Expression Profiling of Adipogenic and Anti-Adipogenic MicroRNA Sequences following Methylmercury Exposure in Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. C. elegans Strains and Handling of the Worms
2.3. RNA Isolation, miRNA Expression, and Real-Time qPCR Gene Expression
2.4. Dose–Response Curves
2.5. Nile Red Staining
2.6. Statistics
3. Results
3.1. MeHg Increases Adipogenic miRNA Expression and Decreases Anti-Adipogenic miRNA Expression
3.2. MeHg Decreases the Expression of Anti-Adipogenic Genes Modulated by mir-124 and mir-355
3.3. MeHg Increases the Expression of Adipogenic Genes Modulated by let-7, mir-240, and mir-786
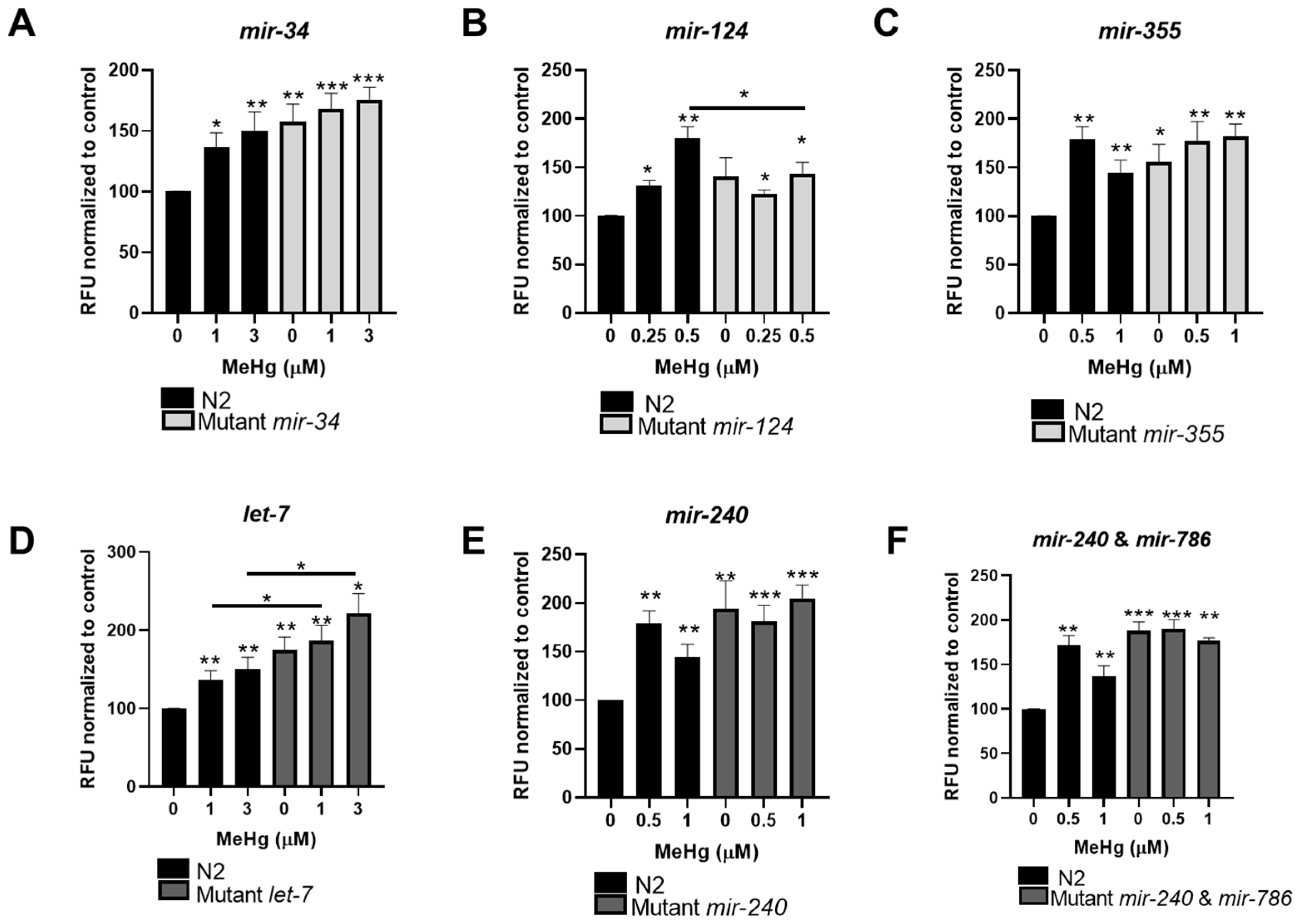
3.4. Reduced miRNA Expression Sensitizes Worms to MeHg Toxicity
3.5. MiRNA Expression Influences MeHg-Induced Lipid Accumulation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chamorro-Garcia, R.; Blumberg, B. Current Research Approaches and Challenges in the Obesogen Field. Front. Endocrinol. 2019, 10, 167. [Google Scholar] [CrossRef]
- Kassotis, C.D.; Stapleton, H.M. Endocrine-Mediated Mechanisms of Metabolic Disruption and New Approaches to Examine the Public Health Threat. Front. Endocrinol. 2019, 10, 39. [Google Scholar] [CrossRef]
- Clarkson, T.W.; Magos, L. The toxicology of mercury and its chemical compounds. Crit. Rev. Toxicol. 2006, 36, 609–662. [Google Scholar] [CrossRef]
- Li, Y.; He, X.; Wang, Y.; Guan, J.; Guo, J.; Xu, B.; Chen, Y.H.; Wang, G. Organic fertilizer amendment increases methylmercury accumulation in rice plants. Chemosphere 2020, 249, 126166. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Zhang, W.; Sun, G.; Feng, Z.; Hurley, J.P.; Yang, L.; Shang, L.; Feng, X. Mercury risk in poultry in the Wanshan Mercury Mine, China. Environ. Pollut. 2017, 230, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Landrigan, P.J.; Sonawane, B.; Butler, R.N.; Trasande, L.; Callan, R.; Droller, D. Early environmental origins of neurodegenerative disease in later life. Environ. Health Perspect. 2005, 113, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- Bulka, C.M.; Persky, V.W.; Daviglus, M.L.; Durazo-Arvizu, R.A.; Argos, M. Multiple metal exposures and metabolic syndrome: A cross-sectional analysis of the National Health and Nutrition Examination Survey 2011–2014. Environ. Res. 2019, 168, 397–405. [Google Scholar] [CrossRef]
- Wang, X.; Mukherjee, B.; Park, S.K. Associations of cumulative exposure to heavy metal mixtures with obesity and its comorbidities among U.S. adults in NHANES 2003–2014. Environ. Int. 2018, 121, 683–694. [Google Scholar] [CrossRef]
- Lee, K. Blood mercury concentration in relation to metabolic and weight phenotypes using the KNHANES 2011–2013 data. Int. Arch. Occup. Environ. Health 2018, 91, 185–193. [Google Scholar] [CrossRef]
- Park, J.S.; Ha, K.H.; He, K.; Kim, D.J. Association between Blood Mercury Level and Visceral Adiposity in Adults. Diabetes Metab. J. 2017, 41, 113–120. [Google Scholar] [CrossRef]
- Park, K.; Seo, E. Association between Toenail Mercury and Metabolic Syndrome Is Modified by Selenium. Nutrients 2016, 8, 424. [Google Scholar] [CrossRef] [PubMed]
- Lacerda Leocadio, P.C.; Dias, R.P.; Pinto, D.V.; Reis, J.M.; Rodrigues Nascimento, J.C.; Anne de Castro Brito, G.; Valenca, J.T., Jr.; Foureaux, G.; Ferreira, A.J.; Windmoller, C.C.; et al. Pollutants and nutrition: Are methylmercury effects on blood pressure and lipoprotein profile comparable to high-fat diet in mice? Ecotoxicol. Environ. Saf. 2020, 204, 111036. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.L.; Leocadio, P.C.L.; Reis, J.M.; Campos, G.P.; Capettini, L.S.A.; Foureaux, G.; Ferreira, A.J.; Windmoller, C.C.; Santos, F.A.; Oria, R.B.; et al. Oral methylmercury intoxication aggravates cardiovascular risk factors and accelerates atherosclerosis lesion development in ApoE knockout and C57BL/6 mice. Toxicol. Res. 2021, 37, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Guaraldi, G.; Lonardo, A.; Maia, L.; Palella, F.J., Jr. Metabolic concerns in aging HIV-infected persons: From serum lipid phenotype to fatty liver. AIDS 2017, 31, S147–S156. [Google Scholar] [CrossRef] [PubMed]
- Alam, R.T.M.; Abu Zeid, E.H.; Khalifa, B.A.; Arisha, A.H.; Reda, R.M. Dietary exposure to methyl mercury chloride induces alterations in hematology, biochemical parameters, and mRNA expression of antioxidant enzymes and metallothionein in Nile tilapia. Environ. Sci. Pollut. Res. Int. 2021, 28, 31391–31402. [Google Scholar] [CrossRef] [PubMed]
- Dutta, H.M.; Haghighi, A.Z. Methylmercuric chloride and serum cholesterol level in the bluegill (Lepomis macrochirus). Bull. Environ. Contam. Toxicol. 1986, 36, 181–185. [Google Scholar] [CrossRef]
- Caito, S.W.; Newell-Caito, J.; Martell, M.; Crawford, N.; Aschner, M. Methylmercury Induces Metabolic Alterations in Caenorhabditis elegans: Role for C/EBP Transcription Factor. Toxicol. Sci. 2020, 174, 112–123. [Google Scholar] [CrossRef]
- Crawford, N.; Martell, M.; Nielsen, T.; Khalil, B.; Imtiaz, F.; Nguidjo, E.; Newell-Caito, J.L.; Bornhorst, J.; Schwerdtle, T.; Caito, S.W. Methylmercury-Induced Metabolic Alterations in Caenorhabditis elegans Are Diet-Dependent. Toxics 2021, 9, 287. [Google Scholar] [CrossRef]
- Nielsen, T.; Crawford, N.; Martell, M.; Khalil, B.; Imtiaz, F.; Newell-Caito, J.L.; Caito, S. MicroRNA Expression Influences Methylmercury-Induced Lipid Accumulation and Mitochondrial Toxicity in Caenorhabditis elegans. Chem. Res. Toxicol. 2022, 35, 77–88. [Google Scholar] [CrossRef]
- Iacomino, G.; Siani, A. Role of microRNAs in obesity and obesity-related diseases. Genes. Nutr. 2017, 12, 23. [Google Scholar] [CrossRef]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Regazzi, R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 2013, 9, 513–521. [Google Scholar] [CrossRef]
- Karolina, D.S.; Tavintharan, S.; Armugam, A.; Sepramaniam, S.; Pek, S.L.; Wong, M.T.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E2271-6. [Google Scholar] [CrossRef]
- Pan, C.H.; Chien, S.C.; Chen, C.J.; Shih, C.M.; Hsieh, M.H.; Huang, C.Y.; Bi, W.F.; Chan, C.S.; Kao, Y.T.; Hsiao, C.Y.; et al. Circulating level of microRNA-142-5p is a potential biomarker for predicting in-stent restenosis: A case-control study. BMC Cardiovasc. Disord. 2021, 21, 77. [Google Scholar] [CrossRef]
- Marsetti, P.S.; Milagro, F.I.; Zulet, M.A.; Martinez, J.A.; Lorente-Cebrian, S. Changes in miRNA expression with two weight-loss dietary strategies in a population with metabolic syndrome. Nutrition 2021, 83, 111085. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; McAnena, O.J.; O’Brien, T.; Kerin, M.J. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J. Clin. Endocrinol. Metab. 2011, 96, E846–E850. [Google Scholar] [CrossRef]
- Ibanez-Ventoso, C.; Vora, M.; Driscoll, M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 2008, 3, e2818. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Sun, T.; Fu, M.; Bookout, A.L.; Kliewer, S.A.; Mangelsdorf, D.J. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol. Endocrinol. 2009, 23, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Frost, R.J.; Olson, E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc. Natl. Acad. Sci. USA 2011, 108, 21075–21080. [Google Scholar] [CrossRef]
- Dowen, R.H.; Breen, P.C.; Tullius, T.; Conery, A.L.; Ruvkun, G. A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes Dev. 2016, 30, 1515–1528. [Google Scholar] [CrossRef]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- Stiernagle, T. Maintenance of C. elegans. In C. elegans: A. Practical. Approach; Hope, I.A., Ed.; Oxford University Press: New York, NY, USA, 1999. [Google Scholar]
- Caito, S.W.; Aschner, M. NAD+ Supplementation Attenuates Methylmercury Dopaminergic and Mitochondrial Toxicity in Caenorhabditis Elegans. Toxicol. Sci. 2016, 151, 139–149. [Google Scholar] [CrossRef]
- Martinez-Finley, E.J.; Chakraborty, S.; Slaughter, J.C.; Aschner, M. Early-life exposure to methylmercury in wildtype and pdr-1/parkin knockout C. elegans. Neurochem. Res. 2013, 38, 1543–1552. [Google Scholar] [CrossRef]
- Chomczynski, P.; Mackey, K. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 1995, 19, 942–945. [Google Scholar]
- Pino, E.C.; Webster, C.M.; Carr, C.E.; Soukas, A.A. Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J. Vis. Exp. 2013, 73, 50180. [Google Scholar]
- Zhi, L.; Yu, Y.; Jiang, Z.; Wang, D. mir-355 Functions as An Important Link between p38 MAPK Signaling and Insulin Signaling in the Regulation of Innate Immunity. Sci. Rep. 2017, 7, 14560. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; An, C.; Wu, X.; Yang, Y.; Xu, J.; Liu, Y.; Wang, C.; Nie, L.; Fang, H.; Yang, Z. MiR-34a regulates mitochondrial content and fat ectopic deposition induced by resistin through the AMPK/PPARalpha pathway in HepG2 cells. Int. J. Biochem. Cell Biol. 2018, 94, 133–145. [Google Scholar] [CrossRef]
- Fang, Q.H.; Shen, Q.L.; Li, J.J.; Yang, Y.; Guo, J.J.; Cheng, Y.; Zhou, H.C.; Niu, W.Y.; Chen, L.M.; Li, C.J.; et al. Inhibition of microRNA-124a attenuates non-alcoholic fatty liver disease through upregulation of adipose triglyceride lipase and the effect of liraglutide intervention. Hepatol. Res. 2019, 49, 743–757. [Google Scholar] [CrossRef]
- Belarbi, Y.; Mejhert, N.; Lorente-Cebrian, S.; Dahlman, I.; Arner, P.; Ryden, M.; Kulyte, A. MicroRNA-193b Controls Adiponectin Production in Human White Adipose Tissue. J. Clin. Endocrinol. Metab. 2015, 100, E1084-8. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xie, H.; Mori, M.A.; Alexander, R.; Yuan, B.; Hattangadi, S.M.; Liu, Q.; Kahn, C.R.; Lodish, H.F. Mir193b-365 is essential for brown fat differentiation. Nat. Cell Biol. 2011, 13, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.T.; Ashrafi, K. Caenorhabditis elegans as an emerging model for studying the basic biology of obesity. Dis. Models Mech. 2009, 2, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Rudgalvyte, M.; Peltonen, J.; Lakso, M.; Wong, G. Chronic MeHg exposure modifies the histone H3K4me3 epigenetic landscape in Caenorhabditis elegans. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2017, 191, 109–116. [Google Scholar] [CrossRef]
- Guida, N.; Valsecchi, V.; Laudati, G.; Serani, A.; Mascolo, L.; Molinaro, P.; Montuori, P.; Di Renzo, G.; Canzoniero, L.M.; Formisano, L. The miR206-JunD Circuit Mediates the Neurotoxic Effect of Methylmercury in Cortical Neurons. Toxicol. Sci. 2018, 163, 569–578. [Google Scholar] [CrossRef]
- Wang, X.; Yan, M.; Zhao, L.; Wu, Q.; Wu, C.; Chang, X.; Zhou, Z. Low-Dose Methylmercury-Induced Genes Regulate Mitochondrial Biogenesis via miR-25 in Immortalized Human Embryonic Neural Progenitor Cells. Int. J. Mol. Sci. 2016, 17, 2058. [Google Scholar] [CrossRef]
- Hu, H.; Shi, Y.; Zhang, Y.; Wu, J.; Asweto, C.O.; Feng, L.; Yang, X.; Duan, J.; Sun, Z. Comprehensive gene and microRNA expression profiling on cardiovascular system in zebrafish co-exposured of SiNPs and MeHg. Sci. Total. Environ. 2017, 607–608, 795–805. [Google Scholar] [CrossRef]
- Lee, J.; Padhye, A.; Sharma, A.; Song, G.; Miao, J.; Mo, Y.Y.; Wang, L.; Kemper, J.K. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 2010, 285, 12604–12611. [Google Scholar] [CrossRef]
- Cornejo, P.J.; Vergoni, B.; Ohanna, M.; Angot, B.; Gonzalez, T.; Jager, J.; Tanti, J.F.; Cormont, M. The Stress-Responsive microRNA-34a Alters Insulin Signaling and Actions in Adipocytes through Induction of the Tyrosine Phosphatase PTP1B. Cells 2022, 11, 2581. [Google Scholar] [CrossRef]
- Dumortier, O.; Hinault, C.; Van Obberghen, E. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 2013, 18, 312–324. [Google Scholar] [CrossRef]
- Fu, T.; Seok, S.; Choi, S.; Huang, Z.; Suino-Powell, K.; Xu, H.E.; Kemper, B.; Kemper, J.K. MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol. Cell Biol. 2014, 34, 4130–4142. [Google Scholar] [CrossRef] [PubMed]
- Qadir, A.S.; Woo, K.M.; Ryoo, H.M.; Baek, J.H. Insulin suppresses distal-less homeobox 5 expression through the up-regulation of microRNA-124 in 3T3-L1 cells. Exp. Cell Res. 2013, 319, 2125–2134. [Google Scholar] [CrossRef]
- Lovis, P.; Gattesco, S.; Regazzi, R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol. Chem. 2008, 389, 305–312. [Google Scholar] [CrossRef]
- Roy, D.; Modi, A.; Ghosh, R.; Ghosh, R.; Benito-Leon, J. Visceral Adipose Tissue Molecular Networks and Regulatory microRNA in Pediatric Obesity: An In Silico Approach. Int. J. Mol. Sci. 2022, 23, 11036. [Google Scholar] [CrossRef]
- Wang, G.; Zou, H.; Lai, C.; Huang, X.; Yao, Y.; Xiang, G. Repression of MicroRNA-124-3p Alleviates High-Fat Diet-Induced Hepatosteatosis by Targeting Pref-1. Front. Endocrinol. 2020, 11, 589994. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, J.; Liu, Q.; Xiong, X.; Zhang, Z.; Jiao, Y.; Li, X.; Liu, B.; Li, Y.; Lu, Y. MicroRNA-124 promotes hepatic triglyceride accumulation through targeting tribbles homolog 3. Sci. Rep. 2016, 6, 37170. [Google Scholar] [CrossRef] [PubMed]
- Chartoumpekis, D.V.; Zaravinos, A.; Ziros, P.G.; Iskrenova, R.P.; Psyrogiannis, A.I.; Kyriazopoulou, V.E.; Habeos, I.G. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE 2012, 7, e34872. [Google Scholar] [CrossRef]
- Kang, Z.; Zhang, S.; Jiang, E.; Wan, F.; Lan, X.; Liu, M. Mir-193b Regulates the Differentiation, Proliferation, and Apoptosis of Bovine Adipose Cells by Targeting the ACSS2/AKT Axis. Animals 2020, 10, 1265. [Google Scholar] [CrossRef]
- Simino, L.A.P.; Panzarin, C.; Fontana, M.F.; de Fante, T.; Geraldo, M.V.; Ignacio-Souza, L.M.; Milanski, M.; Torsoni, M.A.; Ross, M.G.; Desai, M.; et al. MicroRNA Let-7 targets AMPK and impairs hepatic lipid metabolism in offspring of maternal obese pregnancies. Sci. Rep. 2021, 11, 8980. [Google Scholar] [CrossRef]
- Sun, M.; Yamashita, T.; Shang, J.; Liu, N.; Deguchi, K.; Feng, J.; Abe, K. Time-dependent profiles of microRNA expression induced by ischemic preconditioning in the gerbil hippocampus. Cell Transplant. 2015, 24, 367–376. [Google Scholar] [CrossRef]
- Leavy, A.; Brennan, G.P.; Jimenez-Mateos, E.M. MicroRNA Profiling Shows a Time-Dependent Regulation within the First 2 Months Post-Birth and after Mild Neonatal Hypoxia in the Hippocampus from Mice. Biomedicines 2022, 10, 2740. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, S.B.; Ozturk, B.; Kocak, N.; Unlu, A. Differences of time-dependent microRNA expressions in breast cancer cells. Noncoding RNA Res. 2021, 6, 15–22. [Google Scholar] [CrossRef]
- Li, H.; Lin, L.; Chong, L.; Gu, S.; Wen, S.; Yu, G.; Hu, X.; Dong, L.; Zhang, H.; Li, C. Time-resolved mRNA and miRNA expression profiling reveals crucial coregulation of molecular pathways involved in epithelial-pneumococcal interactions. Immunol. Cell Biol. 2020, 98, 726–742. [Google Scholar] [CrossRef] [PubMed]
- Krishna, M.S.; Aneesh Kumar, A.; Abdul Jaleel, K.A. Time-dependent alterations in mRNA, protein and microRNA during in vitro adipogenesis. Mol. Cell Biochem. 2018, 448, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.M.; Goldstein, L.D.; Tevlin, M.; Tavare, S.; Shaham, S.; Miska, E.A. The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic Acids. Res. 2010, 38, 3780–3793. [Google Scholar] [CrossRef]
- Chen, J.S.; Gumbayan, A.M.; Zeller, R.W.; Mahaffy, J.M. An expanded Notch-Delta model exhibiting long-range patterning and incorporating MicroRNA regulation. PLoS. Comput. Biol. 2014, 10, e1003655. [Google Scholar] [CrossRef]
- Wulczyn, F.G.; Smirnova, L.; Rybak, A.; Brandt, C.; Kwidzinski, E.; Ninnemann, O.; Strehle, M.; Seiler, A.; Schumacher, S.; Nitsch, R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB. J. 2007, 21, 415–426. [Google Scholar] [CrossRef] [PubMed]
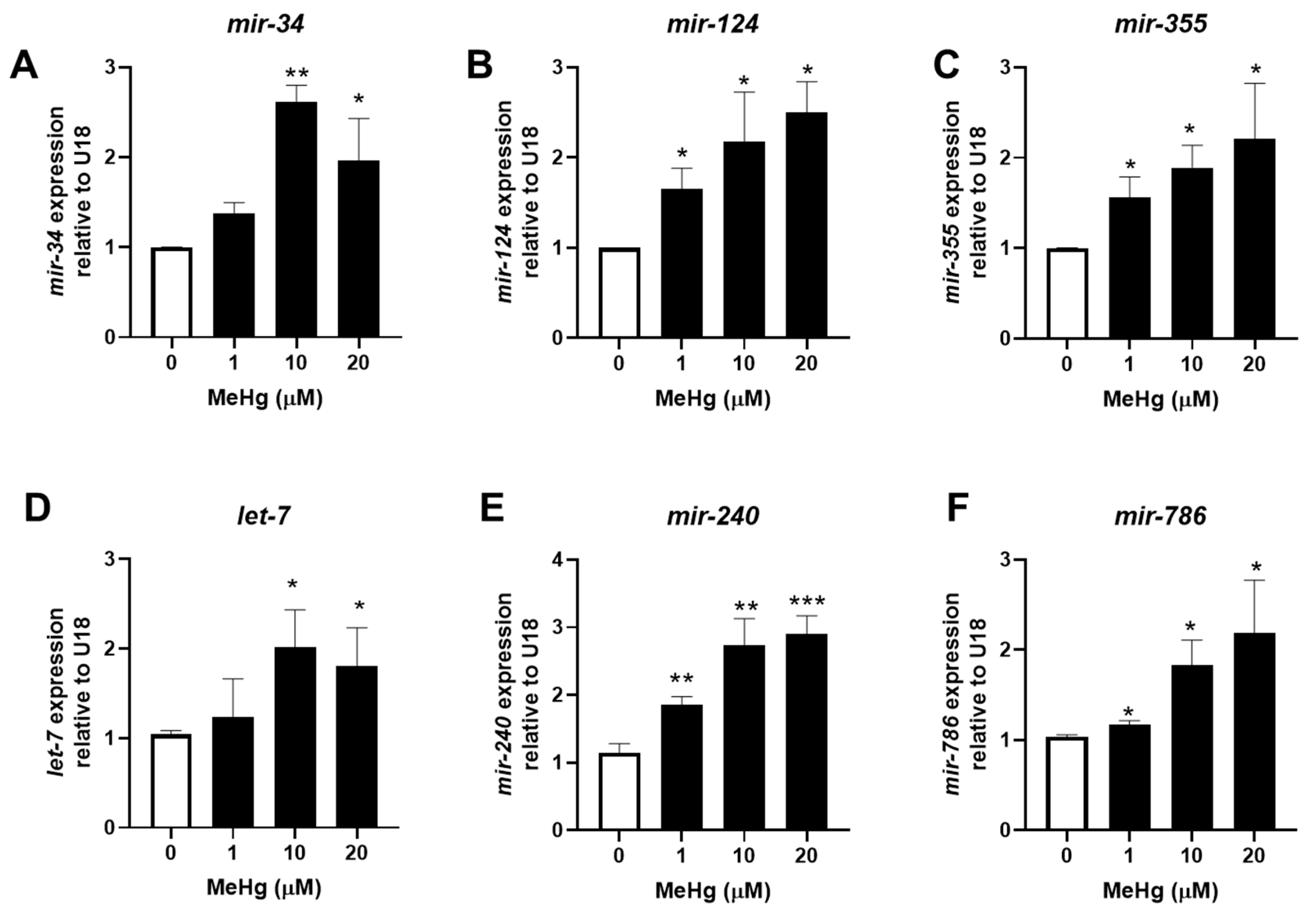

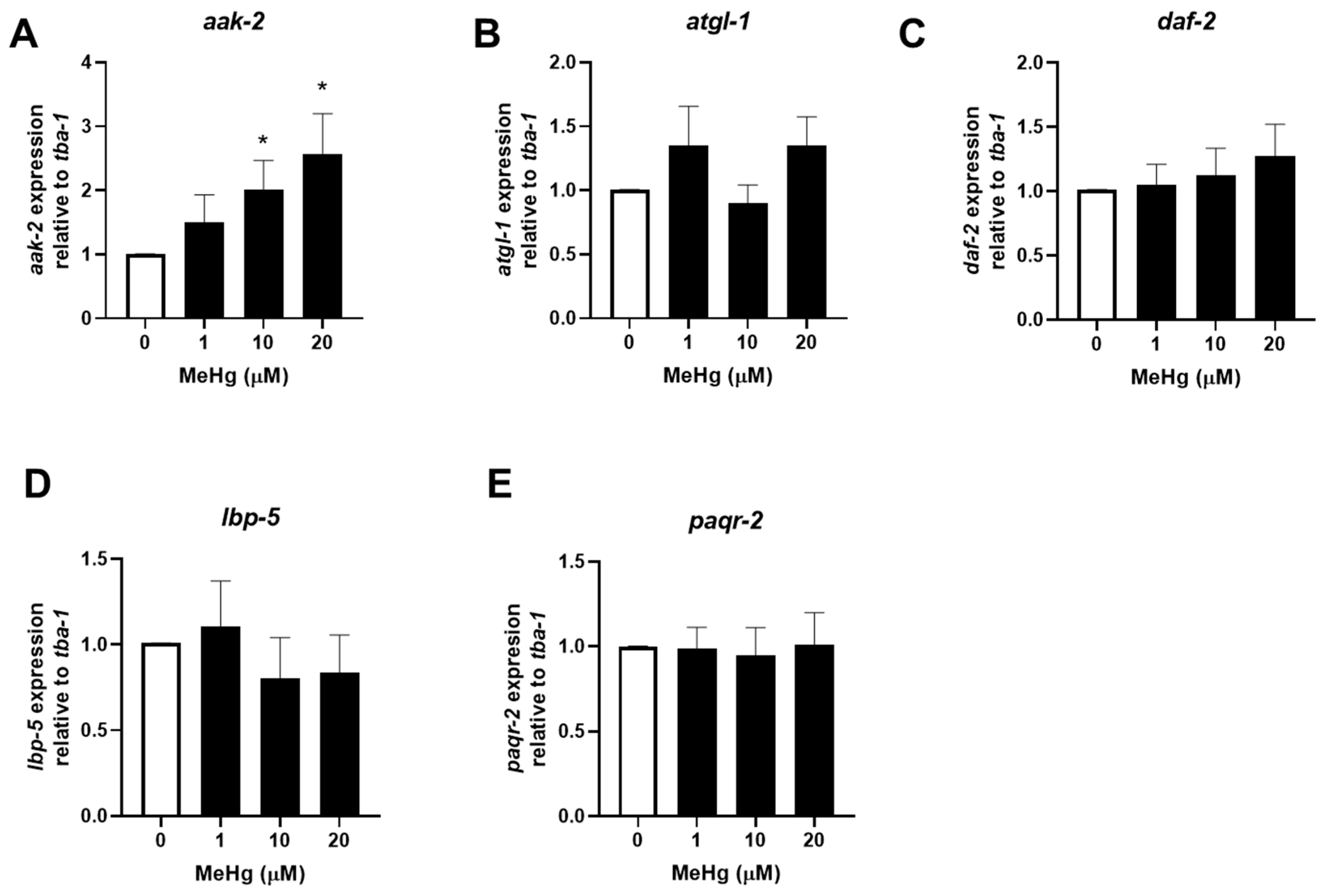


| miRNA | Effect | mRNA Target Gene (Mammalian Homolog) | References |
|---|---|---|---|
| mir-355 | Adipogenic | daf-2 (insulin-like growth factor receptor 1) | [39] |
| mir-34 | Adipogenic | aak-2 (AMP kinase) | [40] |
| mir-124 | Adipogenic | atgl-1 (adipose triglyceride lipase) | [41] |
| mir-240 | Anti-adipogenic | paqr-2 (adiponectin receptor) | [42] |
| mir-786 | Anti-adipogenic | paqr-2 | [43] |
| let-7 | Anti-adipogenic | lbp-5 (Fatty acid-binding protein 4) | [30] |
| Strain | LD50 (μM) | p-Value |
|---|---|---|
| N2 | 20.43 | |
| Mutant mir-34 | 2.53 | <0.0001 |
| Mutant mir-124 | 0.66 | <0.0001 |
| Mutant mir-355 | 1.43 | <0.0001 |
| Mutant let-7 | 3.95 | <0.0001 |
| Mutant mir-240 | 1.77 | <0.0001 |
| Mutant mir-240 and mir-786 | 1.66 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garofalo, G.; Nielsen, T.; Caito, S. Expression Profiling of Adipogenic and Anti-Adipogenic MicroRNA Sequences following Methylmercury Exposure in Caenorhabditis elegans. Toxics 2023, 11, 934. https://doi.org/10.3390/toxics11110934
Garofalo G, Nielsen T, Caito S. Expression Profiling of Adipogenic and Anti-Adipogenic MicroRNA Sequences following Methylmercury Exposure in Caenorhabditis elegans. Toxics. 2023; 11(11):934. https://doi.org/10.3390/toxics11110934
Chicago/Turabian StyleGarofalo, Giancarlo, Tyson Nielsen, and Samuel Caito. 2023. "Expression Profiling of Adipogenic and Anti-Adipogenic MicroRNA Sequences following Methylmercury Exposure in Caenorhabditis elegans" Toxics 11, no. 11: 934. https://doi.org/10.3390/toxics11110934
APA StyleGarofalo, G., Nielsen, T., & Caito, S. (2023). Expression Profiling of Adipogenic and Anti-Adipogenic MicroRNA Sequences following Methylmercury Exposure in Caenorhabditis elegans. Toxics, 11(11), 934. https://doi.org/10.3390/toxics11110934








