Self-Organizing Maps: An AI Tool for Identifying Unexpected Source Signatures in Non-Target Screening Analysis of Urban Wastewater by HPLC-HRMS
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Solutions
2.3. Sample Collection and Processing
2.4. Sample Analysis
2.5. Untargeted Analysis
Annotation Filtering and Manual Supervision
2.6. Multivariate Data Analysis
3. Results and Discussion
- Screening of the compounds for identifying the artifacts (the features that showed a greater peak area in the diluted sample than in the concentrated one have been discarded);
- Identification by HCA of the “unknown common” compounds, i.e., the substances common among the industrial and urban samples;
- Splitting of the “unknown common” dataset in industrial samples and urban samples;
- PCA and SOM model building using the urban subset;
- Projection of the industrial subset into the above-mentioned models;
- Comparison of SOM and PCA outcomes.
3.1. Screening for Artifacts Identification
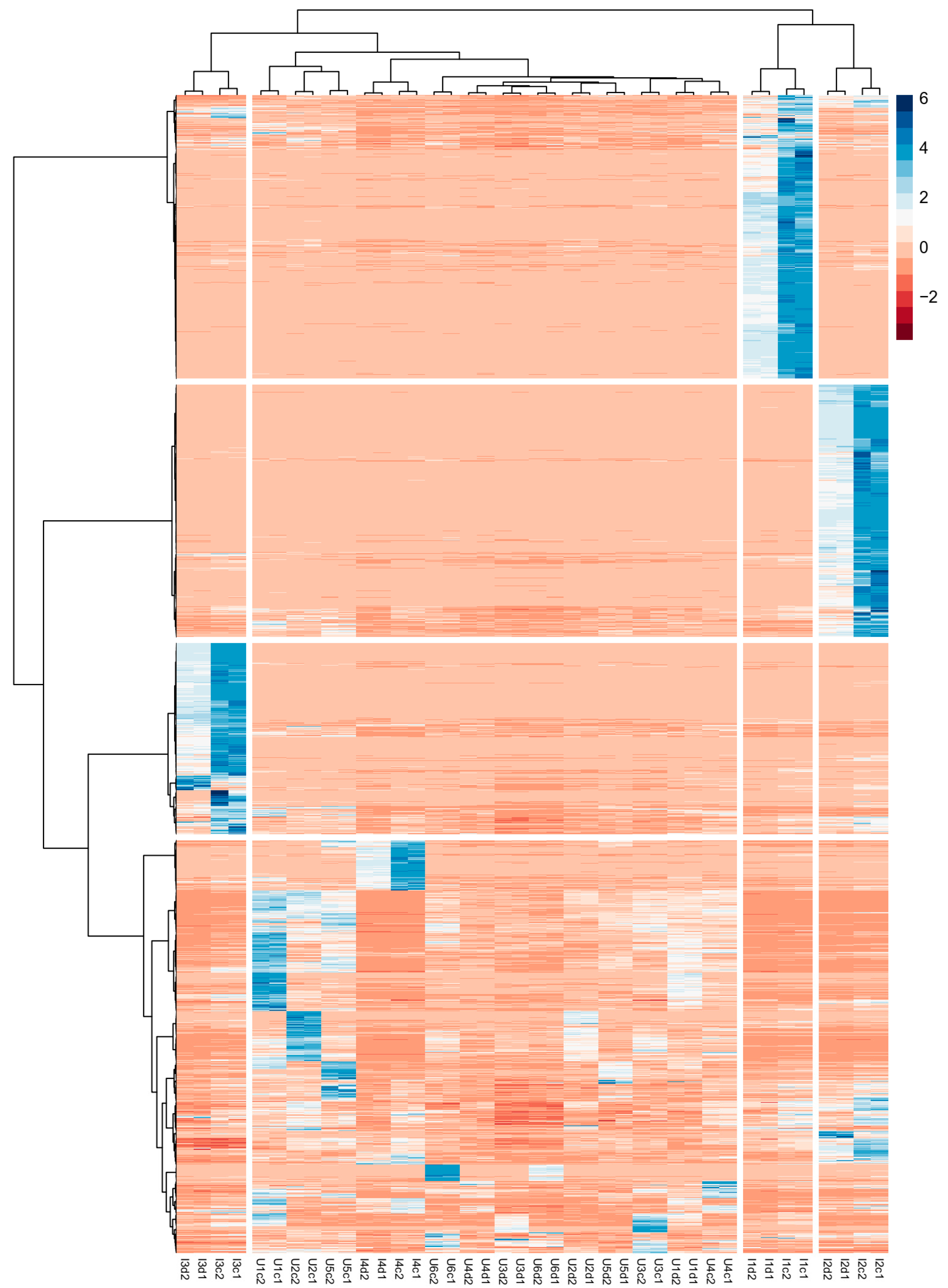
3.2. Selection of “Unknown Common” Compounds
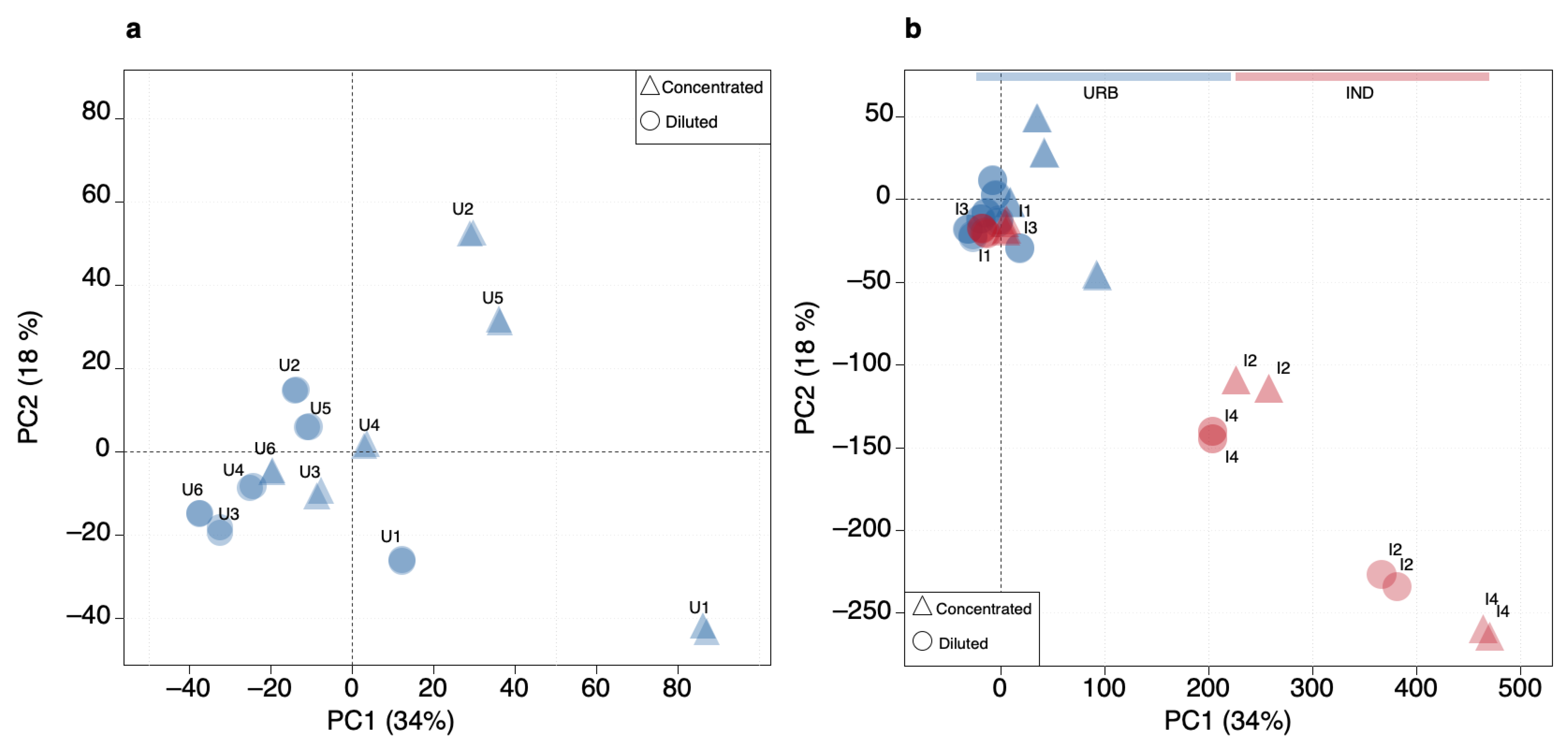
3.3. PCA Model
3.4. SOM Model
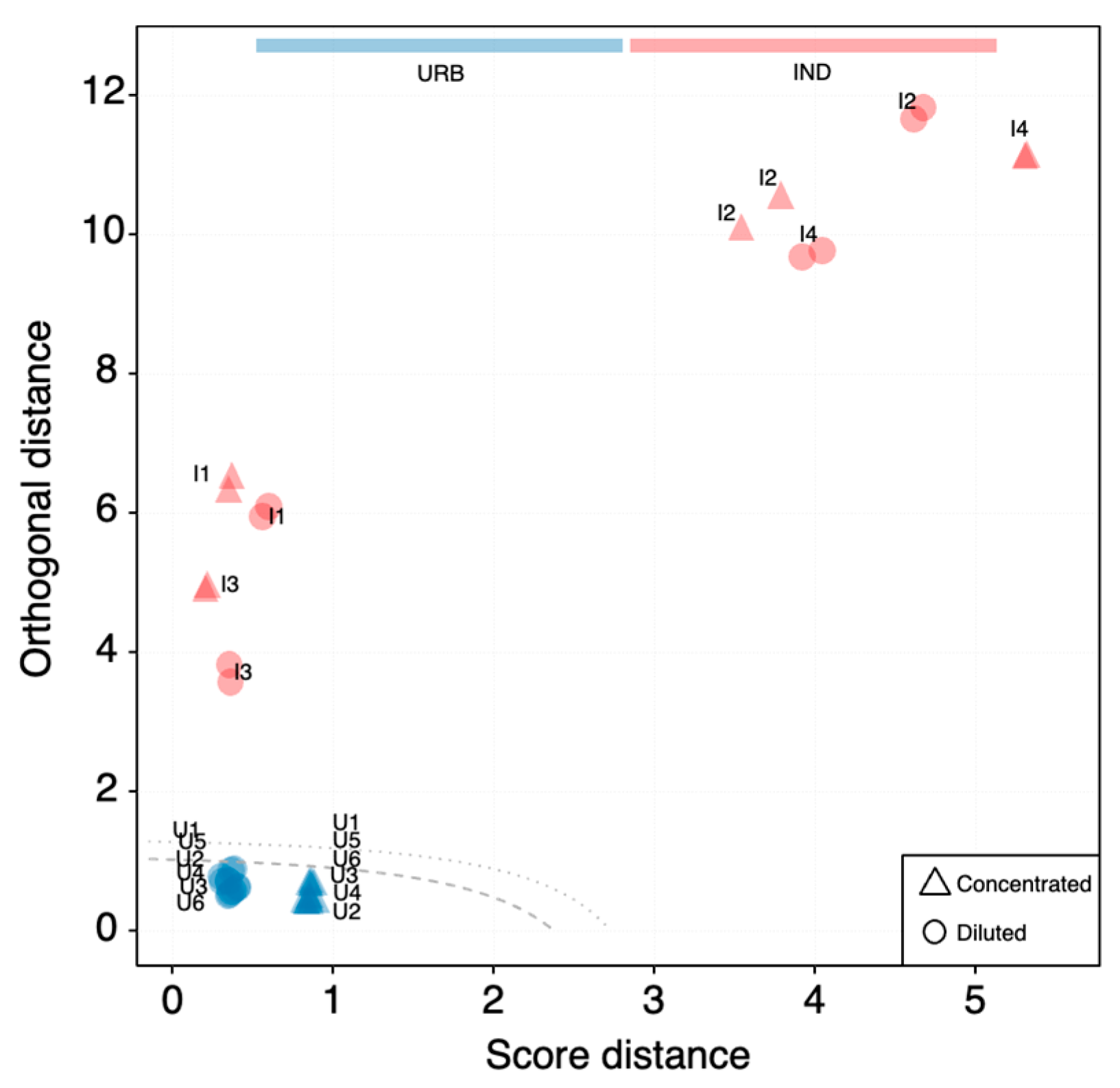
3.5. SOM and PCA Outcome Comparison
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, N.A.; López-Maldonado, E.A.; Majumder, A.; Singh, S.; Varshney, R.; López, J.R.; Méndez, P.F.; Ramamurthy, P.C.; Khan, M.A.; Khan, A.H.; et al. A state-of-art-review on emerging contaminants: Environmental chemistry, health effect, and modern treatment methods. Chemosphere 2023, 344, 140264. [Google Scholar] [CrossRef]
- Rogowska, J.; Cieszynska-Semenowicz, M.; Ratajczyk, W.; Wolska, L. Micropollutants in treated wastewater. Ambio 2020, 49, 487–503. [Google Scholar] [CrossRef] [PubMed]
- Hollender, J.; Schymanski, E.L.; Ahrens, L.; Alygizakis, N.; Béen, F.; Bijlsma, L.; Brunner, A.M.; Celma, A.; Fildier, A.; Fu, Q.; et al. NORMAN guidance on suspect and non-target screening in environmental monitoring. Environ. Sci. Eur. 2023, 35, 75. [Google Scholar] [CrossRef]
- Kiefer, K.; Du, L.; Singer, H.; Hollender, J. Identification of LC-HRMS nontarget signals in groundwater after source related prioritization. Water Res. 2021, 196, 116994. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Peter, K.T.; Gipe, A.D.; Zhao, H.; Hou, F.; Wark, D.A.; Khangaonkar, T.; Kolodziej, E.P.; James, C.A. Suspect and Nontarget Screening for Contaminants of Emerging Concern in an Urban Estuary. Environ. Sci. Technol. 2020, 54, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Bonnefille, B.; Karlsson, O.; Rian, M.B.; Raqib, R.; Parvez, F.; Papazian, S.; Islam, M.S.; Martin, J.W. Nontarget Analysis of Polluted Surface Waters in Bangladesh Using Open Science Workflows. Environ. Sci. Technol. 2023, 57, 6808–6824. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Tian, Z.; Peter, K.T.; Kolodziej, E.P.; Wong, C.S. Developing Unique Nontarget High-Resolution Mass Spectrometry Signatures to Track Contaminant Sources in Urban Waters. Environ. Sci. Technol. Lett. 2020, 7, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Lara-Martín, P.A.; Chiaia-Hernández, A.C.; Biel-Maeso, M.; Baena-Nogueras, R.M.; Hollender, J. Tracing Urban Wastewater Contaminants into the Atlantic Ocean by Nontarget Screening. Environ. Sci. Technol. 2020, 54, 3996–4005. [Google Scholar] [CrossRef]
- Lopez-Herguedas, N.; González-Gaya, B.; Castelblanco-Boyacá, N.; Rico, A.; Etxebarria, N.; Olivares, M.; Prieto, A.; Zuloaga, O. Characterization of the contamination fingerprint of wastewater treatment plant effluents in the Henares River Basin (central Spain) based on target and suspect screening analysis. Sci. Total Environ. 2022, 806, 151262. [Google Scholar] [CrossRef]
- Tisler, S.; Engler, N.; Jørgensen, M.B.; Kilpinen, K.; Tomasi, G.; Christensen, J.H. From data to reliable conclusions: Identification and comparison of persistent micropollutants and transformation products in 37 wastewater samples by non-target screening prioritization. Water Res. 2022, 219, 118599. [Google Scholar] [CrossRef]
- Schollée, J.E.; Hollender, J.; McArdell, C.S. Characterization of advanced wastewater treatment with ozone and activated carbon using LC-HRMS based non-target screening with automated trend assignment. Water Res. 2021, 200, 117209. [Google Scholar] [CrossRef]
- Alygizakis, N.A.; Gago-Ferrero, P.; Hollender, J.; Thomaidis, N.S. Untargeted time-pattern analysis of LC-HRMS data to detect spills and compounds with high fluctuation in influent wastewater. J. Hazard. Mater. 2019, 361, 19–29. [Google Scholar] [CrossRef]
- Carrillo, K.C.; Drouet, J.C.; Rodríguez-Romero, A.; Tovar-Sánchez, A.; Ruiz-Gutiérrez, G.; Viguri Fuente, J.R. Spatial distribution and level of contamination of potentially toxic elements in sediments and soils of a biological reserve wetland, northern Amazon region of Ecuador. J. Environ. Manage. 2021, 289, 112495. [Google Scholar] [CrossRef] [PubMed]
- Purschke, K.; Vosough, M.; Leonhardt, J.; Weber, M.; Schmidt, T.C. Evaluation of Nontarget Long-Term LC-HRMS Time Series Data Using Multivariate Statistical Approaches. Anal. Chem. 2020, 92, 12273–12281. [Google Scholar] [CrossRef]
- Rodrigues, K.L.T.; Sanson, A.L.; de Vasconcelos Quaresma, A.; de Paiva Gomes, R.; da Silva, G.A.Ô.; de Cássia Franco Afonso, R.J. Chemometric approach to optimize the operational parameters of ESI for the determination of contaminants of emerging concern in aqueous matrices by LC-IT-TOF-HRMS. Microchem. J. 2014, 117, 242–249. [Google Scholar] [CrossRef]
- Linghu, K.; Wu, Q.; Zhang, J.; Wang, Z.; Zeng, J.; Gao, S. Occurrence, distribution and ecological risk assessment of antibiotics in Nanming river: Contribution from wastewater treatment plant and implications of urban river syndrome. Process Saf. Environ. Prot. 2023, 169, 428–436. [Google Scholar] [CrossRef]
- Stefano, P.H.P.; Roisenberg, A.; Santos, M.R.; Dias, M.A.; Montagner, C.C. Unraveling the occurrence of contaminants of emerging concern in groundwater from urban setting: A combined multidisciplinary approach and self-organizing maps. Chemosphere 2022, 299, 134395. [Google Scholar] [CrossRef]
- Himberg, J.; Ahola, J.; Alhoniemi, E.; Vesanto, J.; Simula, O. The Self-Organizing Map as a Tool in Knowledge Engineering. In Pattern Recognition in Soft Computing Paradigm; World Scientific: Singapore, 2001; pp. 38–65. [Google Scholar]
- Kohonen, T. Essentials of the self-organizing map. Neural Netw. 2013, 37, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Licen, S.; Astel, A.; Tsakovski, S. Self-organizing map algorithm for assessing spatial and temporal patterns of pollutants in environmental compartments: A review. Sci. Total Environ. 2023, 878, 163084. [Google Scholar] [CrossRef] [PubMed]
- Song, X.H.; Hopke, P.K. Kohonen neural network as a pattern recognition method based on the weight interpretation. Anal. Chim. Acta 1996, 334, 57–66. [Google Scholar] [CrossRef]
- Kohonen, T. Self-Organizing Maps; Springer Series in Information Sciences; Springer: Berlin/Heidelberg, Germany, 2001. [Google Scholar]
- Kohonen, T. Self-organized formation of topologically correct feature maps. Biol Cybern 1982, 43, 59–69. [Google Scholar] [CrossRef]
- Vesanto, J. SOM-based data visualization methods. Intell. Data Anal. 1999, 3, 111–126. [Google Scholar] [CrossRef]
- Russell, S.; Norvig, P. Artificial Intelligence A Modern Approach, 4th ed.; University of California: Berkeley, CA, USA, 2021. [Google Scholar]
- Loos, M.; Singer, H. Nontargeted homologue series extraction from hyphenated high resolution mass spectrometry data. J. Cheminform. 2017, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Aalizadeh, R.; Alygizakis, N.A.; Schymanski, E.L.; Krauss, M.; Schulze, T.; Ibáñez, M.; McEachran, A.D.; Chao, A.; Williams, A.J.; Gago-Ferrero, P.; et al. Development and Application of Liquid Chromatographic Retention Time Indices in HRMS-Based Suspect and Nontarget Screening. Anal. Chem. 2021, 93, 11601–11611. [Google Scholar] [CrossRef] [PubMed]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Hornik, K. The Comprehensive R Archive Network. Wiley Interdiscip. Rev. Comput. Stat. 2012, 4, 394–398. [Google Scholar] [CrossRef]
- Kolde, R. Package “Pheatmap”: Pretty Heatmaps. R Package. 2022. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 17 December 2023).
- Kucheryavskiy, S. mdatools—R package for chemometrics. Chemom. Intell. Lab. Syst. 2020, 198, 103937. [Google Scholar] [CrossRef]
- Licen, S.; Franzon, M.; Rodani, T.; Barbieri, P. SOMEnv: An R package for mining environmental monitoring datasets by Self-Organizing Map and k-means algorithms with a graphical user interface. Microchem. J. 2021, 165, 106181. [Google Scholar] [CrossRef]
- Di Guida, R.; Engel, J.; Allwood, J.W.; Weber, R.J.M.; Jones, M.R.; Sommer, U.; Viant, M.R.; Dunn, W.B. Non-targeted UHPLC-MS metabolomic data processing methods: A comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics 2016, 12, 93. [Google Scholar] [CrossRef]
- Rodionova, O.; Kucheryavskiy, S.; Pomerantsev, A. Efficient tools for principal component analysis of complex data— a tutorial. Chemom. Intell. Lab. Syst. 2021, 213, 104304. [Google Scholar] [CrossRef]
- Clark, S.; Sisson, S.A.; Sharma, A. Tools for enhancing the application of self-organizing maps in water resources research and engineering. Adv. Water Resour. 2020, 143, 103676. [Google Scholar] [CrossRef]
- Muñoz, A.; Muruzábal, J. Self-organizing maps for outlier detection. Neurocomputing 1998, 18, 33–60. [Google Scholar] [CrossRef]
- Licen, S.; Cozzutto, S.; Barbieri, G.; Crosera, M.; Adami, G.; Barbieri, P. Characterization of variability of air particulate matter size profiles recorded by optical particle counters near a complex emissive source by use of Self-Organizing Map algorithm. Chemom. Intell. Lab. Syst. 2019, 190, 48–54. [Google Scholar] [CrossRef]
- Licen, S.; Di Gilio, A.; Palmisani, J.; Petraccone, S.; de Gennaro, G.; Barbieri, P. Pattern recognition and anomaly detection by self-organizing maps in a multi month e-nose survey at an industrial site. Sensors 2020, 20, 1887. [Google Scholar] [CrossRef]
- Gago-Ferrero, P.; Schymanski, E.L.; Hollender, J.; Thomaidis, N.S. Nontarget Analysis of Environmental Samples Based on Liquid Chromatography Coupled to High Resolution Mass Spectrometry (LC-HRMS). In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2016; pp. 381–403. [Google Scholar] [CrossRef]
- Want, E.J. LC-MS untargeted analysis. Methods Mol. Biol. 2018, 1738, 99–116. [Google Scholar] [CrossRef] [PubMed]

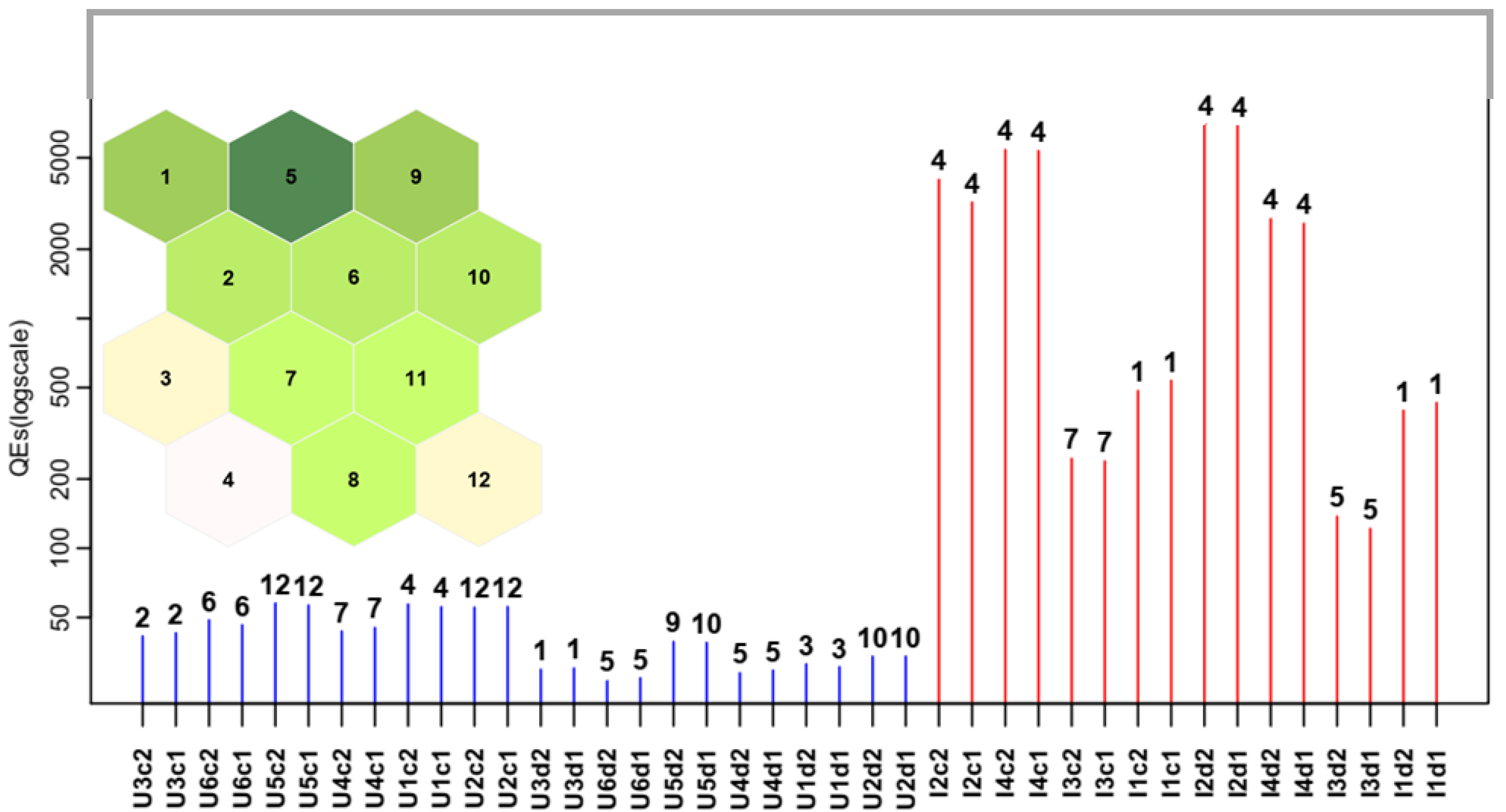
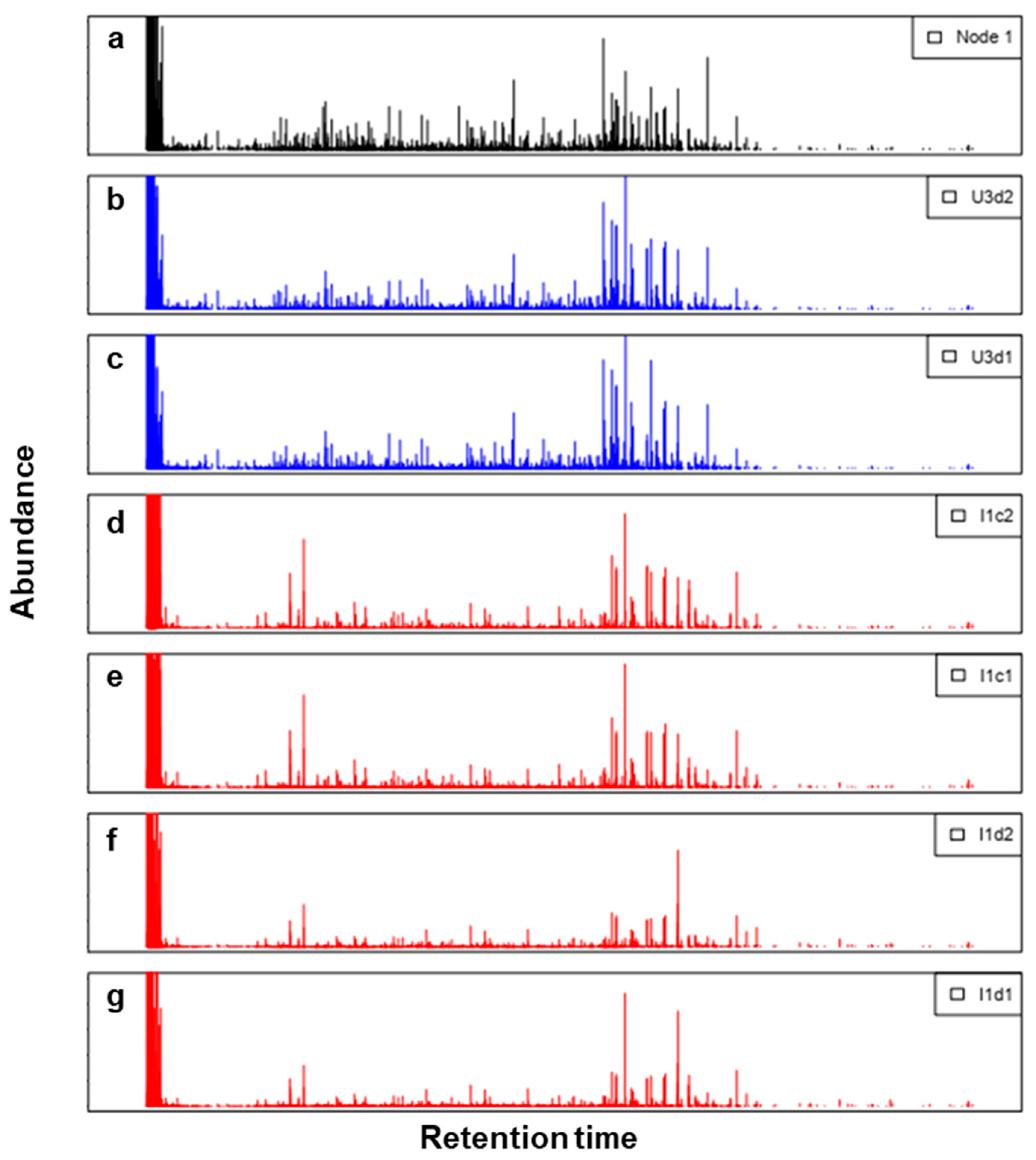
| Map Size | Dimension Ratio | Nodes without Hits | DME | QE | TE |
|---|---|---|---|---|---|
| 6 × 4 (“regular”) | 1.5 | 13 (54%) | 0 | 32.8 | 0 |
| 4 × 2 (“small”) | 2.0 | 0 (0%) | 0 | 43.9 | 0 |
| 5 × 3 | 1.7 | 4 (27%) | 0 | 38.9 | 0 |
| 4 × 3 | 1.3 | 2 (17%) | 0 | 41.6 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gelao, V.; Fornasaro, S.; Briguglio, S.C.; Mattiussi, M.; De Martin, S.; Astel, A.M.; Barbieri, P.; Licen, S. Self-Organizing Maps: An AI Tool for Identifying Unexpected Source Signatures in Non-Target Screening Analysis of Urban Wastewater by HPLC-HRMS. Toxics 2024, 12, 113. https://doi.org/10.3390/toxics12020113
Gelao V, Fornasaro S, Briguglio SC, Mattiussi M, De Martin S, Astel AM, Barbieri P, Licen S. Self-Organizing Maps: An AI Tool for Identifying Unexpected Source Signatures in Non-Target Screening Analysis of Urban Wastewater by HPLC-HRMS. Toxics. 2024; 12(2):113. https://doi.org/10.3390/toxics12020113
Chicago/Turabian StyleGelao, Vito, Stefano Fornasaro, Sara C. Briguglio, Michele Mattiussi, Stefano De Martin, Aleksander M. Astel, Pierluigi Barbieri, and Sabina Licen. 2024. "Self-Organizing Maps: An AI Tool for Identifying Unexpected Source Signatures in Non-Target Screening Analysis of Urban Wastewater by HPLC-HRMS" Toxics 12, no. 2: 113. https://doi.org/10.3390/toxics12020113
APA StyleGelao, V., Fornasaro, S., Briguglio, S. C., Mattiussi, M., De Martin, S., Astel, A. M., Barbieri, P., & Licen, S. (2024). Self-Organizing Maps: An AI Tool for Identifying Unexpected Source Signatures in Non-Target Screening Analysis of Urban Wastewater by HPLC-HRMS. Toxics, 12(2), 113. https://doi.org/10.3390/toxics12020113