Daphnia magna Multigeneration Exposure to Carbendazim: Gene Transcription Responses
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Organism
2.2. Test Chemical
2.3. Multigenerational Experimental Setup
2.4. Gene Transcription Analysis
2.4.1. RNA Extraction
2.4.2. Gene Expression mRNA Microarrays
2.4.3. Microarray Data Extraction and Analysis
2.4.4. Microarray Data Submission
3. Results and Discussion
3.1. Chemical Analyses
3.2. Multigenerational Responses
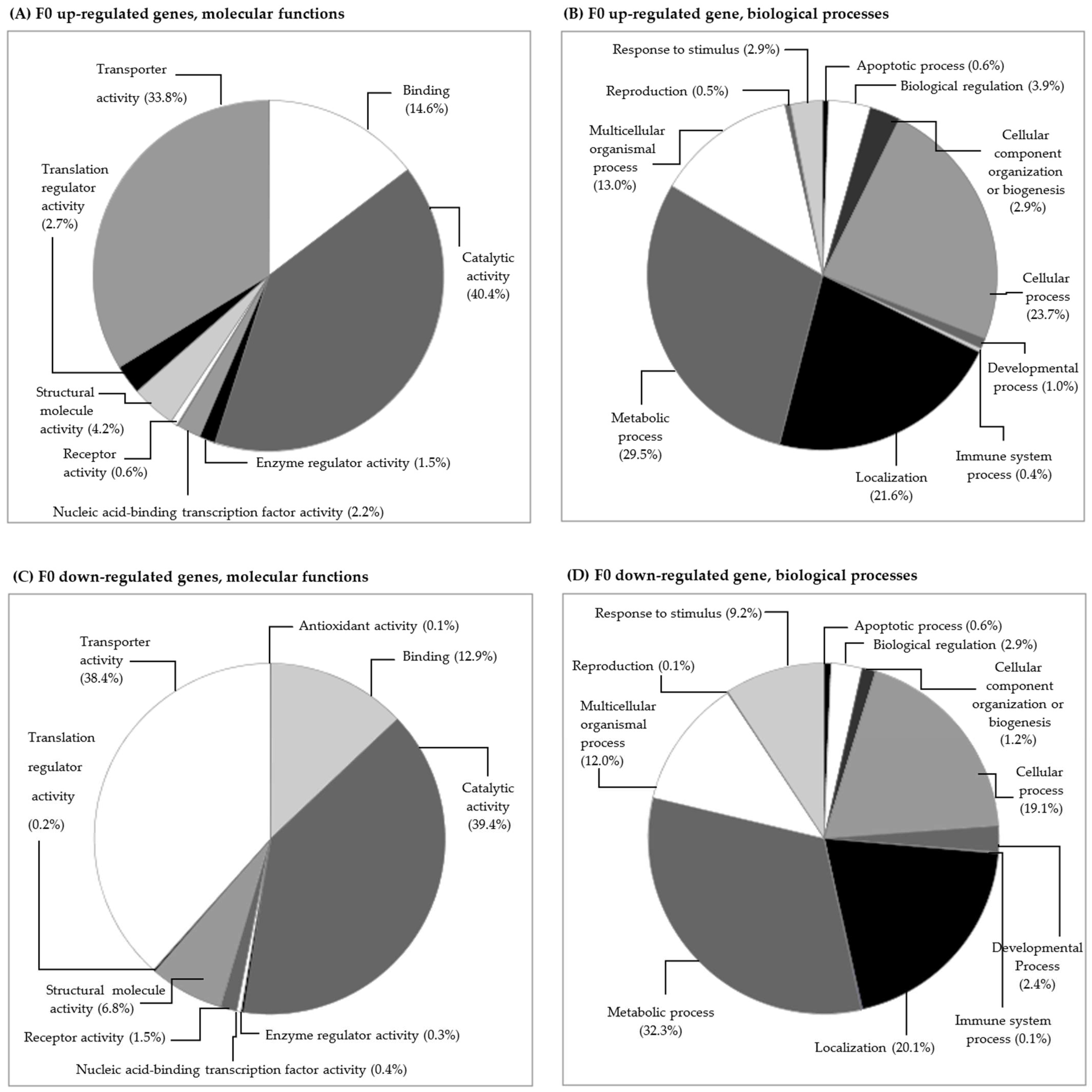
3.2.1. Gene Transcription in the F0 Generation (Clean Medium versus Carbendazim)
Gene Transcription and Its Relation to Different Subcellular Endpoints
Gene Transcription and Its Relation to Individual/Populational Endpoints
3.2.2. Gene Transcription in the F12 Generation (Clean Medium versus Carbendazim)
Gene Transcription and Its Relation to Different Subcellular Endpoints
Gene Transcription and Its Relation to Individual/Populational Endpoints
4. Conclusions
- (i)
- These transcriptional changes were mostly in genes involved in the response to stress, DNA replication/repair, neurotransmission, protein biosynthesis, ATP production, and lipid and carbohydrate metabolism.
- (ii)
- (iii)
- These changes were not maintained over time, since a lower number of differentially transcribed genes was observed after twelve generations of daphnids exposed to carbendazim, and the pathways were differentially affected in the F0 and F12 generations, possibly showing some acclimation.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campos, E.V.; Oliveira, J.L.; da Silva, C.M.; Pascoli, M.; Pasquoto, T.; Lima, R.; Abhilash, P.C.; Fernandes Fraceto, L. Polymeric and Solid Lipid Nanoparticles for Sustained Release of Carbendazim and Tebuconazole in Agricultural Applications. Sci. Rep. 2015, 5, 13809. [Google Scholar] [CrossRef] [PubMed]
- Davidse, L.G. Biochemical aspects of benzimidazole fungicides-action and resistance. In Modern Selective Fungicide-Properties, Applications, Mechanisms of Action; Lyr, H., Ed.; Longman: London, UK, 1987; Volume 24, pp. 43–65. [Google Scholar]
- Selmanoğlu, G.; Barlas, N.; Songür, S.; Koçkaya, E.A. Carbendazim-induced haematological, biochemical and histopathological changes to the liver and kidney of male rats. Hum. Exp. Toxicol. 2001, 20, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Chatupote, W.; Panapitukkul, N. Regional Assessment of Nutrient and Pesticide Leaching in the Vegetable Production Area of Rattaphum Catchment, Thailand. Water Air Soil Pollut. Focus 2005, 5, 165–173. [Google Scholar] [CrossRef]
- Masia, A.; Campo, J.; Vazquez-Roig, P.; Blasco, C.; Pico, Y. Screening of currently used pesticides in water, sediments and biota of the Guadalquivir River Basin (Spain). J. Hazard Mater. 2013, 263 Pt 1, 95–104. [Google Scholar] [CrossRef]
- Palma, G.; Sánchez, A.; Olave, Y.; Encina, F.; Palma, R.; Barra, R. Pesticide levels in surface waters in an agricultural–forestry basin in Southern Chile. Chemosphere 2004, 57, 763–770. [Google Scholar] [CrossRef]
- Acayaba, R.D.A.; de Albuquerque, A.F.; Ribessi, R.L.; Umbuzeiro, G.D.A.; Montagner, C.C. Occurrence of pesticides in waters from the largest sugar cane plantation region in the world. Environ. Sci. Pollut. Res. 2021, 28, 9824–9835. [Google Scholar] [CrossRef]
- Alygizakis, N.A.; Besselink, H.; Paulus, G.K.; Oswald, P.; Hornstra, L.M.; Oswaldova, M.; Medema, G.; Thomaidis, N.S.; Behnisch, P.A.; Slobodnik, J. Characterization of wastewater effluents in the Danube River Basin with chemical screening, in vitro bioassays and antibiotic resistant genes analysis. Environ. Int. 2019, 127, 420–429. [Google Scholar] [CrossRef]
- Casado, J.; Brigden, K.; Santillo, D.; Johnston, P. Screening of pesticides and veterinary drugs in small streams in the European Union by liquid chromatography high resolution mass spectrometry. Sci. Total Environ. 2019, 670, 1204–1225. [Google Scholar] [CrossRef]
- Kong, L.; Kadokami, K.; Duong, H.T.; Chau, H.T.C. Screening of 1300 organic micro-pollutants in groundwater from Beijing and Tianjin, North China. Chemosphere 2016, 165, 221–230. [Google Scholar] [CrossRef]
- Lopez-Pacheco, I.Y.; Silva-Nunez, A.; Salinas-Salazar, C.; Arevalo-Gallegos, A.; Lizarazo-Holguin, L.A.; Barcelo, D.; Iqbal, H.M.N.; Parra-Saldivar, R. Anthropogenic contaminants of high concern: Existence in water resources and their adverse effects. Sci. Total Environ. 2019, 690, 1068–1088. [Google Scholar] [CrossRef]
- Lundqvist, J.; von Brömssen, C.; Rosenmai, A.K.; Ohlsson, Å.; Le Godec, T.; Jonsson, O.; Kreuger, J.; Oskarsson, A. Assessment of pesticides in surface water samples from Swedish agricultural areas by integrated bioanalysis and chemical analysis. Environ. Sci. Eur. 2019, 31, 31–53. [Google Scholar] [CrossRef]
- Merel, S.; Benzing, S.; Gleiser, C.; Di Napoli-Davis, G.; Zwiener, C. Occurrence and overlooked sources of the biocide carbendazim in wastewater and surface water. Environ. Pollut. 2018, 239, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Smit, C.E.; Dang, Z.C. Environmental Risk Limits for Carbendazim; RIVM Letter Report; National Institute for Public Health and the Environment (RIVM): Bilthoven, The Netherlands, 2008; Volume 33. [Google Scholar]
- Zhou, T.; Guo, T.; Wang, Y.; Wang, A.; Zhang, M. Carbendazim: Ecological risks, toxicities, degradation pathways and potential risks to human health. Chemosphere 2023, 314, 137723. [Google Scholar] [CrossRef]
- EU Pesticide Database, EU Pesticide Database, European Comission. 2020. Available online: https://ec.europa.eu/food/plant/pesticides/eu-pesticides-database/start/screen/active-substances/details/506 (accessed on 5 September 2023).
- Test No. 211: Daphnia magna Reproduction Test; OECD Guidelines for Testing of Chemicals, Part II; Organization for Economic Cooperation and Development Publishing: Paris, France, 2012.
- Vandegehuchte, M.B.; De Coninck, D.; Vandenbrouck, T.; De Coen, W.M.; Janssen, C.R. Gene transcription profiles, global DNA methylation and potential transgenerational epigenetic effects related to Zn exposure history in Daphnia magna. Environ. Pollut. 2010, 158, 3323–3329. [Google Scholar] [CrossRef]
- Soetaert, A.; Moens, L.N.; Van der Ven, K.; Van Leemput, K.; Naudts, B.; Blust, R.; De Coen, W.M. Molecular impact of propiconazole on Daphnia magna using a reproduction-related cDNA array. Comparative biochemistry and physiology. Toxicol. Pharmacol. 2006, 142, 66–76. [Google Scholar]
- Vandenbrouck, T.; Dom, N.; Novais, S.; Soetaert, A.; Ferreira, A.L.; Loureiro, S.; Soares, A.M.; De Coen, W. Nickel response in function of temperature differences: Effects at different levels of biological organization in Daphnia magna. Comp. Biochem. Physiology. Part D Genom. Proteom. 2011, 6, 271–281. [Google Scholar] [CrossRef]
- Colbourne, J.K.; Pfrender, M.E.; Gilbert, D.; Thomas, W.K.; Tucker, A.; Oakley, T.H.; Tokishita, S.; Aerts, A.; Arnold, G.J.; Basu, M.K.; et al. The ecoresponsive genome of Daphnia pulex. Science 2011, 331, 555–561. [Google Scholar] [CrossRef]
- Silva, A.R.R.; Cardoso, D.N.; Cruz, A.; Lourenco, J.; Mendo, S.; Soares, A.M.V.M.; Loureiro, S. Ecotoxicity and genotoxicity of a binary combination of triclosan and carbendazim to Daphnia magna. Ecotoxicol. Environ. Saf. 2015, 115, 279–290. [Google Scholar] [CrossRef]
- Silva, A.R.R.; Cardoso, D.N.; Cruz, A.; Pestana, J.L.; Mendo, S.; Soares, A.M.V.M.; Loureiro, S. Multigenerational effects of carbendazim in Daphnia magna. Environ. Toxicol. Chem. 2017, 36, 383–394. [Google Scholar] [CrossRef]
- Silva, A.R.R.; Santos, C.; Ferreira, N.G.C.; Morgado, R.; Cardoso, D.N.; Cruz, A.; Mendo, S.; Soares, A.; Loureiro, S. Multigenerational effects of carbendazim in Daphnia magna: From a subcellular to a population level. Environ Toxicol Chem. 2019, 8, 412–422. [Google Scholar] [CrossRef]
- ASTM. Standard Practice for Conducting Acute Toxicity Tests with Fishes, Macroinvertebrates and Amphibians; Report E-729-80; American Standards for Testing and Materials: Philadelphia, PA, USA, 1980. [Google Scholar]
- Widianarko, B.; Van Straalen, N.M. Toxicokinetics-based survival analysis in bioassays using nonpersistent chemicals. Environ. Toxicol. Chem. 1996, 15, 402–406. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Wong, H.; Wang, Y.; Garza, M.; Weitman, S.D. Carbendazim: Disposition, cellular permeability, metabolite identification, and pharmacokinetic comparison with its nanoparticle. J. Pharm. Sci. 2003, 92, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Thiaré, D.D.; Khonté, A.; Diop, A.; Mendy, A.; Coly, A.; Delattre, F.; Tine, A. Spectrofluorimetric analysis of the fungicide carbendazim and its metabolite 2-aminobenzimidazole in natural water. Am. J. Anal. Chem. 2015, 6, 767–775. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Somero, G.N. Changes in gene expression associated with acclimation to constant temperatures and fluctuating daily temperatures in an annual killifish Austrofundulus limnaeus. J. Exp. Biol. 2004, 207, 2237–2254. [Google Scholar] [CrossRef]
- Vandegehuchte, M.B.; Vandenbrouck, T.; De Coninck, D.; De Coen, W.M.; Janssen, C.R. Can metal stress induce transferable changes in gene transcription in Daphnia magna? Aquat. Toxicol. 2010, 97, 188–195. [Google Scholar] [CrossRef]
- Soares, A.M.; Baird, D.J.; Calow, P. Interclonal variation in the performance of Daphnia magna Straus in chronic bioassays. Environ. Toxicol. Chem. 1992, 11, 1477–1483. [Google Scholar]
- JanakiDevi, V.; Nagarani, N.; YokeshBabu, M.; Kumaraguru, A.K.; Ramakritinan, C.M. A study of proteotoxicity and genotoxicity induced by the pesticide and fungicide on marine invertebrate (Donax faba). Chemosphere 2013, 90, 1158–1166. [Google Scholar] [CrossRef]
- Singh, P.; Srivastava, A.K.; Singh, A.K. Cell cycle stage specific application of cypermethrin and carbendazim to assess the genotoxicity in somatic cells of Hordeum vulgare L. Bull. Environ. Contam. Toxicol. 2008, 81, 258–261. [Google Scholar] [CrossRef]
- Al-Hakim, A.; Escribano-Diaz, C.; Landry, M.C.; O’Donnell, L.; Panier, S.; Szilard, R.K.; Durocher, D. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair 2010, 9, 1229–1240. [Google Scholar] [CrossRef]
- Yan, S.; Gao, S.; Zhou, P. Multi-functions of exonuclease 1 in DNA damage response and cancer susceptibility. Radiat. Med. Prot. 2021, 2, 146–154. [Google Scholar] [CrossRef]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Maheshwari, N.; Khan, F.H.; Mahmood, R. Carbendazim toxicity in different cell lines and mammalian tissues. J. Biochem. Mol. Toxicol. 2022, 36, e23194. [Google Scholar] [CrossRef] [PubMed]
- Pursell, Z.F.; Kunkel, T.A. DNA polymerase epsilon: A polymerase of unusual size (and complexity). Prog. Nucleic Acid Res. Mol. Biol. 2008, 82, 101–145. [Google Scholar] [PubMed]
- Novais, S.; de Coen, W.; Amorim, M.J.B. Gene Expression Responses Linked to Reproduction Effect Concentrations (EC10, 20, 50, 90) of Dimethoate, Atrazine and Carbendazim, in Enchytraeus albidus. PLoS ONE 2012, 7, e36068. [Google Scholar] [CrossRef]
- Rohde, M.; Daugaard, M.; Jensen, M.H.; Helin, K.; Nylandsted, J.; Jäättelä, M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005, 19, 570–582. [Google Scholar] [CrossRef]
- Morano, K.A. New tricks for an old dog: The evolving world of Hsp70. Ann. N. Y. Acad. Sci. 2007, 1113, 1–14. [Google Scholar] [CrossRef]
- Henson, K.L.; Stauffer, G.; Gallagher, E.P. Induction of glutathione S-transferase activity and protein expression in brown bullhead (Ameiurus nebulosus) liver by ethoxyquin. Toxicol. Sci. 2001, 62, 54–60. [Google Scholar] [CrossRef][Green Version]
- Hyne, R.V.; Maher, W.A. Invertebrate biomarkers: Links to toxicosis that predict population decline. Ecotox. Environ. Safe. 2003, 54, 366–374. [Google Scholar] [CrossRef]
- Livingstone, D.R. Oxidative stress in aquatic organisms in relation to pollution and aquaculture. Rev. De Médecine Vétérinaire 2003, 156, 427–430. [Google Scholar]
- Kittler, J.T.; Moss, S.J. Modulation of GABAA receptor activity by phosphorylation and receptor trafficking: Implications for the efficacy of synaptic inhibition. Curr. Opin. Neurobiol. 2003, 13, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Carhan, A.; Reeve, S.; Dee, C.T.; Baines, R.A.; Moffat, K.G. Mutation in slowmo causes defects in Drosophila larval locomotor behaviour. Invertebr. Neurosci. 2003, 5, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Fonnum, F. Glutamate: A neurotransmitter in mammalian brain. J. Neurochem. 1984, 42, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Goulden, C.E.; Henry, L.; Berrigan, D. Egg size, postembryonic yolk, and survival ability. Oecologia 1987, 72, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, F.; Ferreira, N.C.G.; Ferreira, A.; Soares, A.M.V.M.; Loureiro, S. Is ultraviolet radiation a synergistic stressor in combined exposures? The case study of Daphnia magna exposure to UV and carbendazim. Aquat. Toxicol. 2011, 102, 114–122. [Google Scholar] [CrossRef]
- Gu, Z.; Reynolds, E.M.; Song, J.; Lei, H.; Feijen, A.; Yu, L.; He, W.; MacLaughlin, D.T.; van den Eijnden-van Raaij, J.; Donahoe, P.K.; et al. The type I serine/threonine kinase receptor ActRIA (ALK2) is required for gastrulation of the mouse embryo. Development 1999, 126, 2551–2561. [Google Scholar] [CrossRef]
- Davidse, L.C. Benzimidazole fungicides: Mechanism of action and Biological impact. Annu. Rev. Phytopathol. 1986, 24, 43–65. [Google Scholar] [CrossRef]
- Takahashi, M.; Yang, X.J.; Lavery, T.T.; Furge, K.A.; Williams, B.O.; Tretiakova, M.; Montag, A.; Vogelzang, N.J.; Re, G.G.; Garvin, A.J.; et al. Gene Expression Profiling of Favorable Histology Wilms Tumors and Its Correlation with Clinical Features. Cancer Resarch 2002, 62, 6598–6605. [Google Scholar]
- Canton, J. The toxicity of benomyl, thiophanate-methyl, and BCM to four freshwater organisms. Bull. Environ. Contam. Toxicol. 1976, 16, 214–218. [Google Scholar] [CrossRef]
- Vandegehuchte, M.B.; Vandenbrouck, T.; De Coninck, D.; De Coen, W.M.; Janssen, C.R. Gene transcription and higher-level effects of multigenerational Zn exposure in Daphnia magna. Chemosphere 2010, 80, 1014–1020. [Google Scholar] [CrossRef]
- Takahata, T.; Hashikawa, T.; Tochitani, S.; Yamamori, T. Differential expression patterns of OCC1-related, extracellular matrix proteins in the lateral geniculate nucleus of macaque monkeys. J. Chem. Neuroanat. 2010, 40, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, T.; Tatematsu, C.; Nakanishi, Y. Double-stranded RNA-activated protein kinase interacts with apoptosis signal-regulating kinase 1. Eur. J. Biochem. 2002, 269, 6126–6132. [Google Scholar] [CrossRef]
- Chan, S.-L.; Tan, K.-O.; Zhang, L.; Yee, K.S.Y.; Ronca, F.; Chan, M.Y.; Yu, V.C. F1Aα, a death receptor-binding protein homologus to the Caenorhabditis elegans sex-determining protein, FEM-1, is a caspase substrate that mediates apoptosis. Biol. Chem. 1999, 274, 32461–32468. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, S.; Wu, C.; An, X.; Cai, L.; Zhao, X. Embryonic exposure to carbendazim induces the transcription of genes related to apoptosis, immunotoxicity and endocrine disruption in zebrafish (Danio rerio). Fish Shellfish. Immunol. 2014, 41, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J.; Harris, A.L.; Aherne, G.W. Mechanism of cell death following thymidylate synthase inhibition: 2′-deox-yuridine-5′-triphosphate accumulation, DNA damage, and growth inhibition following exposure to CB3717 and dipyridamole. Cancer Res. 1991, 51, 2346–2352. [Google Scholar] [PubMed]
- Caspary, T.; Garcia-Garcia, M.J.; Huangfu, D.; Eggenschwiler, J.T.; Wyler, M.R.; Rakeman, A.S.; Alcorn, H.L.; Anderson, K.V. Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Curr. Biol. 2002, 12, 1628–1632. [Google Scholar] [CrossRef]
- Gordon, S.; Akopyan, G.; Garban, H.; Bonavida, B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene 2006, 25, 1125–1142. [Google Scholar] [CrossRef]
- Gronroos, E.; Terentiev, A.A.; Punga, T.; Ericsson, J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. USA 2004, 101, 12165–12170. [Google Scholar] [CrossRef]
- Kannan, R.R.; Vincent, S.G.P. ELISA based quantification of Pax6 expression in the developing Zebrafish embryos. Ann. Neurosci. 2015, 22, 171–175. [Google Scholar] [CrossRef][Green Version]
- Tran, U.; Zakin, L.; Schweickert, A.; Agrawal, R.; Doger, R.; Blum, M.; De Robertis, E.M.; Wessely, O. The RNA-binding protein bicaudal C regulates polycystin 2 in the kidney by antagonizing miR-17 activity. Development 2010, 137, 1107–1116. [Google Scholar] [CrossRef]


| Gene ID | Gene Description [Species] | |
|---|---|---|
| Up-regulated genes | YP_548045 | hypothetical protein Bpro_1196 [Polaromonas sp.] |
| AAY54998 | IP06749p [Drosophila melanogaster] | |
| XP_001069615 | PREDICTED: similar to YY1 transcription factor [Rattus norvegicus] | |
| AAY66970 | secreted protein [Ixodes scapularis] | |
| XP_785823 | PREDICTED: similar to dispatched homolog 1 [Strongylocentrotus purpuratus] | |
| Down-regulated genes | XP_678020 | hypothetical protein [Plasmodium berghei] |
| CAE73165 | hypothetical protein [Caenorhabditis briggsae] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, A.R.R.; Silva, P.V.; Soares, A.R.; González-Alcaraz, M.N.; van Gestel, C.A.M.; Roelofs, D.; Moura, G.; Soares, A.M.V.M.; Loureiro, S. Daphnia magna Multigeneration Exposure to Carbendazim: Gene Transcription Responses. Toxics 2023, 11, 918. https://doi.org/10.3390/toxics11110918
Silva ARR, Silva PV, Soares AR, González-Alcaraz MN, van Gestel CAM, Roelofs D, Moura G, Soares AMVM, Loureiro S. Daphnia magna Multigeneration Exposure to Carbendazim: Gene Transcription Responses. Toxics. 2023; 11(11):918. https://doi.org/10.3390/toxics11110918
Chicago/Turabian StyleSilva, Ana Rita R., Patrícia V. Silva, Ana Raquel Soares, M. Nazaret González-Alcaraz, Cornelis A. M. van Gestel, Dick Roelofs, Gabriela Moura, Amadeu M. V. M. Soares, and Susana Loureiro. 2023. "Daphnia magna Multigeneration Exposure to Carbendazim: Gene Transcription Responses" Toxics 11, no. 11: 918. https://doi.org/10.3390/toxics11110918
APA StyleSilva, A. R. R., Silva, P. V., Soares, A. R., González-Alcaraz, M. N., van Gestel, C. A. M., Roelofs, D., Moura, G., Soares, A. M. V. M., & Loureiro, S. (2023). Daphnia magna Multigeneration Exposure to Carbendazim: Gene Transcription Responses. Toxics, 11(11), 918. https://doi.org/10.3390/toxics11110918








