| Driver | NVIDIA-550.90.07 |
| Pytorch | 2.2.0 |
| Model | mIoU | F1 | Inference Time (ms) | Params (M) | Flops (B) |
|---|---|---|---|---|---|
| FCN | 0.572 | 0.612 | 1.292 | 17.780 | 11.957 |
| BiSeNetV1 | 0.549 | 0.578 | 2.029 | 12.657 | 1.744 |
| BiSeNetV2 | 0.594 | 0.633 | 2.825 | 3.190 | 1.449 |
| DeepLabV3+(R) | 0.571 | 0.604 | 4.556 | 56.487 | 10.378 |
| DeepLabV3+(M) | 0.636 | 0.677 | 2.917 | 7.207 | 3.968 |
| DeepLabV3+(X) | 0.525 | 0.556 | 6.531 | 51.987 | 6.329 |
| HRNet | 0.574 | 0.609 | 13.115 | 9.191 | 2.189 |
| U-Net | 0.422 | 0.445 | 1.298 | 29.600 | 25.668 |
| Nested U-Net | 0.543 | 0.580 | 2.225 | 34.933 | 65.007 |
| Swin U-Net | 0.397 | 0.416 | 6.525 | 39.427 | 3.487 |
| MNv4-Conv-M-fpn | 0.690 | 0.733 | 4.537 | 18.787 | 1.283 |
| Model | Fiber | Film | Foam | Fragment | Sphere |
|---|---|---|---|---|---|
| FCN | 0.640∣0.698 | 0.625∣0.685 | 0.657∣0.731 | 0.700∣0.741 | 0.720∣0.775 |
| BiSeNetV1 | 0.596∣0.647 | 0.640∣0.713 | 0.659∣0.739 | 0.644∣0.680 | 0.705∣0.764 |
| BiSeNetV2 | 0.635∣0.695 | 0.643∣0.713 | 0.663∣0.741 | 0.672∣0.713 | 0.715∣0.777 |
| DeepLabV3+(R) | 0.610∣0.664 | 0.639∣0.710 | 0.661∣0.740 | 0.665∣0.703 | 0.707∣0.766 |
| DeepLabV3+(M) | 0.647∣0.710 | 0.647∣0.717 | 0.664∣0.742 | 0.706∣0.747 | 0.731∣0.791 |
| DeepLabV3+(X) | 0.598∣0.648 | 0.635∣0.705 | 0.660∣0.740 | 0.645∣0.682 | 0.678∣0.743 |
| HRNet | 0.614∣0.669 | 0.642∣0.714 | 0.663∣0.742 | 0.667∣0.706 | 0.710∣0.768 |
| U-Net | 0.581∣0.623 | 0.638∣0.710 | 0.655∣0.734 | 0.582∣0.611 | 0.643∣0.691 |
| Nested U-Net | 0.622∣0.678 | 0.629∣0.695 | 0.651∣0.727 | 0.670∣0.709 | 0.711∣0.768 |
| Swin U-Net | 0.580∣0.623 | 0.638∣0.710 | 0.655∣0.734 | 0.554∣0.585 | 0.622∣0.682 |
| MNv4-Conv-M-fpn | 0.684∣0.750 | 0.649∣0.719 | 0.667∣0.744 | 0.733∣0.777 | 0.741∣0.802 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.; Xu, W.; Fan, H. A Deep Learning Approach for Microplastic Segmentation in Microscopic Images. Toxics 2025, 13, 1018. https://doi.org/10.3390/toxics13121018
Yao Y, Xu W, Fan H. A Deep Learning Approach for Microplastic Segmentation in Microscopic Images. Toxics. 2025; 13(12):1018. https://doi.org/10.3390/toxics13121018
Chicago/Turabian StyleYao, Yuan, Wending Xu, and Haoxin Fan. 2025. "A Deep Learning Approach for Microplastic Segmentation in Microscopic Images" Toxics 13, no. 12: 1018. https://doi.org/10.3390/toxics13121018
APA StyleYao, Y., Xu, W., & Fan, H. (2025). A Deep Learning Approach for Microplastic Segmentation in Microscopic Images. Toxics, 13(12), 1018. https://doi.org/10.3390/toxics13121018







