In Vivo Investigation of the Effect of Dietary Acrylamide and Evaluation of Its Clinical Relevance in Colon Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vivo Experiment
2.2. Bioinformatics Analysis
2.3. Protein–Protein Network
2.4. TNMplot
2.5. The Cancer Genome Atlas (TCGA) Data Validation
2.6. Human Protein Atlas Database for Protein Expression
2.7. Kaplan–Meier Plotter Analysis
3. Results
3.1. Differential Gene Expression and Pathway Analysis in AA-Exposed Colon Tissue
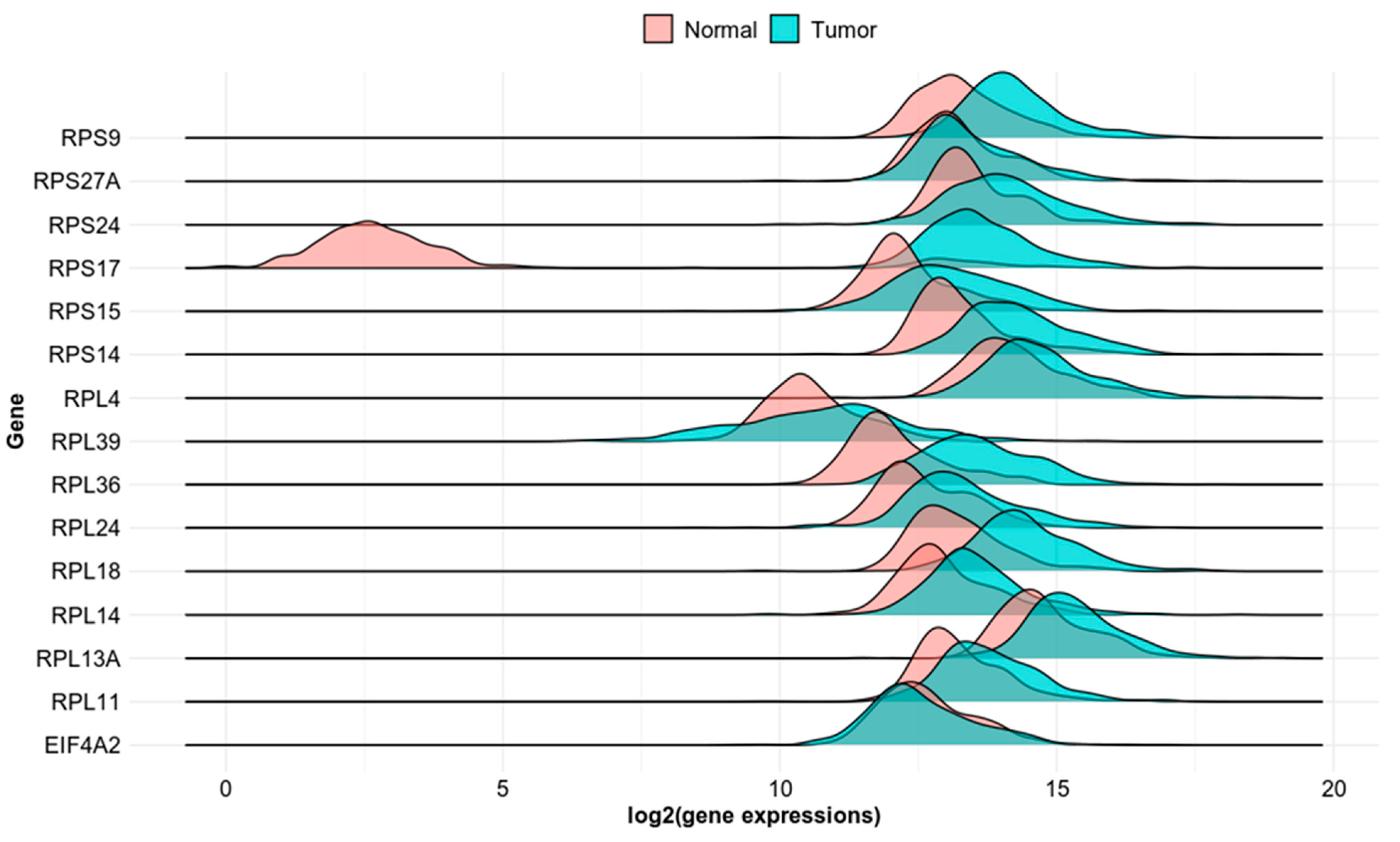
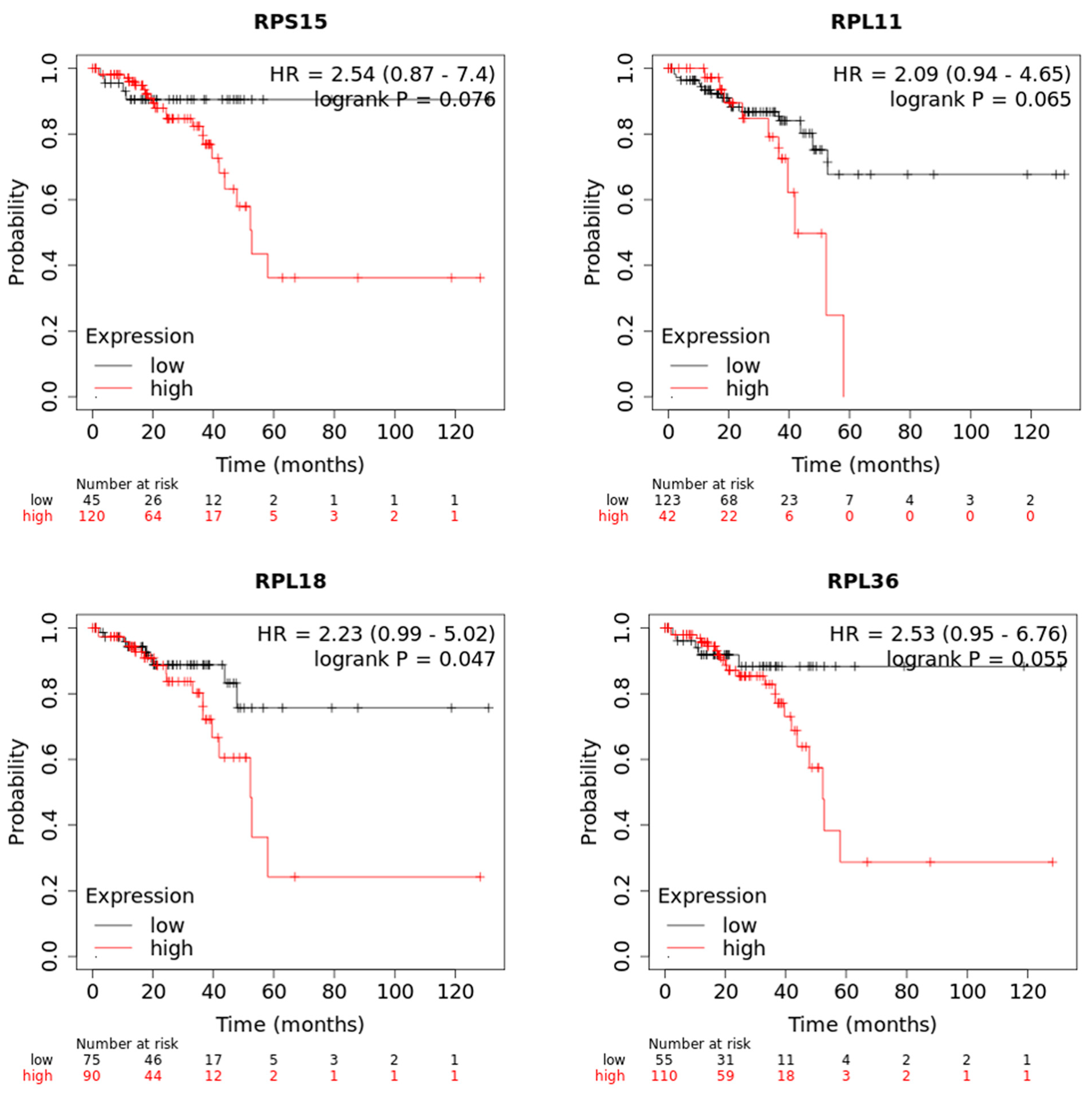
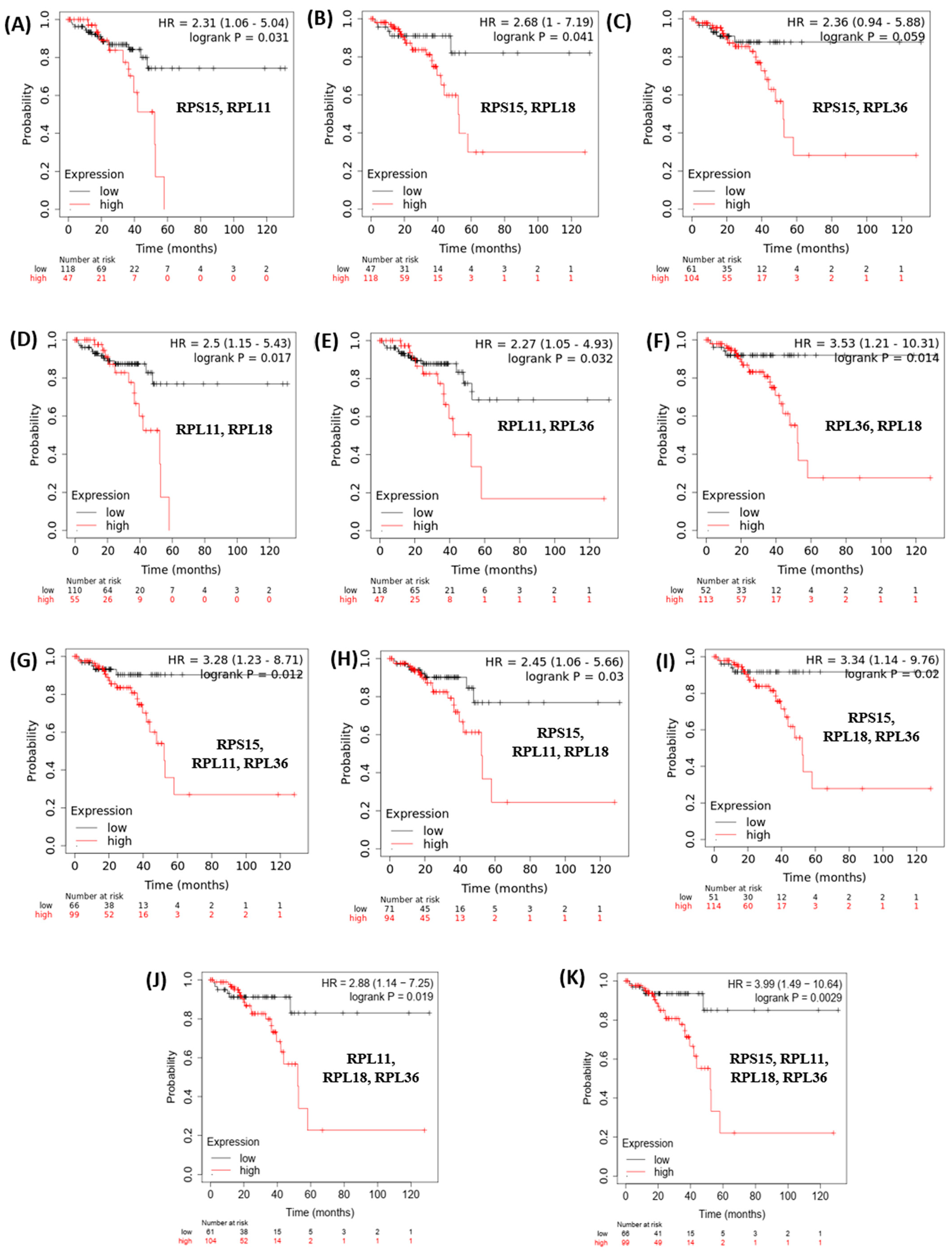
3.2. Evaluation of Clinical Relevance of Affected Genes Using Publicly Available Human Datasets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koszucka, A.; Nowak, A.; Nowak, I.; Motyl, I. Acrylamide in human diet, its metabolism, toxicity, inactivation and the associated European Union legal regulations in food industry. Crit. Rev. Food Sci. Nutr. 2020, 60, 1677–1692. [Google Scholar] [CrossRef]
- Exon, J.H. A review of the toxicology of acrylamide. J. Toxicol. Environ. Health B Crit. Rev. 2006, 9, 397–412. [Google Scholar] [CrossRef]
- Zamani, E.; Shokrzadeh, M.; Fallah, M.; Shaki, F. A review of acrylamide toxicity and its mechanism. Pharm. Biomed. Res. 2017, 3, 1–7. [Google Scholar] [CrossRef]
- Obon-Santacana, M.; Lujan-Barroso, L.; Travis, R.C.; Freisling, H.; Ferrari, P.; Severi, G.; Baglietto, L.; Boutron-Ruault, M.C.; Fortner, R.T.; Ose, J.; et al. Acrylamide and Glycidamide Hemoglobin Adducts and Epithelial Ovarian Cancer: A Nested Case-Control Study in Nonsmoking Postmenopausal Women from the EPIC Cohort. Cancer Epidemiol. Biomark. Prev. 2016, 25, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Rosen, J.; Hellenas, K.E. Analysis of acrylamide in cooked foods by liquid chromatography tandem mass spectrometry. Analyst 2002, 127, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Tareke, E.; Rydberg, P.; Karlsson, P.; Eriksson, S.; Tornqvist, M. Analysis of acrylamide, a carcinogen formed in heated foodstuffs. J. Agric. Food Chem. 2002, 50, 4998–5006. [Google Scholar] [CrossRef]
- Esposito, F.; Squillante, J.; Nolasco, A.; Montuori, P.; Macri, P.G.; Cirillo, T. Acrylamide levels in smoke from conventional cigarettes and heated tobacco products and exposure assessment in habitual smokers. Environ. Res. 2022, 208, 112659. [Google Scholar] [CrossRef] [PubMed]
- Swaen, G.M.; Haidar, S.; Burns, C.J.; Bodner, K.; Parsons, T.; Collins, J.J.; Baase, C. Mortality study update of acrylamide workers. Occup. Environ. Med. 2007, 64, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Marsh, G.M.; Youk, A.O.; Buchanich, J.M.; Kant, I.J.; Swaen, G. Mortality patterns among workers exposed to acrylamide: Updated follow up. J. Occup. Environ. Med. 2007, 49, 82–95. [Google Scholar] [CrossRef]
- Kadry, A.M.; Friedman, M.A.; Abdel-Rahman, M.S. Pharmacokinetics of acrylamide after oral administration in male rats. Environ. Toxicol. Pharmacol. 1999, 7, 127–133. [Google Scholar] [CrossRef]
- Friedman, M. Chemistry, biochemistry, and safety of acrylamide. A review. J. Agric. Food Chem. 2003, 51, 4504–4526. [Google Scholar] [CrossRef] [PubMed]
- Gamboa da Costa, G.; Churchwell, M.I.; Hamilton, L.P.; Von Tungeln, L.S.; Beland, F.A.; Marques, M.M.; Doerge, D.R. DNA adduct formation from acrylamide via conversion to glycidamide in adult and neonatal mice. Chem. Res. Toxicol. 2003, 16, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Besaratinia, A.; Pfeifer, G.P. A review of mechanisms of acrylamide carcinogenicity. Carcinogenesis 2007, 28, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, G.S.; Hogervorst, J.G.; Schouten, L.J.; Konings, E.J.; Goldbohm, R.A.; van den Brandt, P.A. Dietary acrylamide intake and estrogen and progesterone receptor-defined postmenopausal breast cancer risk. Breast Cancer Res. Treat. 2010, 122, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Hirvonen, T.; Kontto, J.; Jestoi, M.; Valsta, L.; Peltonen, K.; Pietinen, P.; Virtanen, S.M.; Sinkko, H.; Kronberg-Kippila, C.; Albanes, D.; et al. Dietary acrylamide intake and the risk of cancer among Finnish male smokers. Cancer Causes Control 2010, 21, 2223–2229. [Google Scholar] [CrossRef] [PubMed]
- Hogervorst, J.G.; Schouten, L.J.; Konings, E.J.; Goldbohm, R.A.; van den Brandt, P.A. Lung cancer risk in relation to dietary acrylamide intake. J. Natl. Cancer Inst. 2009, 101, 651–662. [Google Scholar] [CrossRef]
- Duda-Chodak, A.; Wajda, L.; Tarko, T.; Sroka, P.; Satora, P. A review of the interactions between acrylamide, microorganisms and food components. Food Funct. 2016, 7, 1282–1295. [Google Scholar] [CrossRef]
- Obon-Santacana, M.; Slimani, N.; Lujan-Barroso, L.; Travier, N.; Hallmans, G.; Freisling, H.; Ferrari, P.; Boutron-Ruault, M.C.; Racine, A.; Clavel, F.; et al. Dietary intake of acrylamide and pancreatic cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort. Ann. Oncol. 2013, 24, 2645–2651. [Google Scholar] [CrossRef]
- Hogervorst, J.G.; Schouten, L.J.; Konings, E.J.; Goldbohm, R.A.; van den Brandt, P.A. Dietary acrylamide intake is not associated with gastrointestinal cancer risk. J. Nutr. 2008, 138, 2229–2236. [Google Scholar] [CrossRef]
- Doerge, D.R.; Young, J.F.; McDaniel, L.P.; Twaddle, N.C.; Churchwell, M.I. Toxicokinetics of acrylamide and glycidamide in B6C3F1 mice. Toxicol. Appl. Pharmacol. 2005, 202, 258–267. [Google Scholar] [CrossRef]
- Doerge, D.R.; Young, J.F.; McDaniel, L.P.; Twaddle, N.C.; Churchwell, M.I. Toxicokinetics of acrylamide and glycidamide in Fischer 344 rats. Toxicol. Appl. Pharmacol. 2005, 208, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Galaxy, C. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res 2022, 50, W345–W351. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Gyorffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef]
- Nagy, A.; Munkacsy, G.; Gyorffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Liu, R.; Sobue, T.; Kitamura, T.; Kitamura, Y.; Ishihara, J.; Kotemori, A.; Zha, L.; Ikeda, S.; Sawada, N.; Iwasaki, M.; et al. Dietary Acrylamide Intake and Risk of Esophageal, Gastric, and Colorectal Cancer: The Japan Public Health Center-Based Prospective Study. Cancer Epidemiol Biomark. Prev 2019, 28, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Mucci, L.A.; Adami, H.O.; Wolk, A. Prospective study of dietary acrylamide and risk of colorectal cancer among women. Int. J. Cancer 2006, 118, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Larsson, S.C.; Akesson, A.; Bergkvist, L.; Wolk, A. Dietary acrylamide intake and risk of colorectal cancer in a prospective cohort of men. Eur. J. Cancer 2009, 45, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Hogervorst, J.G.; de Bruijn-Geraets, D.; Schouten, L.J.; van Engeland, M.; de Kok, T.M.; Goldbohm, R.A.; van den Brandt, P.A.; Weijenberg, M.P. Dietary acrylamide intake and the risk of colorectal cancer with specific mutations in KRAS and APC. Carcinogenesis 2014, 35, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Employment, Social Affairs & Inclusion. Recommendation from the Scientific Committee on Occupational Exposure Limits for Acrylamide; European Commission: Brussels, Belgium, 2012. [Google Scholar]
- Svensson, K.; Abramsson, L.; Becker, W.; Glynn, A.; Hellenas, K.E.; Lind, Y.; Rosen, J. Dietary intake of acrylamide in Sweden. Food Chem. Toxicol. 2003, 41, 1581–1586. [Google Scholar] [CrossRef]
- Joint FAO/WHO Expert Committee on Food Additives. Summary and Conclusions of the Seventy-Second Meeting, Expert Committee on Food Additives; WHO: Geneva, Switzerland, 2016; Available online: https://www.fao.org/3/bl839e/bl839e.pdf (accessed on 16 June 2023).
- Konings, E.J.; Baars, A.J.; van Klaveren, J.D.; Spanjer, M.C.; Rensen, P.M.; Hiemstra, M.; van Kooij, J.A.; Peters, P.W. Acrylamide exposure from foods of the Dutch population and an assessment of the consequent risks. Food Chem. Toxicol. 2003, 41, 1569–1579. [Google Scholar] [CrossRef]
- Basaran, B.; Cuvalci, B.; Kaban, G. Dietary Acrylamide Exposure and Cancer Risk: A Systematic Approach to Human Epidemiological Studies. Foods 2023, 12, 346. [Google Scholar] [CrossRef]
- Ehlers, A.; Lenze, D.; Broll, H.; Zagon, J.; Hummel, M.; Lampen, A. Dose dependent molecular effects of acrylamide and glycidamide in human cancer cell lines and human primary hepatocytes. Toxicol. Lett. 2013, 217, 111–120. [Google Scholar] [CrossRef]
- Bergmark, E.; Calleman, C.J.; Costa, L.G. Formation of hemoglobin adducts of acrylamide and its epoxide metabolite glycidamide in the rat. Toxicol. Appl. Pharmacol. 1991, 111, 352–363. [Google Scholar] [CrossRef]
- Nogueira, G.; Fernandes, R.; Garcia-Moreno, J.F.; Romao, L. Nonsense-mediated RNA decay and its bipolar function in cancer. Mol. Cancer 2021, 20, 72. [Google Scholar] [CrossRef]
- Bokhari, A.; Jonchere, V.; Lagrange, A.; Bertrand, R.; Svrcek, M.; Marisa, L.; Buhard, O.; Greene, M.; Demidova, A.; Jia, J.; et al. Targeting nonsense-mediated mRNA decay in colorectal cancers with microsatellite instability. Oncogenesis 2018, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Hardiman, G.; Savage, S.J.; Hazard, E.S.; da Silveira, W.A.; Morgan, R.; Harris, A.; Jefferson, M.S.; Wilson, R.C.; Caulder, S.; Ambrose, L.; et al. A Systems Approach to Interrogate Gene Expression Patterns in African American Men Presenting with Clinically Localized Prostate Cancer. Cancers 2021, 13, 5143. [Google Scholar] [CrossRef] [PubMed]
- Faoro, C.; Ataide, S.F. Noncanonical Functions and Cellular Dynamics of the Mammalian Signal Recognition Particle Components. Front. Mol. Biosci. 2021, 8, 679584. [Google Scholar] [CrossRef] [PubMed]
- Dolezal, J.M.; Dash, A.P.; Prochownik, E.V. Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer 2018, 18, 275. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, W.; Nasr, Z. Deregulation of ribosomal proteins in human cancers. Biosci. Rep. 2021, 41, BSR20211577. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.D.; Xu, J. Ribosomal proteins and colorectal cancer. Curr. Genom. 2007, 8, 43–49. [Google Scholar] [CrossRef]
- Shi, D.; Liu, J. RPS15a Silencing Suppresses Cell Proliferation and Migration of Gastric Cancer. Yonsei Med. J. 2018, 59, 1166–1173. [Google Scholar] [CrossRef]
- Liu, C.; He, X.; Liu, X.; Yu, J.; Zhang, M.; Yu, F.; Wang, Y. RPS15A promotes gastric cancer progression via activation of the Akt/IKK-beta/NF-kappaB signalling pathway. J. Cell. Mol. Med. 2019, 23, 2207–2218. [Google Scholar] [CrossRef]
- Chen, J.; Lei, C.; Zhang, H.; Huang, X.; Yang, Y.; Liu, J.; Jia, Y.; Shi, H.; Zhang, Y.; Zhang, J.; et al. RPL11 promotes non-small cell lung cancer cell proliferation by regulating endoplasmic reticulum stress and cell autophagy. BMC Mol. Cell Biol. 2023, 24, 7. [Google Scholar] [CrossRef]
- Kumar, K.U.; Srivastava, S.P.; Kaufman, R.J. Double-stranded RNA-activated protein kinase (PKR) is negatively regulated by 60S ribosomal subunit protein L18. Mol. Cell. Biol. 1999, 19, 1116–1125. [Google Scholar] [CrossRef]
- Hos, B.J.; Camps, M.G.M.; van den Bulk, J.; Tondini, E.; van den Ende, T.C.; Ruano, D.; Franken, K.; Janssen, G.M.C.; Ru, A.; Filippov, D.V.; et al. Identification of a neo-epitope dominating endogenous CD8 T cell responses to MC-38 colorectal cancer. Oncoimmunology 2019, 9, 1673125. [Google Scholar] [CrossRef] [PubMed]
- Song, M.J.; Jung, C.K.; Park, C.H.; Hur, W.; Choi, J.E.; Bae, S.H.; Choi, J.Y.; Choi, S.W.; Han, N.I.; Yoon, S.K. RPL36 as a prognostic marker in hepatocellular carcinoma. Pathol. Int. 2011, 61, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Zoncu, R.; Efeyan, A.; Sabatini, D.M. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011, 12, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Buxade, M.; Parra-Palau, J.L.; Proud, C.G. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front. Biosci. 2008, 13, 5359–5373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Yang, D.Q. Phosphorylation of eIF-4E positively regulates formation of the eIF-4F translation initiation complex following DNA damage. Biochem. Biophys. Res. Commun. 2008, 367, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, A.; Loiarro, M.; Bielli, P.; Busa, R.; Paronetto, M.P.; Loreni, F.; Geremia, R.; Sette, C. Phosphorylation of eIF4E by MNKs supports protein synthesis, cell cycle progression and proliferation in prostate cancer cells. Carcinogenesis 2008, 29, 2279–2288. [Google Scholar] [CrossRef]
- Konicek, B.W.; Stephens, J.R.; McNulty, A.M.; Robichaud, N.; Peery, R.B.; Dumstorf, C.A.; Dowless, M.S.; Iversen, P.W.; Parsons, S.; Ellis, K.E.; et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 2011, 71, 1849–1857. [Google Scholar] [CrossRef]
- Atala, A. Re: eIF4E Phosphorylation Promotes Tumorigenesis and is Associated With Prostate Cancer Progression. J. Urol. 2011, 185, 1533. [Google Scholar] [CrossRef]
- Ueda, T.; Sasaki, M.; Elia, A.J.; Chio, I.I.; Hamada, K.; Fukunaga, R.; Mak, T.W. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc. Natl. Acad. Sci. USA 2010, 107, 13984–13990. [Google Scholar] [CrossRef]
- Graff, J.R.; Konicek, B.W.; Lynch, R.L.; Dumstorf, C.A.; Dowless, M.S.; McNulty, A.M.; Parsons, S.H.; Brail, L.H.; Colligan, B.M.; Koop, J.W.; et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009, 69, 3866–3873. [Google Scholar] [CrossRef]
- Yoshizawa, A.; Fukuoka, J.; Shimizu, S.; Shilo, K.; Franks, T.J.; Hewitt, S.M.; Fujii, T.; Cordon-Cardo, C.; Jen, J.; Travis, W.D. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non-small cell lung cancer. Clin. Cancer Res. 2010, 16, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Lyu, S.; Lu, J.; Chen, W.; Huang, W.; Huang, H.; Xi, S.; Yan, S. High expression of eIF4A2 is associated with a poor prognosis in esophageal squamous cell carcinoma. Oncol. Lett. 2020, 20, 177. [Google Scholar] [CrossRef] [PubMed]
- Dave, B.; Granados-Principal, S.; Zhu, R.; Benz, S.; Rabizadeh, S.; Soon-Shiong, P.; Yu, K.D.; Shao, Z.; Li, X.; Gilcrease, M.; et al. Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 8838–8843. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sui, J.; Li, X.; Cao, F.; He, J.; Yang, B.; Zhu, X.; Sun, Y.; Pu, Y.D. RPS24 knockdown inhibits colorectal cancer cell migration and proliferation in vitro. Gene 2015, 571, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cai, Y.; Fu, X.; Chen, L. High RPS27A Expression Predicts Poor Prognosis in Patients With HPV Type 16 Cervical Cancer. Front. Oncol. 2021, 11, 752974. [Google Scholar] [CrossRef]
- Wang, H.; Xie, B.; Kong, Y.; Tao, Y.; Yang, G.; Gao, M.; Xu, H.; Zhan, F.; Shi, J.; Zhang, Y.; et al. Overexpression of RPS27a contributes to enhanced chemoresistance of CML cells to imatinib by the transactivated STAT3. Oncotarget 2016, 7, 18638–18650. [Google Scholar] [CrossRef]
- Knight, J.R.; Vlahov, N.; Gay, D.M.; Ridgway, R.A.; Faller, W.J.; Proud, C.; Mallucci, G.R.; von der Haar, T.; Smales, C.M.; Willis, A.E.; et al. Rpl24(Bst) mutation suppresses colorectal cancer by promoting eEF2 phosphorylation via eEF2K. Elife 2021, 10, e69729. [Google Scholar] [CrossRef]
- Wilson-Edell, K.A.; Kehasse, A.; Scott, G.K.; Yau, C.; Rothschild, D.E.; Schilling, B.; Gabriel, B.S.; Yevtushenko, M.A.; Hanson, I.M.; Held, J.M.; et al. RPL24: A potential therapeutic target whose depletion or acetylation inhibits polysome assembly and cancer cell growth. Oncotarget 2014, 5, 5165–5176. [Google Scholar] [CrossRef]
- Lin, Z.; Peng, R.; Sun, Y.; Zhang, L.; Zhang, Z. Identification of ribosomal protein family in triple-negative breast cancer by bioinformatics analysis. Biosci. Rep. 2021, 41, BSR20200869. [Google Scholar] [CrossRef]
- Bian, Z.; Yu, Y.; Quan, C.; Guan, R.; Jin, Y.; Wu, J.; Xu, L.; Chen, F.; Bai, J.; Sun, W.; et al. RPL13A as a reference gene for normalizing mRNA transcription of ovarian cancer cells with paclitaxel and 10-hydroxycamptothecin treatments. Mol. Med. Rep. 2015, 11, 3188–3194. [Google Scholar] [CrossRef]
- Meng, J.; Guan, Y.; Wang, B.; Chen, L.; Chen, J.; Zhang, M.; Liang, C. Risk subtyping and prognostic assessment of prostate cancer based on consensus genes. Commun. Biol. 2022, 5, 233. [Google Scholar] [CrossRef] [PubMed]
- Fahrmann, J.F.; Grapov, D.; Phinney, B.S.; Stroble, C.; DeFelice, B.C.; Rom, W.; Gandara, D.R.; Zhang, Y.; Fiehn, O.; Pass, H.; et al. Proteomic profiling of lung adenocarcinoma indicates heightened DNA repair, antioxidant mechanisms and identifies LASP1 as a potential negative predictor of survival. Clin. Proteom. 2016, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Hori, K.; Tanaka-Okamoto, M.; Higashiyama, M.; Itoh, Y.; Inoue, M.; Morinaka, S.; Miyoshi, J. Decreased expression of LMO7 and its clinicopathological significance in human lung adenocarcinoma. Exp. Ther. Med. 2011, 2, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.; Ou, Y.; Wang, G.; Chen, J.; Li, D.; Zhan, Q. miR-875-5p exerts tumor-promoting function via down-regulation of CAPZA1 in esophageal squamous cell carcinoma. PeerJ 2021, 9, e10020. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Xu, C.; Holck, A.L.; Liu, R. Acrylamide inhibits autophagy, induces apoptosis and alters cellular metabolic profiles. Ecotoxicol. Environ. Saf. 2021, 208, 111543. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, P.; Xu, C.; Shan, X.; Feng, Q. Acrylamide induces HepG2 cell proliferation through upregulation of miR-21 expression. J. Biomed. Res. 2019, 33, 181–191. [Google Scholar] [CrossRef]
- Chidawanyika, T.; Sergison, E.; Cole, M.; Mark, K.; Supattapone, S. SEC24A identified as an essential mediator of thapsigargin-induced cell death in a genome-wide CRISPR/Cas9 screen. Cell Death Discov. 2018, 4, 115. [Google Scholar] [CrossRef]
- Chiarini, A.; Liu, D.; Armato, U.; Dal Pra, I. Bcl10 crucially nucleates the pro-apoptotic complexes comprising PDK1, PKCzeta and caspase-3 at the nuclear envelope of etoposide-treated human cervical carcinoma C4-I cells. Int. J. Mol. Med. 2015, 36, 845–856. [Google Scholar] [CrossRef][Green Version]
- Fuhr, U.; Boettcher, M.I.; Kinzig-Schippers, M.; Weyer, A.; Jetter, A.; Lazar, A.; Taubert, D.; Tomalik-Scharte, D.; Pournara, P.; Jakob, V.; et al. Toxicokinetics of acrylamide in humans after ingestion of a defined dose in a test meal to improve risk assessment for acrylamide carcinogenicity. Cancer Epidemiol. Biomark. Prev. 2006, 15, 266–271. [Google Scholar] [CrossRef]
- Nixon, B.J.; Stanger, S.J.; Nixon, B.; Roman, S.D. Chronic exposure to acrylamide induces DNA damage in male germ cells of mice. Toxicol. Sci. 2012, 129, 135–145. [Google Scholar] [CrossRef]
- Lopachin, R.M.; Gavin, T. Acrylamide-induced nerve terminal damage: Relevance to neurotoxic and neurodegenerative mechanisms. J. Agric. Food Chem. 2008, 56, 5994–6003. [Google Scholar] [CrossRef] [PubMed]
- Katen, A.L.; Chambers, C.G.; Nixon, B.; Roman, S.D. Chronic Acrylamide Exposure in Male Mice Results in Elevated DNA Damage in the Germline and Heritable Induction of CYP2E1 in the Testes. Biol. Reprod. 2016, 95, 86. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]



| Gene | Functional Role 1 | Fold Change |
|---|---|---|
| Upregulated | ||
| Rps9 | Component of the 40S subunit | 2.6 |
| Rps14 | 2.4 | |
| Rps15 | 2.0 | |
| Rps17 | 3.0 | |
| Rps24 | 2.3 | |
| Rps27a | 2.8 | |
| Rpl4 | Component of the 60S subunit | 2.3 |
| Rpl11 | 3.7 | |
| Rpl13a | 2.9 | |
| Rpl14 | 2.5 | |
| Rpl18 | 2.7 | |
| Rpl24 | 2.2 | |
| Rpl36 | 2.2 | |
| Rpl39 | 2.1 | |
| Eif4a2 |
| 2.5 |
| Downregulated | ||
| Upf3a |
| 0.4 |
| Smg6 | Component of the telomerase ribonucleoprotein complex | 0.48 |
| Ddx23 | Translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly | 0.38 |
| Ppie | Accelerates the folding of proteins. | 0.46 |
| Sptb |
| 0.43 |
| St3gal3 |
| 0.47 |
| Gnai2 | Hormonal regulation of adenylate cyclase | 0.48 |
| Lmo7 | Signaling by ALK in cancer | 0.41 |
| Capza1 |
| 0.49 |
| Sec24a | Mediates protein transport from the endoplasmic reticulum | 0.49 |
| Hnrnpd | Influences pre-mRNA processing and other aspects of mRNA metabolism and transport | 0.40 |
| Furin | Processes protein and peptide precursors trafficking through regulated or constitutive branches of the secretory pathway | 0.42 |
| Bcl10 | Induces apoptosis and to activate NF-kappaB | 0.47 |
| Dcun1d5 |
| 0.31 |
| Spsb2 |
| 0.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neophytou, C.M.; Katsonouri, A.; Christodoulou, M.-I.; Papageorgis, P. In Vivo Investigation of the Effect of Dietary Acrylamide and Evaluation of Its Clinical Relevance in Colon Cancer. Toxics 2023, 11, 856. https://doi.org/10.3390/toxics11100856
Neophytou CM, Katsonouri A, Christodoulou M-I, Papageorgis P. In Vivo Investigation of the Effect of Dietary Acrylamide and Evaluation of Its Clinical Relevance in Colon Cancer. Toxics. 2023; 11(10):856. https://doi.org/10.3390/toxics11100856
Chicago/Turabian StyleNeophytou, Christiana M., Andromachi Katsonouri, Maria-Ioanna Christodoulou, and Panagiotis Papageorgis. 2023. "In Vivo Investigation of the Effect of Dietary Acrylamide and Evaluation of Its Clinical Relevance in Colon Cancer" Toxics 11, no. 10: 856. https://doi.org/10.3390/toxics11100856
APA StyleNeophytou, C. M., Katsonouri, A., Christodoulou, M.-I., & Papageorgis, P. (2023). In Vivo Investigation of the Effect of Dietary Acrylamide and Evaluation of Its Clinical Relevance in Colon Cancer. Toxics, 11(10), 856. https://doi.org/10.3390/toxics11100856










