Proteomic Analysis of the Mitochondrial Responses in P19 Embryonic Stem Cells Exposed to Florfenicol
Abstract
1. Introduction
2. Results
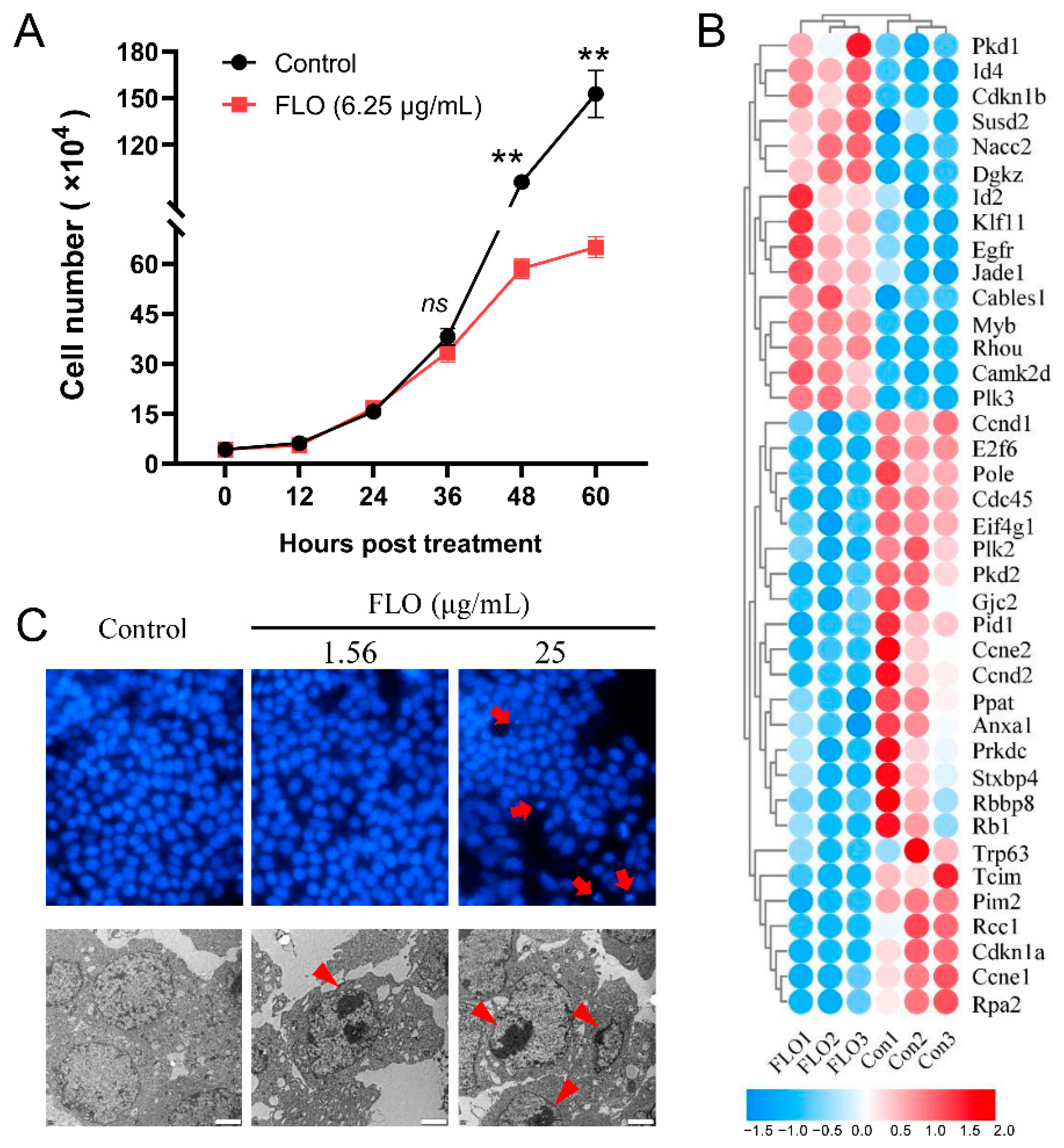
2.1. FLO Inhibits P19SC Proliferation by Perturbing G1/S Phase Transition
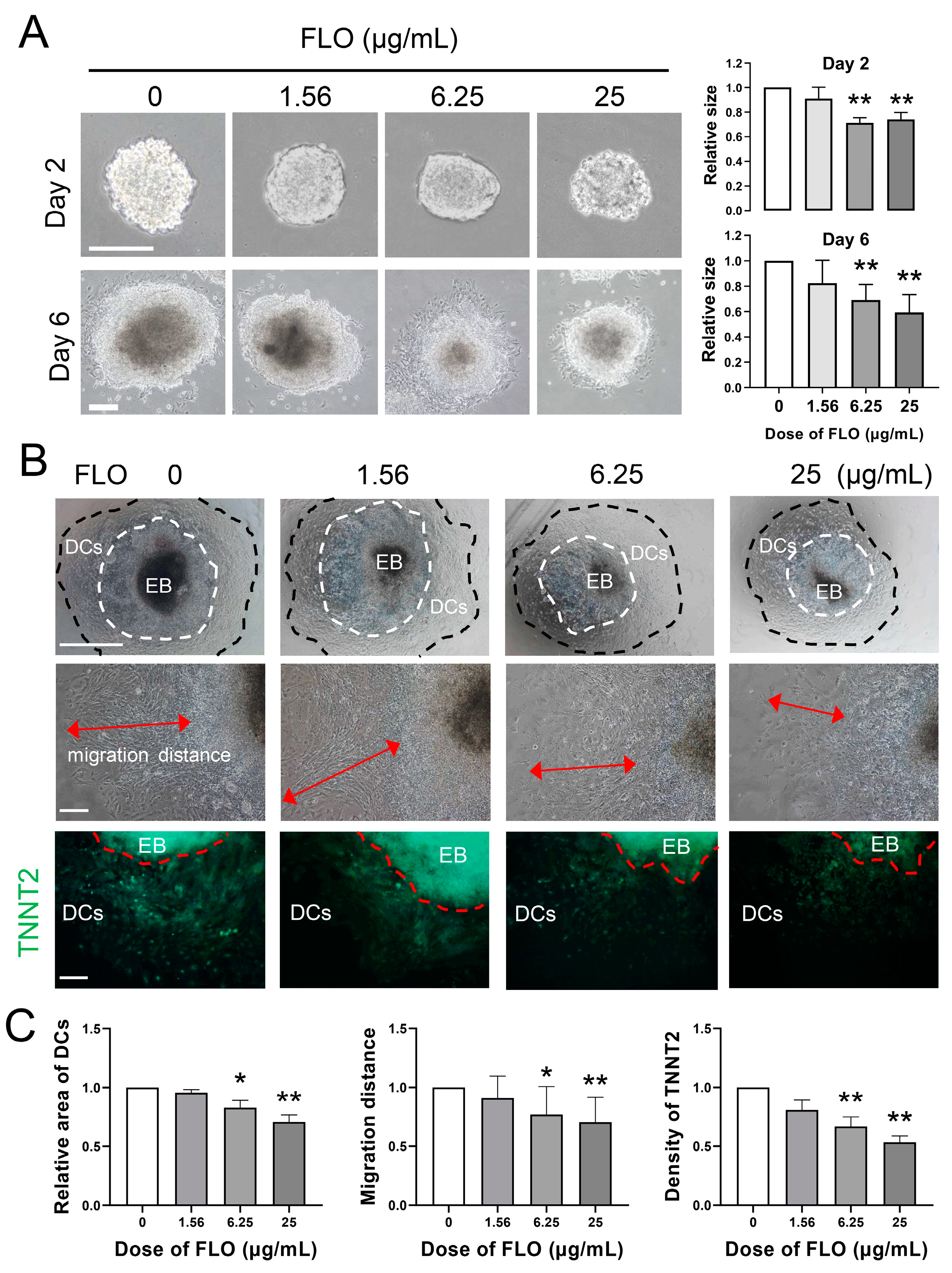
2.2. FLO Inhibits Cardiac Differentiation of P19SCs
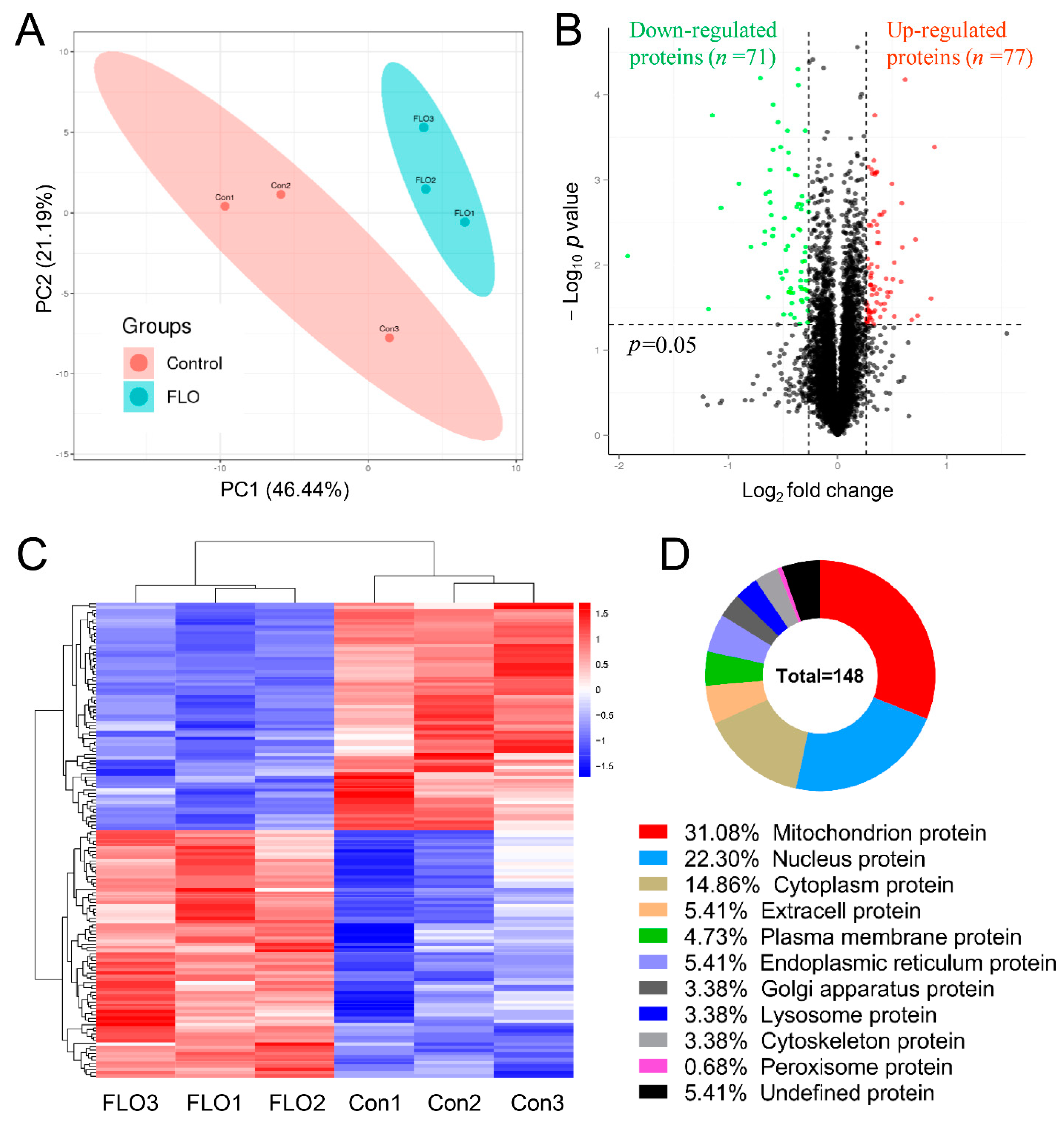
2.3. Overview of Proteome Alterations Induced by FLO Treatment
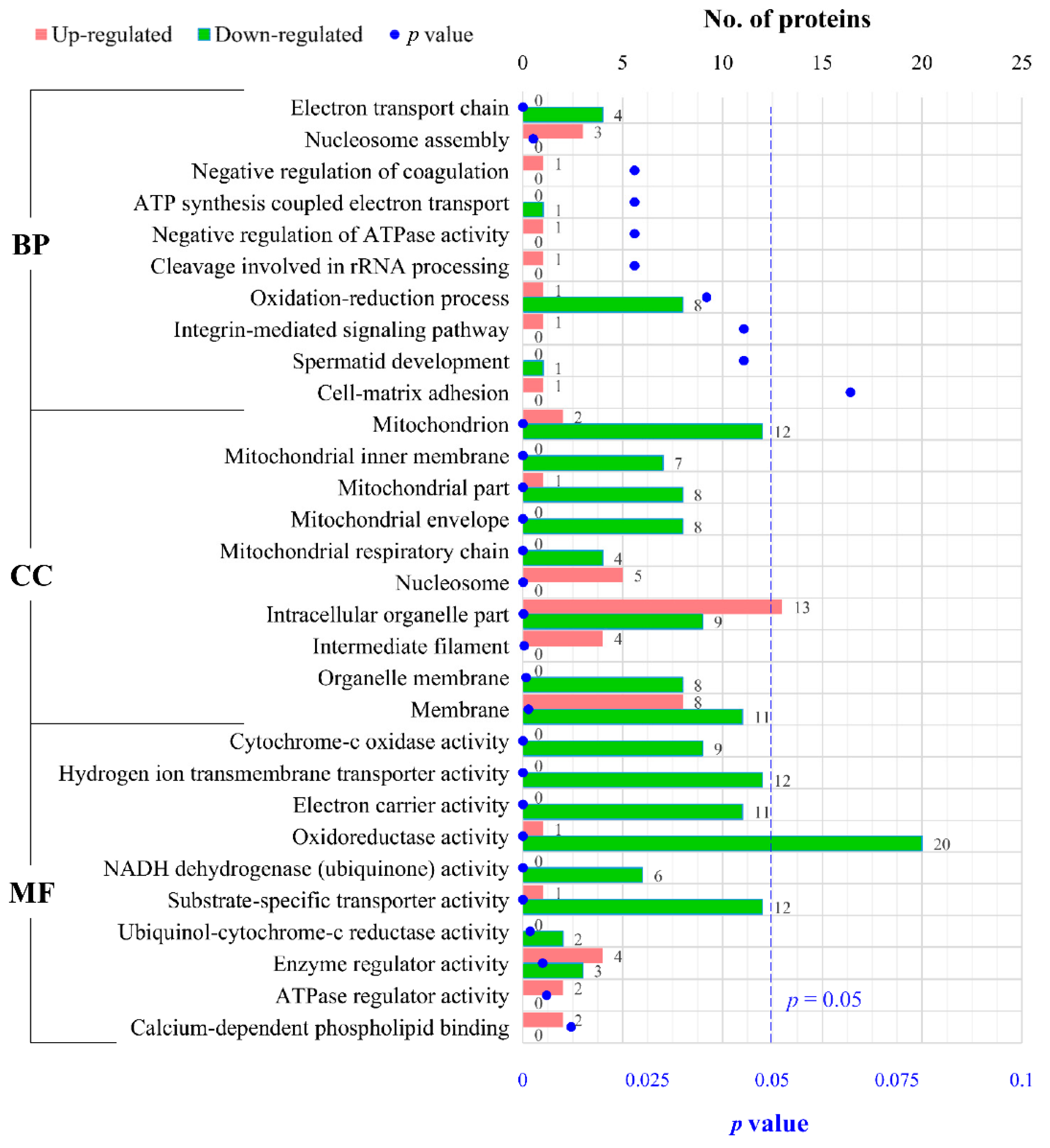
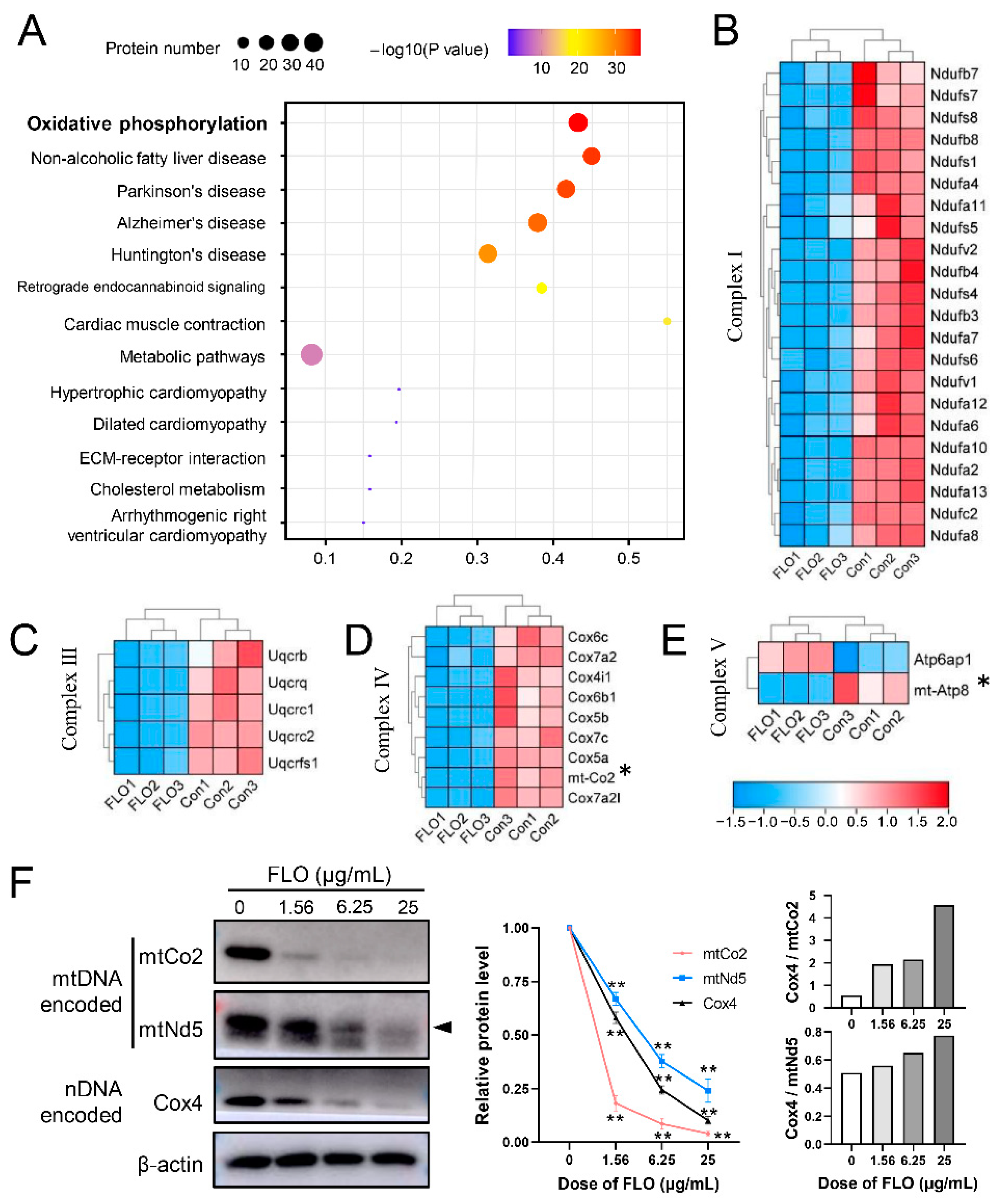
2.4. GO Functional Analysis of DEPs
2.5. KEGG Pathway Analysis of DEPs
2.6. Validation of Proteomic Results
2.7. FLO Induced Mitochondrial Hyperfusion and Unfolded Protein Response
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture and Drug Treatment
4.3. Differentiation of P19SCs
4.4. Cell Growth Curve and Cell Cycle-Related Genes Analysis
4.5. Apoptosis Assay
4.6. Transmission Electron Microscope
4.7. Immunofluorescence Staining
4.8. Western Blot Analysis
4.9. iTRAQ Quantification and Bioinformatics Analysis
4.10. ROS Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaudias, J. Antibiotic prophylaxis in orthopedics-traumatology. Orthop. Traumatol. Surg. Res. 2020, 107, 102751. [Google Scholar] [CrossRef] [PubMed]
- Guidi, L.R.; Tette, P.A.; Fernandes, C.; Silva, L.H.; Gloria, M.B.A. Advances on the chromatographic determination of amphenicols in food. Talanta 2017, 162, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Xia, Y.; Wei, W.; Ni, B.-J. Accelerated spread of antibiotic resistance genes (ARGs) induced by non-antibiotic conditions: Roles and mechanisms. Water Res. 2022, 224, 119060. [Google Scholar] [CrossRef] [PubMed]
- Apreja, M.; Sharma, A.; Balda, S.; Kataria, K.; Capalash, N.; Sharma, P. Antibiotic residues in environment: Antimicrobial resistance development, ecological risks, and bioremediation. Environ. Sci. Pollut. Res. Int. 2022, 29, 3355–3371. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Müller, M.M.; Nedielkov, R.; Arndt, K.M. Strategies for Enzymatic Inactivation of the Veterinary Antibiotic Florfenicol. Antibiotics 2022, 11, 443. [Google Scholar] [CrossRef]
- Liu, D.; Liu, N.Y.; Chen, L.T.; Shao, Y.; Shi, X.M.; Zhu, D.Y. Perfluorooctane sulfonate induced toxicity in embryonic stem cell-derived cardiomyocytes via inhibiting autophagy-lysosome pathway. Toxicol. In Vitro 2020, 69, 104988. [Google Scholar] [CrossRef]
- Wiest, D.B.; Cochran, J.B.; Tecklenburg, F.W. Chloramphenicol Toxicity Revisited: A 12-Year-Old Patient With a Brain Abscess. J. Pediatr. Pharmacol. Ther. 2012, 17, 182–188. [Google Scholar] [CrossRef]
- Wang, X.; Liu, W.; Liu, Y.; Jiao, Y.; Rong, C.; Liu, Q.; Shi, W. Florfenicol induced renal inflammatory response and apoptosis via cell adhesion molecules signaling pathway. Poult. Sci. 2022, 101, 102152. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, W.; Zhang, D.; Jiao, Y.; Zhao, Q.; Liu, Y.; Shi, W.; Bao, Y. Salvia miltiorrhiza polysaccharides alleviate florfenicol-induced inflammation and oxidative stress in chick livers by regulating phagosome signaling pathway. Ecotoxicol. Environ. Saf. 2023, 249, 114428. [Google Scholar] [CrossRef]
- Zhang, L.; Qiu, J.; Li, Y.; He, L.; Mao, M.; Wang, T.; Pan, Y.; Li, Z.; Mu, X.; Qian, Y. Maternal transfer of florfenicol impacts development and disrupts metabolic pathways in F1 offspring zebrafish by destroying mitochondria. Ecotoxicol. Environ. Saf. 2023, 252, 114597. [Google Scholar] [CrossRef]
- Al-Shahrani, S.; Naidoo, V. Florfenicol induces early embryonic death in eggs collected from treated hens. BMC Vet.-Res. 2015, 11, 213. [Google Scholar] [CrossRef][Green Version]
- Hu, D.; Meng, F.; Cui, Y.; Yin, M.; Ning, H.; Yin, Z.; Chen, L.; Ge, Y.; Liu, S. Growth and cardiovascular development are repressed by florfenicol exposure in early chicken embryos. Poult. Sci. 2020, 99, 2736–2745. [Google Scholar] [CrossRef]
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef]
- Hu, D.; Cao, S.; Zhang, G.; Xiao, Y.; Liu, S.; Shang, Y. Florfenicol-induced Mitochondrial Dysfunction Suppresses Cell Proliferation and Autophagy in Fibroblasts. Sci. Rep. 2017, 7, 13554. [Google Scholar] [CrossRef]
- Kim, Y.S.; Yoon, J.W.; Kim, D.; Choi, S.; Kim, H.K.; Youm, J.B.; Han, J.; Heo, S.C.; Hyun, S.-A.; Seo, J.-W.; et al. Tomatidine-stimulated maturation of human embryonic stem cell-derived cardiomyocytes for modeling mitochondrial dysfunction. Exp. Mol. Med. 2022, 54, 493–502. [Google Scholar] [CrossRef]
- Martins, W.K.; Santos, N.F.; Rocha, C.d.S.; Bacellar, I.O.L.; Tsubone, T.M.; Viotto, A.C.; Matsukuma, A.Y.; Abrantes, A.B.d.P.; Siani, P.; Dias, L.G.; et al. Parallel damage in mitochondria and lysosomes is an efficient way to photoinduce cell death. Autophagy 2019, 15, 259–279. [Google Scholar] [CrossRef]
- Lee, S.; Jeong, Y.; Roe, J.-S.; Huh, H.; Paik, S.H.; Song, J. Mitochondrial dysfunction induced by callyspongiolide promotes autophagy-dependent cell death. BMB Rep. 2021, 54, 227–232. [Google Scholar] [CrossRef]
- Morii, A.; Katayama, S.; Inazu, T. Establishment of a Simple Method for Inducing Neuronal Differentiation of P19 EC Cells without Embryoid Body Formation and Analysis of the Role of Histone Deacetylase 8 Activity in This Differentiation. Biol. Pharm. Bull. 2020, 43, 1096–1103. [Google Scholar] [CrossRef]
- Kanungo, J. Retinoic Acid Signaling in P19 Stem Cell Differentiation. Anti-Cancer Agents Med. Chem. 2017, 17, 1184–1198. [Google Scholar] [CrossRef]
- Liu, F.; Ma, M.; Li, L.; Wang, M.; Qin, Y.; Xu, F. Investigation and health risk assessment of veterinary drug residues in chickens and eggs sold in Ningxia from 2016 to 2020. Wei Sheng Yan Jiu = J. Hyg. Res. 2022, 51, 497–508. [Google Scholar] [CrossRef]
- Zhang, H.; Rong, X.; Wang, C.; Liu, Y.; Lu, L.; Li, Y.; Zhao, C.; Zhou, J. VBP1 modulates Wnt/β-catenin signaling by mediating the stability of the transcription factors TCF/LEFs. J. Biol. Chem. 2020, 295, 16826–16839. [Google Scholar] [CrossRef]
- Guo, F.; Wang, Y.; Peng, J.; Huang, H.; Tu, X.; Zhao, H.; Zhan, N.; Rao, Z.; Zhao, G.; Yang, H. Occurrence, Distribution, and Risk Assessment of Antibiotics in the Aquatic Environment of the Karst Plateau Wetland of Yangtze River Basin, Southwestern China. Int. J. Environ. Res. Public Health 2022, 19, 7211. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Guo, R.; Zhang, Q.; Cao, X.; Suranjana, M.; Liu, Y. Effects of florfenicol on growth, photosynthesis and antioxidant system of the non-target organism Isochrysis galbana. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2020, 233, 108764. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, P.; Wang, M.; Wu, Y.; Sun, Y.; Su, H.; Deng, J. Mixture toxicity effects of chloramphenicol, thiamphenicol, florfenicol in Daphnia magna under different temperatures. Ecotoxicology 2021, 30, 31–42. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X.; Liu, Y.; Fang, S.; Wu, Z.; Han, C.; Shi, W.; Bao, Y. Effects of early florfenicol exposure on glutathione signaling pathway and PPAR signaling pathway in chick liver. Ecotoxicol. Environ. Saf. 2022, 237, 113529. [Google Scholar] [CrossRef]
- Mu, Y.; Lan, M.; Li, Y.; Zhang, Z.; Guan, Y. Effects of florfenicol on the antioxidant and immune systems of Chinese soft-shelled turtle (Pelodiscus sinensis). Fish Shellfish. Immunol. 2023, 140, 108991. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Ma, X.; Hu, H. The Influence of Cell Cycle Regulation on Chemotherapy. Int. J. Mol. Sci. 2021, 22, 6923. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, B.; Suo, Y.; Li, Z.; Wan, Z.; Zhao, W.; Chen, L.; Yin, Z.; Ning, H.; Ge, Y.; et al. Molecular Mechanisms Underlying the Inhibition of Proliferation and Differentiation by Florfenicol in P19 Stem Cells: Transcriptome Analysis. Front. Pharmacol. 2022, 13, 779664. [Google Scholar] [CrossRef]
- Paccola, C.; Souza, G.; Freitas, I.; Souza, J.; Martins, L.; Vendramini, V.; Miraglia, S. Does maternal exposure to nicotine affect the oocyte quality and reproductive capacity in adult offspring? Toxicol. Appl. Pharmacol. 2021, 426, 115638. [Google Scholar] [CrossRef]
- Khacho, M.; Clark, A.; Svoboda, D.S.; MacLaurin, J.G.; Lagace, D.C.; Park, D.S.; Slack, R.S. Mitochondrial dysfunction underlies cognitive defects as a result of neural stem cell depletion and impaired neurogenesis. Hum. Mol. Genet. 2017, 26, 3327–3341. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sumberaz, P. Mitochondrial DNA Damage: Prevalence, Biological Consequence, and Emerging Pathways. Chem. Res. Toxicol. 2020, 33, 2491–2502. [Google Scholar] [CrossRef] [PubMed]
- Houtkooper, R.H.; Mouchiroud, L.; Ryu, D.; Moullan, N.; Katsyuba, E.; Knott, G.; Williams, R.W.; Auwerx, J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 2013, 497, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Tournaire, G.; Loopmans, S.; Stegen, S.; Rinaldi, G.; Eelen, G.; Torrekens, S.; Moermans, K.; Carmeliet, P.; Ghesquière, B.; Thienpont, B.; et al. Skeletal progenitors preserve proliferation and self-renewal upon inhibition of mitochondrial respiration by rerouting the TCA cycle. Cell Rep. 2022, 40, 111105. [Google Scholar] [CrossRef]
- Mottis, A.; Li, T.Y.; El Alam, G.; Rapin, A.; Katsyuba, E.; Liaskos, D.; D’amico, D.; Harris, N.L.; Grier, M.C.; Mouchiroud, L.; et al. Tetracycline-induced mitohormesis mediates disease tolerance against influenza. J. Clin. Investig. 2022, 132, e151540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fang, Q.; Wang, H.; Qi, J.; Sun, S.; Liao, M.; Wu, Y.; Hu, Y.; Jiang, P.; Cheng, C.; et al. Increased mitophagy protects cochlear hair cells from aminoglycoside-induced damage. Autophagy 2023, 19, 75–91. [Google Scholar] [CrossRef]
- Adebayo, M.; Singh, S.; Singh, A.P.; Dasgupta, S. Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. FASEB J. 2021, 35, e21620. [Google Scholar] [CrossRef]
- Yapa, N.M.; Lisnyak, V.; Reljic, B.; Ryan, M.T. Mitochondrial dynamics in health and disease. FEBS Lett. 2021, 595, 1184–1204. [Google Scholar] [CrossRef]
- Wakabayashi, J.; Zhang, Z.; Wakabayashi, N.; Tamura, Y.; Fukaya, M.; Kensler, T.W.; Iijima, M.; Sesaki, H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 2009, 186, 805–816. [Google Scholar] [CrossRef]
- De Palma, C.; Falcone, S.; Pisoni, S.; Cipolat, S.; Panzeri, C.; Pambianco, S.; Pisconti, A.; Allevi, R.; Bassi, M.T.; Cossu, G.; et al. Nitric oxide inhibition of Drp1-mediated mitochondrial fission is critical for myogenic differentiation. Cell Death Differ. 2010, 17, 1684–1696. [Google Scholar] [CrossRef]
- Kasahara, A.; Cipolat, S.; Chen, Y.; Dorn, G.W., 2nd; Scorrano, L. Mitochondrial Fusion Directs Cardiomyocyte Differentiation via Calcineurin and Notch Signaling. Science 2013, 342, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Yan, S.; Yu, Q.; Chen, D.; Yan, S.S. Mfn2 is Required for Mitochondrial Development and Synapse Formation in Human Induced Pluripotent Stem Cells/hiPSC Derived Cortical Neurons. Sci. Rep. 2016, 6, 31462. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Gao, K.; Liu, Y. UPR mt coordinates immunity to maintain mitochondrial homeostasis and animal fitness. Mitochondrion 2018, 41, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Z.; Hou, X.; Wang, X.; Shen, Z.; Pang, H.; Chen, L.; Yin, Z.; Ren, F.; Li, W.; Ge, Y.; et al. Proteomic Analysis of the Mitochondrial Responses in P19 Embryonic Stem Cells Exposed to Florfenicol. Toxics 2023, 11, 992. https://doi.org/10.3390/toxics11120992
Dong Z, Hou X, Wang X, Shen Z, Pang H, Chen L, Yin Z, Ren F, Li W, Ge Y, et al. Proteomic Analysis of the Mitochondrial Responses in P19 Embryonic Stem Cells Exposed to Florfenicol. Toxics. 2023; 11(12):992. https://doi.org/10.3390/toxics11120992
Chicago/Turabian StyleDong, Zhihua, Xueke Hou, Xueying Wang, Zihui Shen, Huiqing Pang, Lingli Chen, Zhihong Yin, Fei Ren, Weiguo Li, Yaming Ge, and et al. 2023. "Proteomic Analysis of the Mitochondrial Responses in P19 Embryonic Stem Cells Exposed to Florfenicol" Toxics 11, no. 12: 992. https://doi.org/10.3390/toxics11120992
APA StyleDong, Z., Hou, X., Wang, X., Shen, Z., Pang, H., Chen, L., Yin, Z., Ren, F., Li, W., Ge, Y., Ning, H., & Hu, D. (2023). Proteomic Analysis of the Mitochondrial Responses in P19 Embryonic Stem Cells Exposed to Florfenicol. Toxics, 11(12), 992. https://doi.org/10.3390/toxics11120992




