Molecular Determinants of Per- and Polyfluoroalkyl Substances Binding to Estrogen Receptors
Abstract
1. Introduction
2. Methods and Materials
2.1. Receptor Preparation
2.2. Preparation of PFAS
2.3. Molecular Docking
2.4. Descriptor Selection
2.5. Generation of QSPR Datasets with Descriptors
2.6. Development of QSPR/QSAR Model
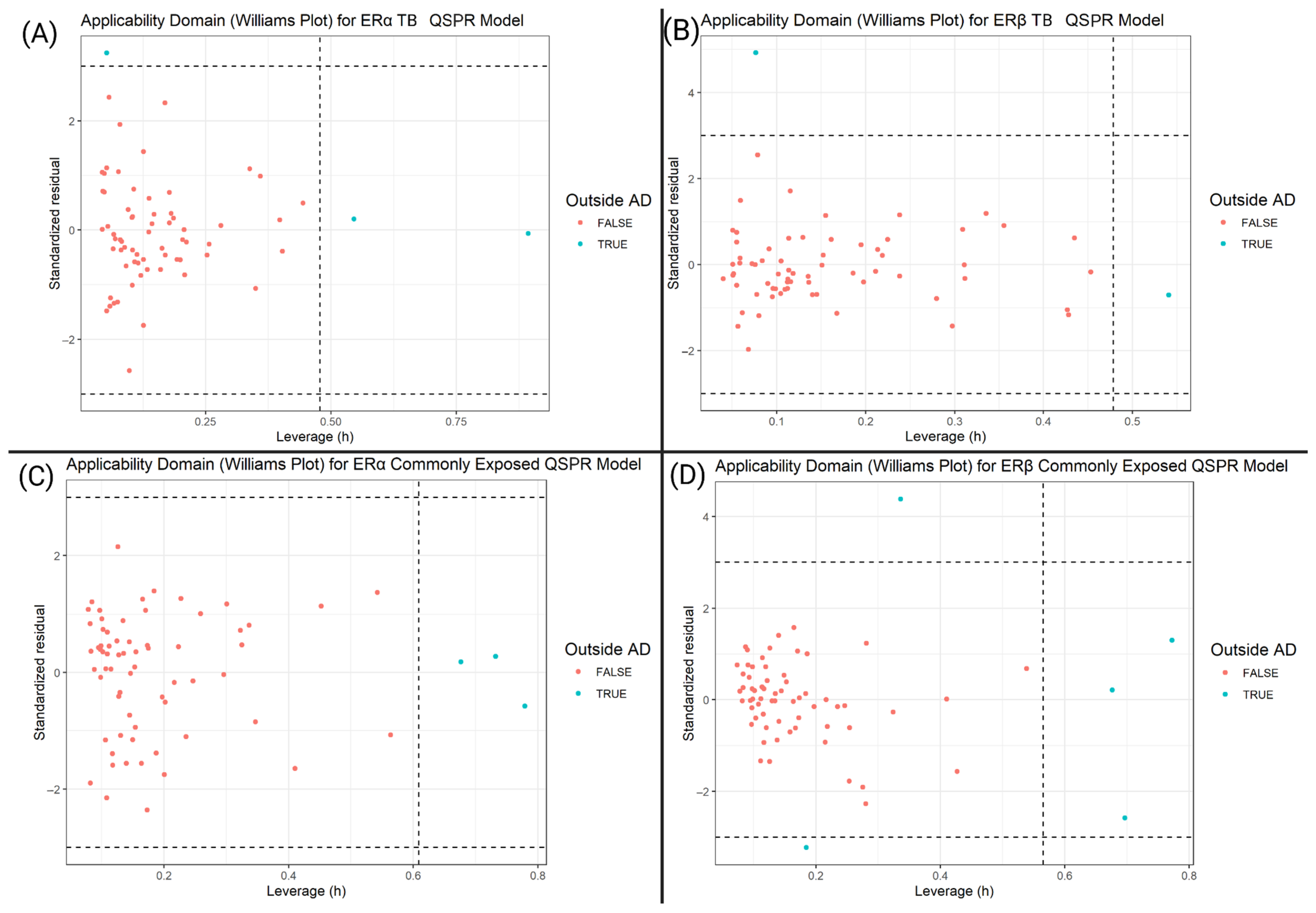
2.7. Applicability Domain Determination
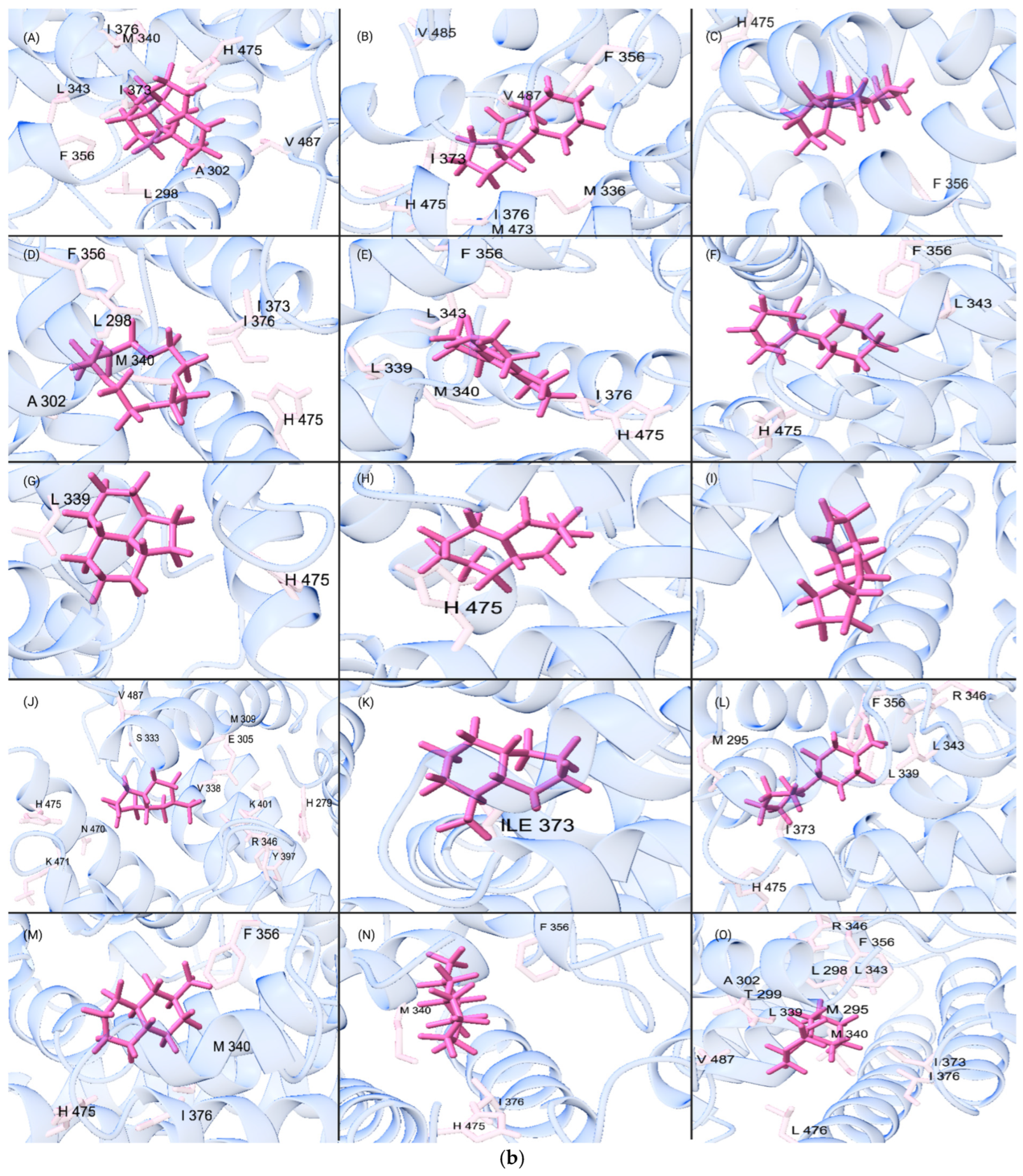
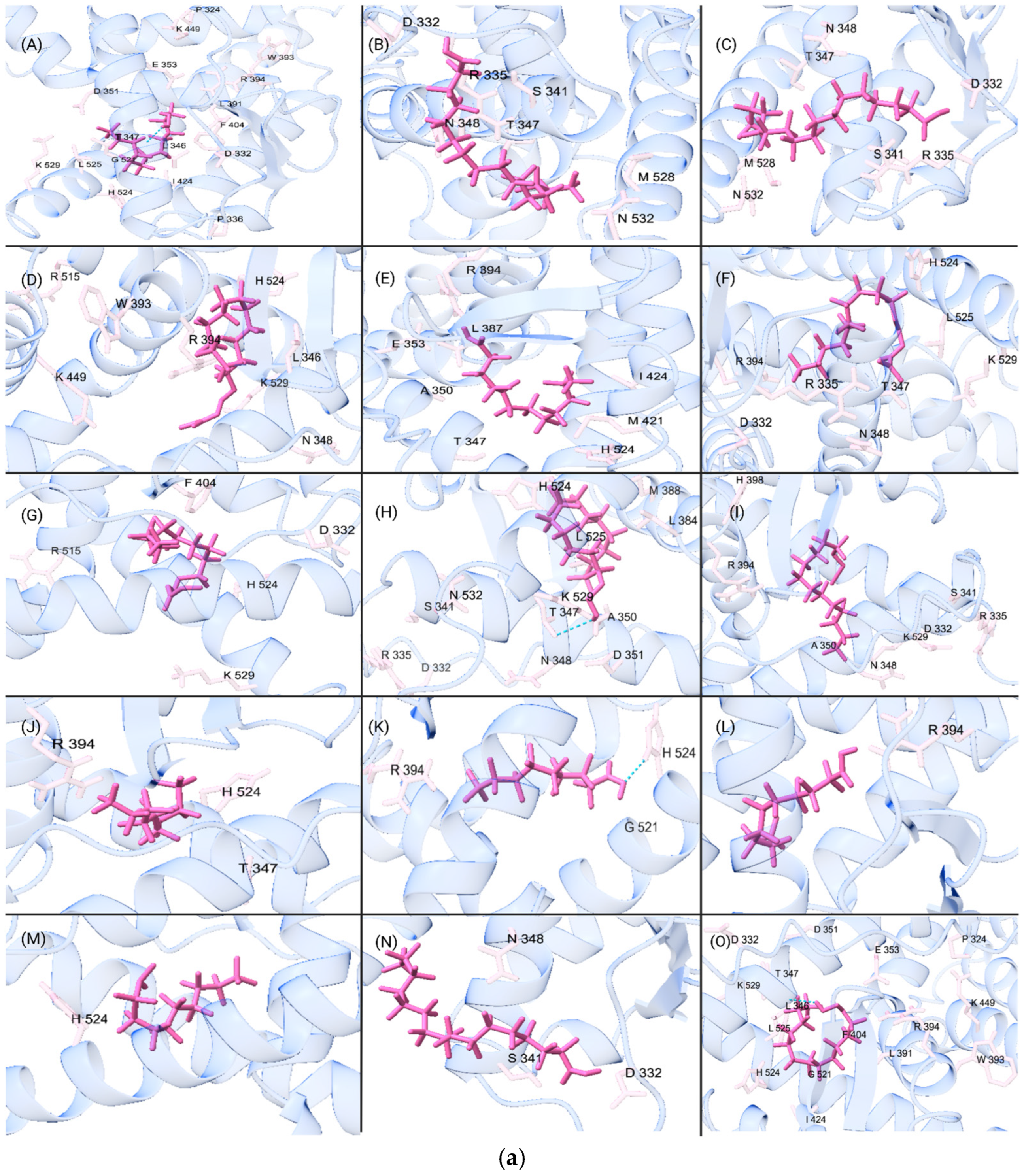
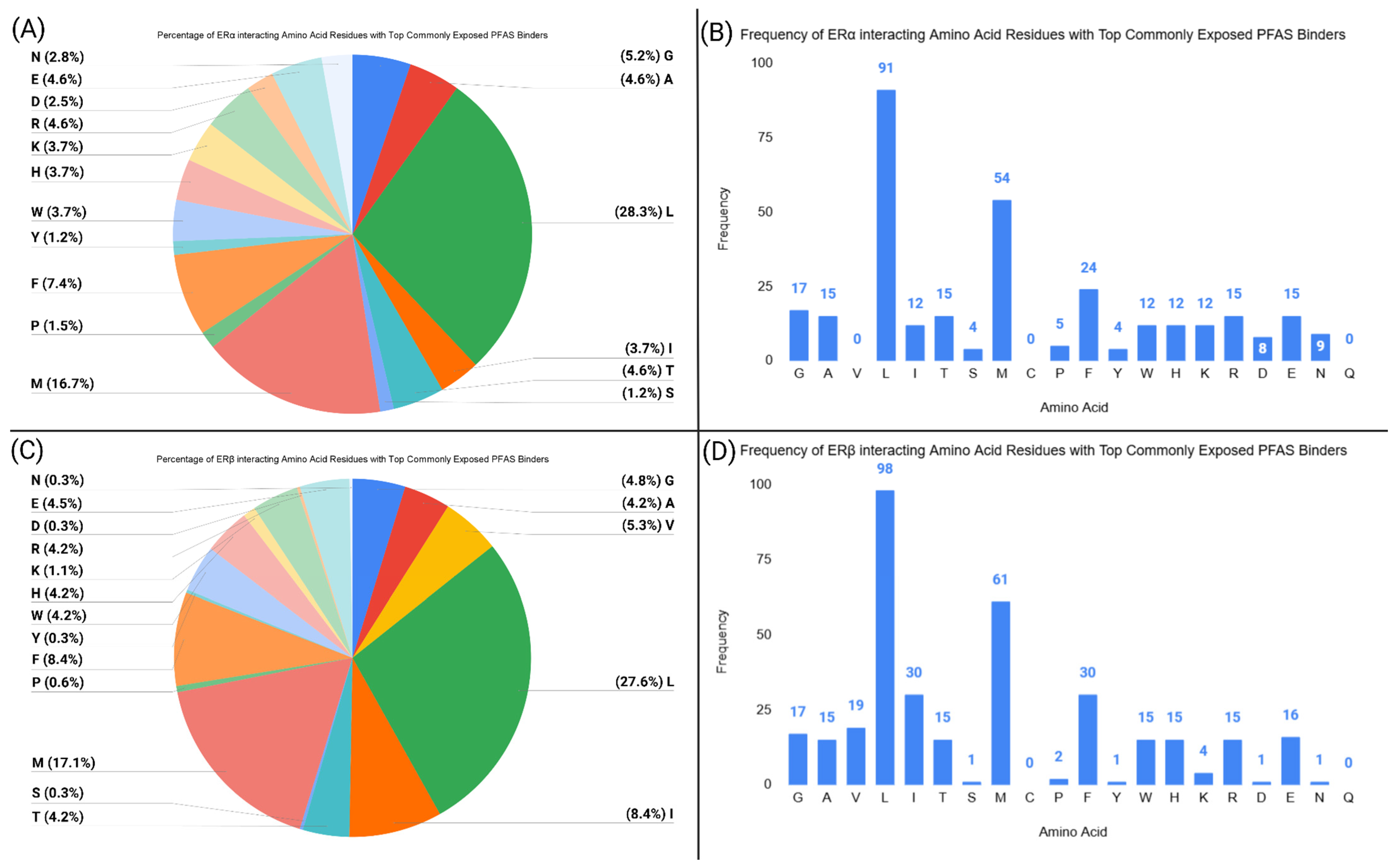
2.8. Assessment of Amino Acid Residue Interactions
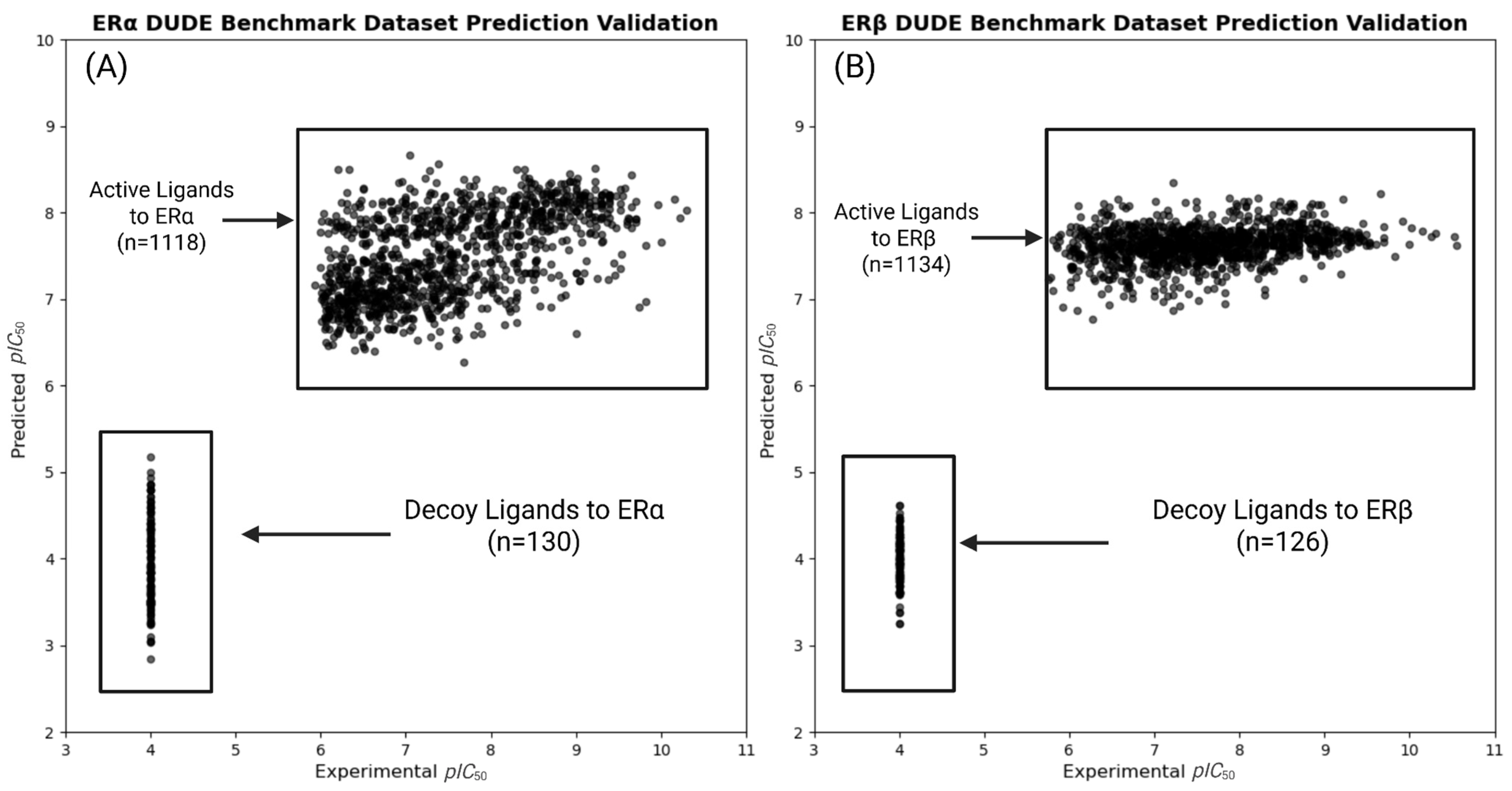
2.9. DUD-E Benchmarks for QSAR Model Validation
2.10. Development of QSAR Models for QSPR Validation and Affinity Prediction
2.11. Large-Scale QSAR Prediction Models
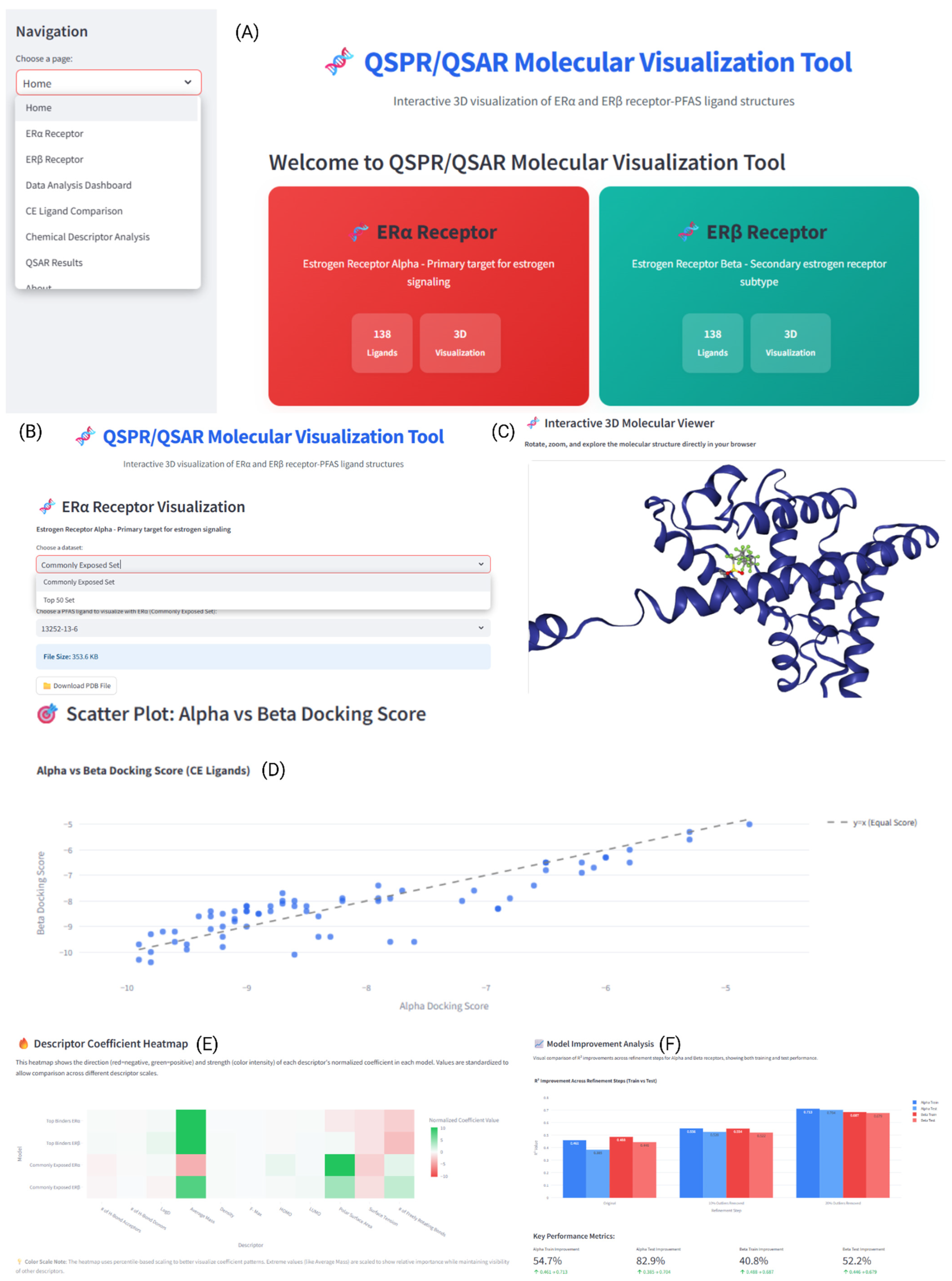
2.12. Development of GUI for QSAR Molecular Visualization
2.13. Workflow Overview
3. Results and Discussion
3.1. QSPR Equations and Molecular Descriptor Coefficients
3.2. Applicability Domain and Williams Plot
3.3. Analysis of QSPR Results
3.3.1. The Effect of HOMO and LUMO Energies on Binding Affinity
3.3.2. The Effect of Hydrogen Bonding and LogD on Binding Affinity
3.3.3. Impact of Surface Tension on Binding Affinity
3.3.4. Impact of Flexibility on Binding Strength
3.3.5. The Effect of Density on Binding Affinity
3.3.6. Binding Affinity Correlations with Fukui Index and Polarity
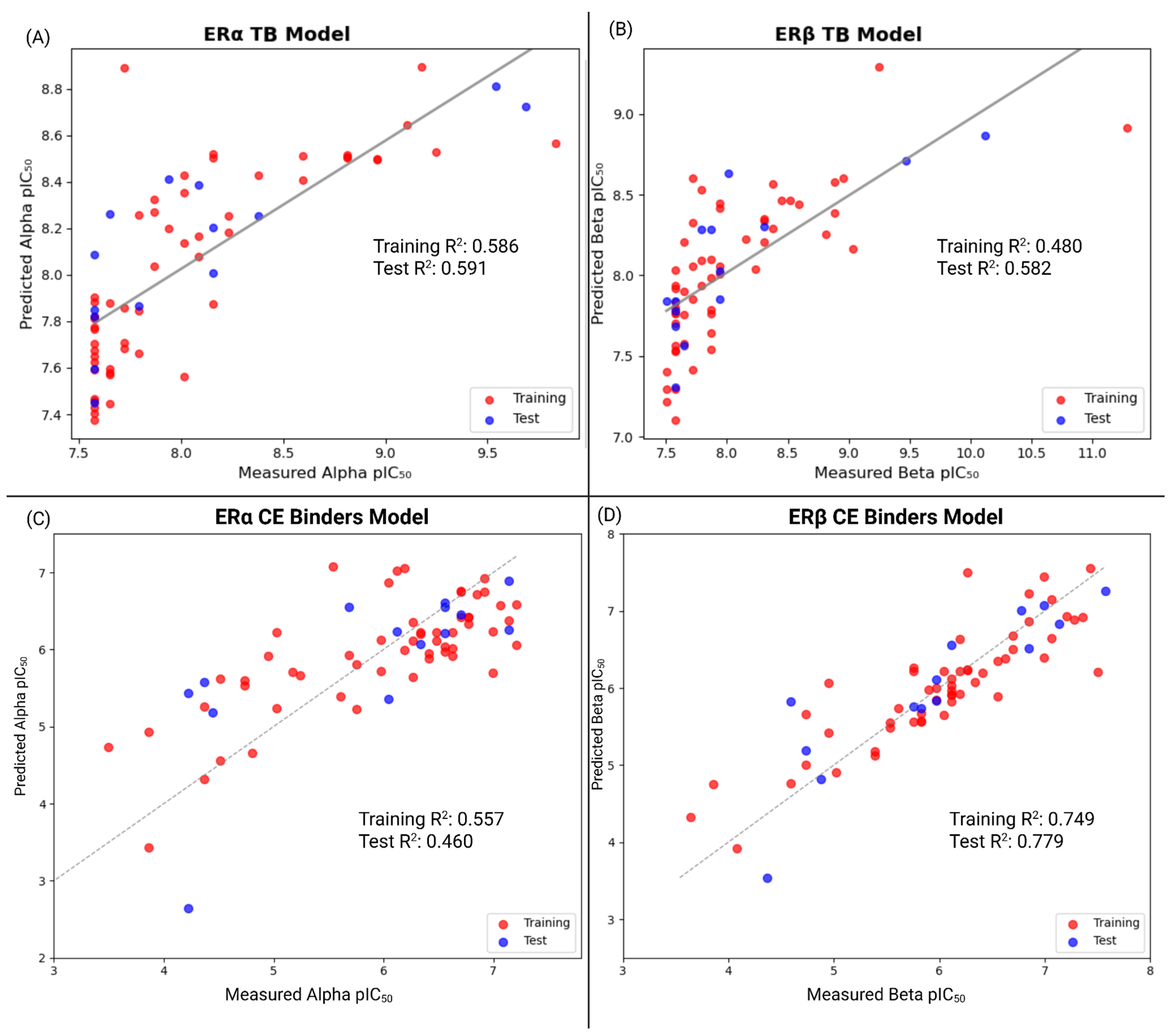
3.4. QSAR Prediction Models
3.4.1. QSAR DUD-E Benchmark Validation
3.4.2. QSAR Analysis for TB PFAS Combined with Experimental PFAS Affinities
3.4.3. QSAR Binding Prediction for Large-Scale PFAS Molecules
4. Strengths and Limitations
5. Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Multi-Industry Per- and Polyfluoroalkyl Substances (PFAS). Available online: https://www.epa.gov/system/files/documents/2021-09/multi-industry-pfas-study_preliminary-2021-report_508_2021.09.08.pdf (accessed on 20 June 2024).
- Starnes, H.M.; Rock, K.D.; Jackson, T.W.; Belcher, S.M. A Critical Review and Meta-Analysis of Impacts of Per- and Polyfluorinated Substances on the Brain and Behavior. Front. Toxicol. 2022, 4, 881584. [Google Scholar] [CrossRef] [PubMed]
- Brunn, H.; Arnold, G.; Körner, W.; Rippen, G.; Steinhäuser, K.G.; Valentin, I. PFAS: Forever chemicals—Persistent, bioaccumulative and mobile. Reviewing the status and the need for their phase out and remediation of contaminated sites. Environ. Sci. Eur. 2023, 35, 20. [Google Scholar] [CrossRef]
- Guidugli, L.F.; Reza, T. Fundamental insight on how carbon chain length affects per-and polyfluoroalkyl substances adsorption onto hydrophobic deep eutectic solvents. J. Mol. Liq. 2024, 398, 124238. [Google Scholar] [CrossRef]
- Jian, J.M.; Chen, D.; Han, F.J.; Guo, Y.; Zeng, L.; Lu, X.; Wang, F. A short review on human exposure to and tissue distribution of per-and polyfluoroalkyl substances (PFASs). Sci. Total Environ. 2018, 636, 1058–1069. [Google Scholar] [CrossRef] [PubMed]
- Fenton, S.E.; Ducatman, A.; Boobis, A.; DeWitt, J.C.; Lau, C.; Ng, C.; Smith, J.S.; Roberts, S.M. Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research. Environ. Toxicol. Chem. 2021, 40, 606–630. [Google Scholar] [CrossRef]
- Kirk, A.B.; Michelsen-Correa, S.; Rosen, C.; Martin, C.F.; Blumberg, B. PFAS and Potential Adverse Effects on Bone and Adipose Tissue Through Interactions with PPARγ. Endocrinology 2021, 162, bqab194. [Google Scholar] [CrossRef]
- Azhagiya Singam, E.R.; Durkin, K.A.; La Merrill, M.A.; Furlow, J.D.; Wang, J.C.; Smith, M.T. Prediction of the interactions of a large number of per-and poly-fluoroalkyl substances with ten nuclear receptors. Environ. Sci. Technol. 2024, 58, 4487–4499. [Google Scholar] [CrossRef]
- Villeneuve, D.L.; Blackwell, B.R.; Cavallin, J.E.; Collins, J.; Hoang, J.X.; Hofer, R.N.; Houck, K.A.; Jensen, K.M.; Kahl, M.D.; Kutsi, R.N.; et al. Verification of In Vivo Estrogenic Activity for Four Per- and Polyfluoroalkyl Substances (PFAS) Identified as Estrogen Receptor Agonists via New Approach Methodologies. Environ. Sci. Technol. 2023, 57, 3794–3803. [Google Scholar] [CrossRef]
- Fischer, F.C.; Ludtke, S.; Thackray, C.; Pickard, H.M.; Haque, F.; Dassuncao, C.; Endo, S.; Schaider, L.; Sunderland, E.M. Binding of Per- and Polyfluoroalkyl Substances (PFAS) to Serum Proteins: Implications for Toxicokinetics in Humans. Environ. Sci. Technol. 2024, 58, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, B.; Ou-Yang, L. Role of estrogen receptors in health and disease. Front. Endocrinol. 2022, 13, 839005. [Google Scholar] [CrossRef]
- Paterni, I.; Granchi, C.; Katzenellenbogen, J.A.; Minutolo, F. Estrogen receptors alpha (ERα) and beta (ERβ): Subtype-selective ligands and clinical potential. Steroids 2014, 90, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Deroo, B.J.; Korach, K.S. Estrogen receptors and human disease. J. Clin. Investig. 2006, 116, 561–570. [Google Scholar] [CrossRef]
- Bhattacharjee, A.K.; Kyle, D.E.; Vennerstrom, J.L.; Milhous, W.K. A 3D QSAR Pharmacophore model and quantum chemical structure− activity analysis of chloroquine (CQ)-resistance reversal. J. Chem. Inf. Comput. Sci. 2002, 42, 1212–1220. [Google Scholar] [CrossRef]
- Votano, J.R.; Parham, M.; Hall, L.M.; Hall, L.H.; Kier, L.B.; Oloff, S.; Tropsha, A. QSAR modeling of human serum protein binding with several modeling techniques utilizing structure− information representation. J. Med. Chem. 2006, 49, 7169–7181. [Google Scholar] [CrossRef] [PubMed]
- Fjodorova, N.; Vracko, M.; Novic, M.; Roncaglioni, A.; Benfenati, E. New public QSAR model for carcinogenicity. Chem. Cent. J. 2010, 4 (Suppl. S1), S3. [Google Scholar] [CrossRef]
- Marques, E.; Pfohl, M.; Wei, W.; Tarantola, G.; Ford, L.; Amaeze, O.; Alesio, J.; Ryu, S.; Jia, X.; Zhu, H.; et al. Replacement per- and polyfluoroalkyl substances (PFAS) are potent modulators of lipogenic and drug metabolizing gene expression signatures in primary human hepatocytes. Toxicol. Appl. Pharmacol. 2022, 442, 115991. [Google Scholar] [CrossRef]
- Kowalska, D.; Sosnowska, A.; Bulawska, N.; Stępnik, M.; Besselink, H.; Behnisch, P.; Puzyn, T. How the Structure of Per- and Polyfluoroalkyl Substances (PFAS) Influences Their Binding Potency to the Peroxisome Proliferator-Activated and Thyroid Hormone Receptors—An In Silico Screening Study. Molecules 2023, 28, 479. [Google Scholar] [CrossRef]
- Sosnowska, A.; Mudlaff, M.; Gorb, L.; Bulawska, N.; Zdybel, S.; Bakker, M.; Peijnenburg, W.; Puzyn, T. Expanding the applicability domain of QSPRs for predicting water solubility and vapor pressure of PFAS. Chemosphere 2023, 340, 139965. [Google Scholar] [CrossRef]
- Tanenbaum, D.M.; Wang, Y.; Williams, S.P.; Sigler, P.B. Crystallographic comparison of the estrogen and progesterone receptors ligand binding domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5998–6003. [Google Scholar] [CrossRef]
- Shiau, A.K.; Barstad, D.; Radek, J.T.; Meyers, M.J.; Nettles, K.W.; Katzenellenbogen, B.S.; Katzenellenbogen, J.A.; Agard, D.A.; Greene, G.L. Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism. Nat. Struct. Biol. 2002, 9, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Singh, U.C.; Kollman, P.A. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 1984, 5, 129–145. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- U.S. EPA CompTox Chemicals Dashboard. Navigation Panel to PFAS Structure Lists. Available online: https://comptox.epa.gov/dashboard/chemical-lists/PFASSTRUCT (accessed on 15 July 2024).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- RCSB PDB. 3ERT: Human Estrogen Receptor Alpha Ligand-Binding Domain. Available online: https://www.rcsb.org/structure/3ERT (accessed on 24 June 2024).
- Bafna, D.; Ban, F.; Rennie, P.S.; Singh, K.; Cherkasov, A. Computer-Aided Ligand Discovery for Estrogen Receptor Alpha. Int. J. Mol. Sci. 2020, 21, 4193. [Google Scholar] [CrossRef]
- Pike, A.C.; Brzozowski, A.M.; Hubbard, R.E.; Bonn, T.; Thorsell, A.G.; Engström, O.; Ljunggren, J.; Gustafsson, J.A.; Carlquist, M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. 1999, 18, 4608–4618. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Pike, A.C.; Dauter, Z.; Hubbard, R.E.; Bonn, T.; Engström, O.; Ohman, L.; Greene, G.L.; Gustafsson, J.A.; Carlquist, M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 1997, 389, 753–758. [Google Scholar] [CrossRef]
- Puranik, N.V.; Srivastava, P.; Bhatt, G.; Mary, D.J.S.J.; Limaye, A.M.; Sivaraman, J. Determination and analysis of agonist and antagonist potential of naturally occurring flavonoids for estrogen receptor (ERα) by various parameters and molecular modelling approach. Sci. Rep. 2019, 9, 7450. [Google Scholar] [CrossRef]
- Royal Society of Chemistry. ChemSpider. Available online: https://www.chemspider.com/ (accessed on 4 September 2024).
- Pourbasheer, E.; Riahi, S.; Ganjali, M.R.; Norouzi, P. Quantitative structure–activity relationship (QSAR) study of interleukin-1 receptor associated kinase 4 (IRAK-4) inhibitor activity by the genetic algorithm and multiple linear regression (GA-MLR) method. J. Enzym. Inhib. Med. Chem. 2010, 25, 844–853. [Google Scholar] [CrossRef]
- Python Software Foundation. The Python Standard Library. Available online: https://docs.python.org/3/library/index.html (accessed on 15 September 2025).
- Reitz, K. Requests: HTTP for HumansTM. Available online: https://requests.readthedocs.io/en/latest/ (accessed on 15 September 2024).
- Pandas Development Team. Pandas Documentation. 2025. Available online: https://pandas.pydata.org/docs/ (accessed on 1 October 2025).
- Richardson, L. Beautiful Soup Documentation. 2025. Available online: https://www.crummy.com/software/BeautifulSoup/bs4/doc/ (accessed on 15 September 2024).
- Landrum, G. RDKit: Open-Source Cheminformatics Software. 2025. Available online: https://www.rdkit.org (accessed on 20 September 2024).
- U.S. Environmental Protection Agency. List of PFAS Added to the TRI by the NDAA. 2024. Available online: https://www.epa.gov/system/files/documents/2024-01/list-of-pfas-added-to-the-tri-by-the-ndaa.pdf (accessed on 25 November 2024).
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Dewar, M.; Stewart, J. MOPAC2016™. Available online: http://openmopac.net/background.html (accessed on 11 December 2024).
- Rowan Scientific. Available online: https://www.rowansci.com (accessed on 14 December 2024).
- Crescent Silico. ChemMaster. 2024. Available online: https://crescent-silico.com/chemmaster/ (accessed on 16 June 2025).
- Rakhimbekova, A.; Madzhidov, T.I.; Nugmanov, R.I.; Gimadiev, T.R.; Baskin, I.I.; Varnek, A. Comprehensive Analysis of Applicability Domains of QSPR Models for Chemical Reactions. Int. J. Mol. Sci. 2020, 21, 5542. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, H.; Moran, J.; Roy, S.; Mathew, J.; Ackah-Blay, J.; Costello, E.; Shan, P.; Dakshanamurthy, S. Impact of Persistent Endocrine-Disrupting Chemicals on Human Nuclear Receptors: Insights from In Silico and Experimental Characterization. Int. J. Mol. Sci. 2025, 26, 2879. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Qu, K.; Luan, F.; Liu, Y.; Zhu, Y.; Yuan, Y.; Li, H.; Zhang, H.; Hai, Y.; Zhao, C. Binding specificities of estrogen receptor with perfluorinated compounds: A cross species comparison. Environ. Int. 2020, 134, 105284. [Google Scholar] [CrossRef]
- Xin, Y.; Ren, X.M.; Wan, B.; Guo, L.H. Comparative in Vitro and in Vivo Evaluation of the Estrogenic Effect of Hexafluoropropylene Oxide Homologues. Environ. Sci. Technol. 2019, 53, 8371–8380. [Google Scholar] [CrossRef]
- Alexander, D.L.J.; Tropsha, A.; Winkler, D.A. Beware of R(2): Simple, unambiguous assessment of the prediction accuracy of QSAR and QSPR models. J. Chem. Inf. Model. 2015, 55, 1316–1322. [Google Scholar] [CrossRef]
- Nettles, K.W.; Sun, J.; Radek, J.T.; Sheng, S.; Rodriguez, A.L.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S.; Greene, G.L. Allosteric control of ligand selectivity between estrogen receptors alpha and beta: Implications for other nuclear receptors. Mol. Cell 2004, 13, 317–327. [Google Scholar] [CrossRef]
- Benninghoff, A.D.; Bisson, W.H.; Koch, D.C.; Ehresman, D.J.; Kolluri, S.K.; Williams, D.E. Estrogen-like activity of perfluoroalkyl acids in vivo and interaction with human and rainbow trout estrogen receptors in vitro. Toxicol. Sci. Off. J. Soc. Toxicol. 2011, 120, 42–58. [Google Scholar] [CrossRef]
- Zhao, C.; Dahlman-Wright, K.; Gustafsson, J.A. Estrogen receptor beta: An overview and update. Nucl. Recept. Signal. 2008, 6, e003. [Google Scholar] [CrossRef] [PubMed]
- Souza, P.C.T.; Textor, L.C.; Melo, D.C.; Nascimento, A.S.; Skaf, M.S.; Polikarpov, I. An alternative conformation of ERβ bound to estradiol reveals H12 in a stable antagonist position. Sci. Rep. 2017, 7, 3509. [Google Scholar] [CrossRef] [PubMed]
- Dandibhotla, S.; Samudrala, M.; Kaneriya, A.; Dakshanamurthy, S. GNNSeq: A Sequence-Based Graph Neural Network for Predicting Protein–Ligand Binding Affinity. Pharmaceuticals 2025, 18, 329. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Liang, Y.; Xu, Q.; Li, H.; Chen, X. A new strategy of outlier detection for QSAR/QSPR. J. Comput. Chem. 2010, 31, 592–602. [Google Scholar] [CrossRef]






| Estrogen Receptor Ligands | PubChem CID | IC50 Values (nM) | pIC50 Values | AutoDock Vina Score (kcal/mol) | AutoDock IC50 Values (nM) | AutoDock pIC50 Values | AutoDock pIC50 Literature pIC50 Difference |
|---|---|---|---|---|---|---|---|
| Hydroxytamoxifen | 449459 | 40 | 7.40 | −9 | 278.38 | 6.60 | −0.844 |
| Caffeic Acid | 689043 | 120,000 | 3.92 | −6.4 | 21,800 | 4.66 | 0.74 |
| Bazedoxifene | 154257 | 26 | 7.59 | −8.5 | 643.90 | 6.19 | −1.39 |
| Lasofoxifene | 216416 | 1.08 | 8.97 | −8.4 | 761.48 | 6.19 | −2.85 |
| Fulvestrant | 104741 | 4.4 | 8.36 | −8.3 | 900.52 | 6.05 | −2.31 |
| Estrogen Receptor Ligands | PubChem CID | IC50 Values (nM) | pIC50 Values | AutoDock Vina Score (kcal/mol) | AutoDock IC50 Values (nM) | AutoDock pIC50 Values | AutoDock pIC50-Literature pIC50 Difference |
|---|---|---|---|---|---|---|---|
| Estradiol | 5757 | 46 | 7.34 | −10 | 52.0 | 7.28 | −0.0535 |
| Raloxifene | 5035 | 7.7 | 8.11 | −11.1 | 8.79 | 8.06 | −0.0575 |
| QYA | 145949437 | 5000 | 5.30 | −6.97 | 127,000 | 3.90 | −1.40 |
| Benzoxazole | 9228 | 5.4 | 8.27 | −11.3 | 6.18 | 8.21 | −0.0586 |
| Hydroxytamoxifen | 449459 | 23,000 | 4.64 | −6.23 | 36,700 | 4.44 | −0.23 |
| ChemSpider | Avogadro and MOPAC | RowanSci |
|---|---|---|
| Density, Number of Hydrogen Bond Acceptors and Donors, Number of Freely Rotating Bonds, LogD (pH 7.4), Polar Surface Area, and Surface Tension | HOMO, LUMO, Average Mass | F+ Max |
| Model | R2 (Train) | RMSE (Train) | MAE (Train) | Q2 (Internal) | R2 (Test) | RMSE (Test) | MAE (Test) |
|---|---|---|---|---|---|---|---|
| TBs (ERα) | 0.586 | 0.351 | 0.253 | 0.307 | 0.591 | 0.432 | 0.332 |
| TBs (ERβ) | 0.480 | 0.459 | 0.295 | 0.191 | 0.582 | 0.485 | 0.356 |
| Commonly Exposed (ERα) | 0.557 | 0.639 | 0.514 | 0.139 | 0.460 | 0.769 | 0.604 |
| Commonly Exposed (ERβ) | 0.749 | 0.435 | 0.315 | 0.456 | 0.779 | 0.465 | 0.332 |
| DUD-E (ERα) | 0.678 | 0.795 | 0.648 | 0.672 | 0.672 | 0.867 | 0.700 |
| DUD-E (ERβ) | 0.618 | 0.845 | 0.678 | 0.611 | 0.672 | 0.823 | 0.662 |
| Large-Scale QSAR (ERα) | 0.461 | 0.506 | 0.388 | 0.455 | 0.385 | 0.525 | 0.387 |
| Large-Scale QSAR (ERβ) | 0.488 | 0.364 | 0.274 | 0.484 | 0.446 | 0.364 | 0.274 |
| a = Average Mass | c = Number of H-Bond Donors | d = Density |
| e = LUMO | f = Number of Freely Rotating Bonds | g = Number of H-Bond Acceptors |
| h = F+ Max | i = Polar Surface Area | j = HOMO |
| k = LogD (pH 7.4) | s = Surface Tension |
| (A) Descriptor Name | Top Binders ERα Coefficients | Top Binders ERβ Coefficients | Commonly Exposed ERα Coefficients | Commonly Exposed ERβ Coefficients |
| Number of H-Bond Acceptors | 0.0154 | 0.0857 | −0.2744 | −0.1780 |
| Number of H-Bond Donors | −0.0312 | 0.0395 | −0.0874 | −0.0343 |
| LogD | 0.0485 | 0.2245 | 0.0483 | −0.0835 |
| Average Mass | 0.2222 | 0.1336 | −0.0198 | 0.0344 |
| Density | 0.0887 | 0.1731 | 0.3900 | 0.3057 |
| F+ Max | 0.0166 | −0.0471 | −0.0485 | 0.0343 |
| HOMO | −0.0198 | −0.0218 | 0.1857 | 0.0242 |
| LUMO | 0.0285 | 0.1132 | 0.0316 | 0.0734 |
| Polar Surface Area | −0.0185 | 0.0069 | 0.4216 | 0.1504 |
| Surface Tension | −0.0821 | −0.0635 | −0.4753 | −0.2855 |
| Number of Freely Rotating Bonds | −0.3843 | −0.4810 | 0.2519 | 0.5858 |
| (B) Descriptor Name | Top Binders ERα Coefficients | Top Binders ERβ Coefficients | Commonly Exposed ERα Coefficients | Commonly Exposed ERβ Coefficients |
| Number of H-Bond Acceptors | 0.0408 | 0.1842 | −0.4964 | −0.3563 |
| Number of H-Bond Donors | −0.0322 | 0.0363 | −0.0487 | −0.0212 |
| LogD | 0.2422 | 0.8252 | 0.1923 | −0.3679 |
| Average Mass | 49.2514 | 30.1649 | −4.1092 | 7.8995 |
| Density | 0.0219 | 0.0420 | 0.0513 | 0.0445 |
| F+ Max | 0.0011 | −0.0048 | −0.0043 | 0.0034 |
| HOMO | −0.0449 | −0.0459 | 0.7999 | 0.1153 |
| LUMO | 0.0434 | 0.1306 | 0.1539 | 0.3954 |
| Polar Surface Area | −0.9347 | 0.2707 | 10.6211 | 4.1924 |
| Surface Tension | −1.3549 | −1.3352 | −2.3411 | −1.5560 |
| Number of Freely Rotating Bonds | −2.3991 | −3.3561 | 1.0370 | 2.6684 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mada, S.; Jordan, S.; Mathew, J.; Loveranes, C.; Moran, J.; Ganesh, H.; Dakshanamurthy, S. Molecular Determinants of Per- and Polyfluoroalkyl Substances Binding to Estrogen Receptors. Toxics 2025, 13, 903. https://doi.org/10.3390/toxics13110903
Mada S, Jordan S, Mathew J, Loveranes C, Moran J, Ganesh H, Dakshanamurthy S. Molecular Determinants of Per- and Polyfluoroalkyl Substances Binding to Estrogen Receptors. Toxics. 2025; 13(11):903. https://doi.org/10.3390/toxics13110903
Chicago/Turabian StyleMada, Sahith, Samuel Jordan, Joshua Mathew, Coby Loveranes, James Moran, Harrish Ganesh, and Sivanesan Dakshanamurthy. 2025. "Molecular Determinants of Per- and Polyfluoroalkyl Substances Binding to Estrogen Receptors" Toxics 13, no. 11: 903. https://doi.org/10.3390/toxics13110903
APA StyleMada, S., Jordan, S., Mathew, J., Loveranes, C., Moran, J., Ganesh, H., & Dakshanamurthy, S. (2025). Molecular Determinants of Per- and Polyfluoroalkyl Substances Binding to Estrogen Receptors. Toxics, 13(11), 903. https://doi.org/10.3390/toxics13110903









