Chemical Mixtures in Household Environments: In Silico Predictions and In Vitro Testing of Potential Joint Action on PPARγ in Human Liver Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Organizing Chemical Exposure Data from CPDat
2.2. Identifying Chemicals That Target PPARγ
2.3. Exposure Co-Occurrence Characterization of Environmental Chemicals
2.4. Selection of High-Interest Chemicals That Co-Occur as Mixtures for In Vitro Testing
2.5. Chemical Procurement for In Vitro Testing
2.6. Cell Culture and Treatment
2.7. Cytotoxicity Assay
2.8. PPARγ and INSR Gene Expression Screening
3. Results
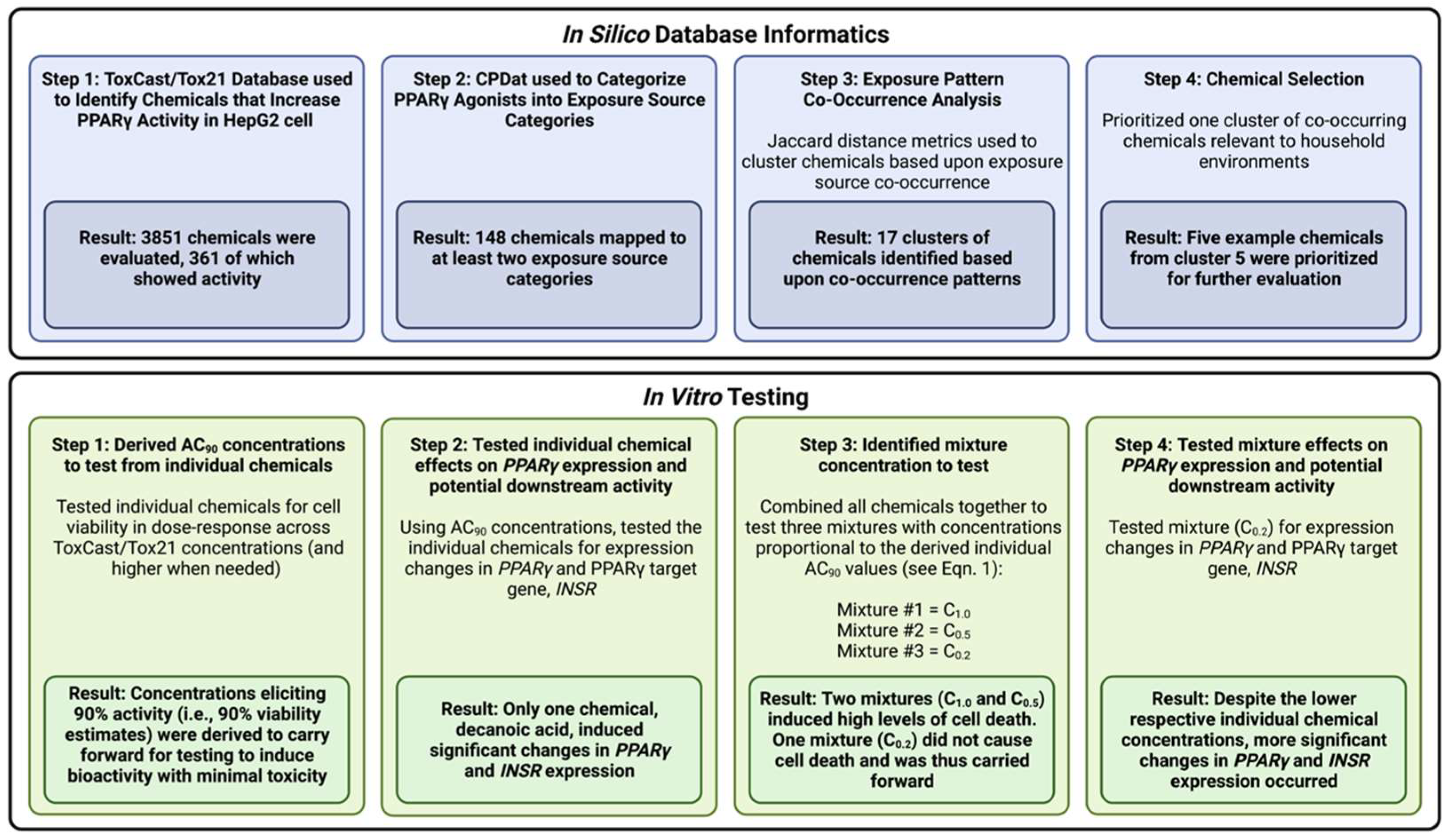
3.1. Study Overview
3.2. Identification of Chemicals That Increase PPARγ Activity in Human Liver Cells
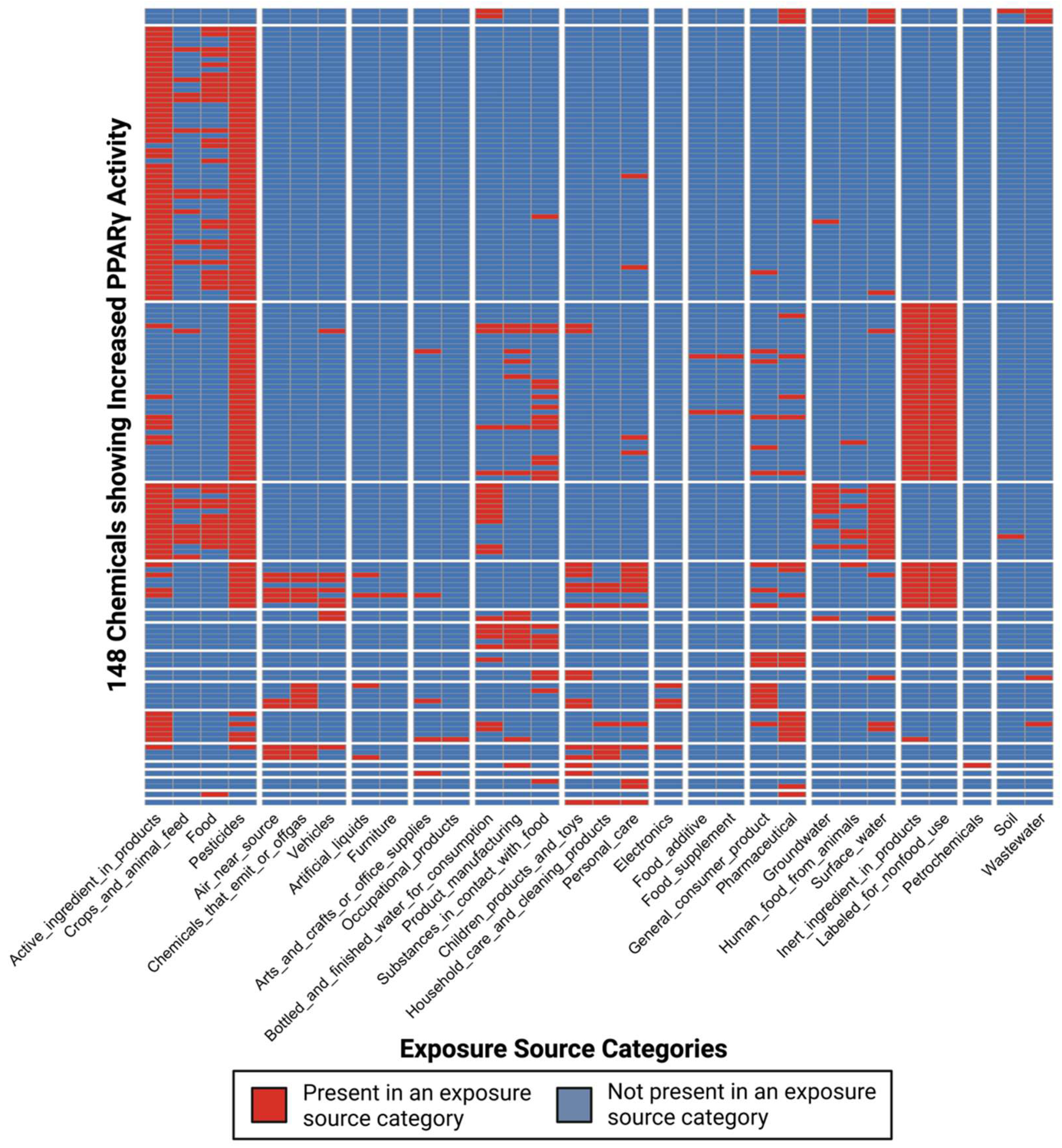
3.3. Dataset of Chemicals with Exposure Information and Evidence of PPARγ Activity Changes
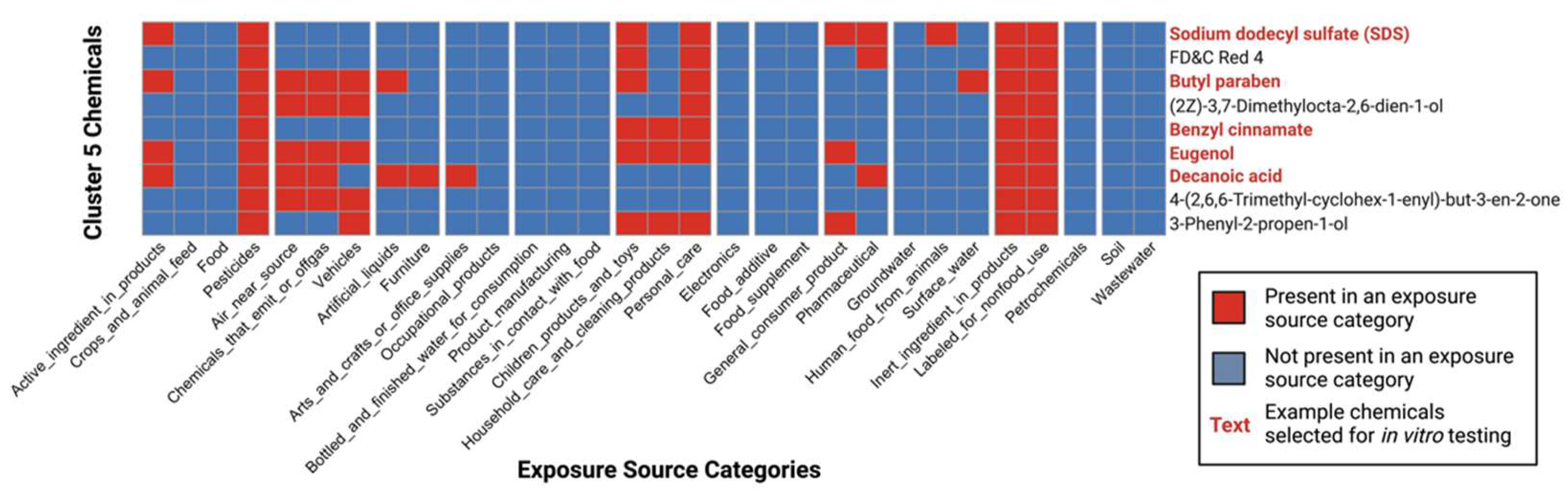
3.4. Characterization of Co-Occurring Chemicals in the Environment That Increase PPARγ Activity
3.5. Selection of High-Interest Household Chemicals That Co-Occur as Mixtures for In Vitro Testing
3.6. Cell Viability in Response to Treatment Conditions
3.7. PPARγ and INSR Expression Changes in Response to Individual Household Chemicals vs. Household Chemical Mixture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rager, J.E.; Strynar, M.; Liang, S.; McMahen, R.L.; Richard, A.M.; Grulke, C.; Wambaugh, J.; Isaacs, K.; Judson, R.; Williams, A.; et al. Linking high resolution mass spectrometry data with exposure and toxicity forecasts to advance high-throughput environmental monitoring. Environ. Int. 2016, 88, 269–280. [Google Scholar] [CrossRef]
- Hoffman, K.; Garantziotis, S.; Birnbaum, L.; Stapleton, H.M. Monitoring Indoor Exposure to Organophosphate Flame Retardants: Hand Wipes and House Dust. Environ. Health Perspect. 2015, 123, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Kannan, K. A Survey of Phthalates and Parabens in Personal Care Products from the United States and Its Implications for Human Exposure. Environ. Sci. Technol. 2013, 47, 14442–14449. [Google Scholar] [CrossRef] [PubMed]
- Rider, C.; Furr, J.R.; Wilson, V.; Gray, L.E., Jr. Cumulative effects of in utero administration of mixtures of reproductive toxicants that disrupt common target tissues via diverse mechanisms of toxicity. Int. J. Androl. 2010, 33, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Rider, C.V.; Carlin, D.J.; DeVito, M.J.; Thompson, C.L.; Walker, N.J. Mixtures research at NIEHS: An evolving program. Toxicology 2013, 313, 94–102. [Google Scholar] [CrossRef]
- Watt, J.; Webster, T.F.; Schlezinger, J.J. Generalized Concentration Addition Modeling Predicts Mixture Effects of Environmental PPARγ Agonists. Toxicol. Sci. 2016, 153, 18–27. [Google Scholar] [CrossRef][Green Version]
- Grygiel-Górniak, B. Peroxisome proliferator-activated receptors and their ligands: Nutritional and clinical implications—A review. Nutr. J. 2014, 13, 17. [Google Scholar] [CrossRef]
- Choi, J.-M.; Bothwell, A.L.M. The nuclear receptor PPARs as important regulators of T-cell functions and autoimmune diseases. Mol. Cells 2012, 33, 217–222. [Google Scholar] [CrossRef]
- Patel, H.; Truant, R.; Rachubinski, R.A.; Capone, J.P. Activity and subcellular compartmentalization of peroxisome proliferator-activated receptor α are altered by the centrosome-associated protein CAP350. J. Cell Sci. 2005, 118, 175–186. [Google Scholar] [CrossRef]
- Toxicology Testing in the 21st Century (Tox21)|Safer Chemicals Research|US EPA. Available online: https://www.epa.gov/chemical-research/toxicology-testing-21st-century-tox21 (accessed on 28 April 2021).
- NCBI. PPARG Peroxisome Proliferator Activated Receptor Gamma. 2021. Available online: https://www.ncbi.nlm.nih.gov/gene/5468 (accessed on 1 October 2021).
- Wang, Y.; Nakajima, T.; Gonzalez, F.J.; Tanaka, N. PPARs as Metabolic Regulators in the Liver: Lessons from Liver-Specific PPAR-Null Mice. Int. J. Mol. Sci. 2020, 21, 2061. [Google Scholar] [CrossRef]
- Ahmed, M.; Lai, T.H.; Hwang, J.S.; Zada, S.; Pham, T.M.; Kim, D.R. Transcriptional Regulation of Autophagy Genes via Stage-Specific Activation of CEBPB and PPARG during Adipogenesis: A Systematic Study Using Public Gene Expression and Transcription Factor Binding Datasets. Cells 2019, 8, 1321. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Kim, H.; Park, J.-M.; Im, S.-S.; Bae, J.-S.; Kim, M.-Y.; Yoon, H.-G.Y.; Cha, J.-Y.; Kim, K.-S.; Ahn, Y.H. Interrelationship between Liver X Receptor α, Sterol Regulatory Element-binding Protein-1c, Peroxisome Proliferator-activated Receptor γ, and Small Heterodimer Partner in the Transcriptional Regulation of Glucokinase Gene Expression in Liver. J. Biol. Chem. 2009, 284, 15071–15083. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Foti, D.; Paonessa, F.; Chiefari, E.; Palaia, L.; Brunetti, G.; Gulletta, E.; Fusco, A.; Brunetti, A. The insulin receptor: A new anticancer target for peroxisome proliferator-activated receptor-g (PPARg) and thiazolidinedione-PPARg agonists. Endocr.-Relat. Cancer 2008, 15, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Selva, D.M.; Hammond, G. Peroxisome-Proliferator Receptor γ Represses Hepatic Sex Hormone-Binding Globulin Expression. Endocrinology 2009, 150, 2183–2189. [Google Scholar] [CrossRef]
- Chi, C.-W.; Hsu, H.-T. Emerging role of the peroxisome proliferator-activated receptor-gamma in hepatocellular carcinoma. J. Hepatocell. Carcinoma 2014, 1, 127–135. [Google Scholar] [CrossRef]
- Di Marzio, D. Peroxisome proliferator-activated receptor-γ agonists and diabetes: Current evidence and future perspectives. Vasc. Health Risk Manag. 2008, 4, 297–304. [Google Scholar] [CrossRef]
- Motawi, T.K.; Shaker, O.G.; Ismail, M.F.; Sayed, N.H. Peroxisome Proliferator-Activated Receptor Gamma in Obesity and Colorectal Cancer: The Role of Epigenetics. Sci. Rep. 2017, 7, 10714. [Google Scholar] [CrossRef]
- González-Castro, T.B.; Tovilla-Zárate, C.A.; Juárez-Rojop, I.E.; Hernández-Díaz, Y.; López-Narváez, M.L.; Rodríguez-Pérez, C.; González-Hernández, Y.K.; Ramos-Méndez, M. Ángel PON2 and PPARG polymorphisms as biomarkers of risk for coronary heart disease. Biomark. Med. 2018, 12, 287–297. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Luo, S.; Zhan, Y.; Lu, Q. The roles of PPARγ and its agonists in autoimmune diseases: A comprehensive review. J. Autoimmun. 2020, 113, 102510. [Google Scholar] [CrossRef]
- Grbić, E.; Peterlin, A.; Kunej, T.; Petrovič, D. PPARγ gene and atherosclerosis: Genetic polymorphisms, epigenetics and therapeutic implications. Balk. J. Med. Genet. 2018, 21, 39–46. [Google Scholar] [CrossRef]
- Dionisio, K.L.; Phillips, K.; Price, P.S.; Grulke, C.M.; Williams, A.; Biryol, D.; Hong, T.; Isaacs, K.K. The Chemical and Products Database, a resource for exposure-relevant data on chemicals in consumer products. Sci. Data 2018, 5, 180125. [Google Scholar] [CrossRef] [PubMed]
- EPA. The Chemical and Products Database (CPDat) MySQL Data File. 2020. Available online: https://epa.figshare.com/articles/dataset/The_Chemical_and_Products_Database_CPDat_MySQL_Data_File/5352997 (accessed on 23 September 2020).
- EPA. Exploring ToxCast Data: Downloadable Data. 2021. Available online: https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data (accessed on 1 June 2021).
- Fang, L.; Zhang, M.; Li, Y.; Liu, Y.; Cui, Q.; Wang, N. PPARgene: A Database of Experimentally Verified and Computationally Predicted PPAR Target Genes. PPAR Res. 2016, 2016, 6042162. [Google Scholar] [CrossRef] [PubMed]
- Judson, R.; Houck, K.; Martin, M.; Richard, A.M.; Knudsen, T.B.; Shah, I.; Little, S.; Wambaugh, J.; Setzer, R.W.; Kothiya, P.; et al. Analysis of the Effects of Cell Stress and Cytotoxicity onIn Vitro Assay Activity Across a Diverse Chemical and Assay Space. Toxicol. Sci. 2016, 153, 409. [Google Scholar] [CrossRef]
- Judson, R.S.; Magpantay, F.M.; Chickarmane, V.; Haskell, C.; Tania, N.; Taylor, J.; Xia, M.; Huang, R.; Rotroff, D.; Filer, D.L.; et al. Integrated Model of Chemical Perturbations of a Biological Pathway Using 18In VitroHigh-Throughput Screening Assays for the Estrogen Receptor. Toxicol. Sci. 2015, 148, 137–154. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, S.; Filer, D.; Reif, D.; Walker, V.; Holloway, A.C.; Schlezinger, J.; Srinivasan, S.; Svoboda, D.; Judson, R.; Bucher, J.R.; et al. Prioritizing Environmental Chemicals for Obesity and Diabetes Outcomes Research: A Screening Approach Using ToxCast™ High-Throughput Data. Environ. Health Perspect. 2016, 124, 1141–1154. [Google Scholar] [CrossRef]
- Rager, J.E.; Ring, C.L.; Fry, R.C.; Suh, M.; Proctor, D.M.; Haws, L.C.; Harris, M.A.; Thompson, C.M. High-Throughput Screening Data Interpretation in the Context of In Vivo Transcriptomic Responses to Oral Cr(VI) Exposure. Toxicol. Sci. 2017, 158, 199–212. [Google Scholar] [CrossRef]
- Ring, C.; Sipes, N.S.; Hsieh, J.-H.; Carberry, C.; Koval, L.E.; Klaren, W.D.; Harris, M.A.; Auerbach, S.S.; Rager, J.E. Predictive modeling of biological responses in the rat liver using in vitro Tox21 bioactivity: Benefits from high-throughput toxicokinetics. Comput. Toxicol. 2021, 18, 100166. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V.; Xiang, H.; Holliday, J.; Buscema, P.M.; Willett, P. Similarity Coefficients for Binary Chemoinformatics Data: Overview and Extended Comparison Using Simulated and Real Data Sets. J. Chem. Inf. Model. 2012, 52, 2884–2901. [Google Scholar] [CrossRef]
- Bajusz, D.; Rácz, A.; Héberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Chemin. 2015, 7, 20. [Google Scholar] [CrossRef]
- Helman, G.; Shah, I.; Williams, A.J.; Edwards, J.; Dunne, J.; Patlewicz, G. Generalized Read-Across (GenRA): A workflow implemented into the EPA CompTox Chemicals Dashboard. ALTEX 2019, 36, 462–465. [Google Scholar] [CrossRef]
- Davis, A.P.; Murphy, C.G.; Saraceni-Richards, C.A.; Rosenstein, M.C.; Wiegers, T.C.; Hampton, T.H.; Mattingly, C.J. GeneComps and ChemComps: A new CTD metric to identify genes and chemicals with shared toxicogenomic profiles. Bioinformation 2009, 4, 173–174. [Google Scholar] [CrossRef] [PubMed]
- Klaren, W.D.; Ring, C.; Harris, M.A.; Thompson, C.M.; Borghoff, S.; Sipes, N.S.; Hsieh, J.-H.; Auerbach, S.S.; Rager, J.E. Identifying Attributes That InfluenceIn Vitro-to-In VivoConcordance by ComparingIn VitroTox21 Bioactivity VersusIn VivoDrugMatrix Transcriptomic Responses Across 130 Chemicals. Toxicol. Sci. 2018, 167, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Rager, J.E.; Clark, J.; Eaves, L.A.; Avula, V.; Niehoff, N.M.; Kim, Y.H.; Jaspers, I.; Gilmour, M.I. Mixtures modeling identifies chemical inducers versus repressors of toxicity associated with wildfire smoke. Sci. Total Environ. 2021, 775, 145759. [Google Scholar] [CrossRef]
- Leydesdorff, L. On the normalization and visualization of author co-citation data: Salton’s Cosine versus the Jaccard index. J. Am. Soc. Inf. Sci. Technol. 2007, 59, 77–85. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ryan, K.R.; Huang, M.C.; Ferguson, S.S.; Waidyanatha, S.; Ramaiahgari, S.; Rice, J.R.; Dunlap, P.E.; Auerbach, S.S.; Mutlu, E.; Cristy, T.; et al. Evaluating Sufficient Similarity of Botanical Dietary Supplements: Combining Chemical and In Vitro Biological Data. Toxicol. Sci. 2019, 172, 316–329. [Google Scholar] [CrossRef]
- Collins, B.J.; Kerns, S.P.; Aillon, K.; Mueller, G.; Rider, C.V.; Derose, E.F.; London, R.E.; Harnly, J.M.; Waidyanatha, S. Comparison of phytochemical composition of Ginkgo biloba extracts using a combination of non-targeted and targeted analytical approaches. Anal. Bioanal. Chem. 2020, 412, 6789–6809. [Google Scholar] [CrossRef]
- Kapraun, D.F.; Wambaugh, J.; Ring, C.; Tornero-Velez, R.; Setzer, R.W. A Method for Identifying Prevalent Chemical Combinations in the U.S. Population. Environ. Health Perspect. 2017, 125, 087017. [Google Scholar] [CrossRef]
- Green, A.J.; Mohlenkamp, M.J.; Das, J.; Chaudhari, M.; Truong, L.; Tanguay, R.L.; Reif, D.M. Leveraging high-throughput screening data, deep neural networks, and conditional generative adversarial networks to advance predictive toxicology. PLoS Comput. Biol. 2021, 17, e1009135. [Google Scholar] [CrossRef]
- Wambaugh, J.F.; Wang, A.; Dionisio, K.L.; Frame, A.; Egeghy, P.; Judson, R.; Setzer, R.W. High Throughput Heuristics for Prioritizing Human Exposure to Environmental Chemicals. Environ. Sci. Technol. 2014, 48, 12760–12767. [Google Scholar] [CrossRef]
- Baker, N.; Knudsen, T.; Williams, A.J. Abstract Sifter: A comprehensive front-end system to PubMed. F1000Research 2017, 6, 2164. [Google Scholar] [CrossRef] [PubMed]
- EPA. Benzyl Cinnamate. 2021. Available online: https://comptox.epa.gov/dashboard/chemical/details/DTXSID3041663 (accessed on 10 December 2021).
- EPA. 4-Hydroxybenzoic Acid Butyl Ester. 94-26-8 | DTXSID3020209. 2021. Available online: https://comptox.epa.gov/dashboard/chemical/details/DTXSID3020209 (accessed on 15 December 2021).
- EPA. Decanoic Acid. 334-48-5 | DTXSID9021554. 2021. Available online: https://comptox.epa.gov/dashboard/chemical/details/DTXSID9021554 (accessed on 15 December 2021).
- Api, A.; Belsito, D.; Biserta, S.; Botelho, D.; Bruze, M.; Burton, G.; Buschmann, J.; Cancellieri, M.; Dagli, M.; Date, M.; et al. RIFM fragrance ingredient safety assessment, decanoic acid, CAS Registry Number 334-48-5. Food Chem. Toxicol. 2020, 144, 111465. [Google Scholar] [CrossRef] [PubMed]
- EPA. Eugenol. 97-53-0 | DTXSID9020617. 2021. Available online: https://comptox.epa.gov/dashboard/chemical/details/DTXSID9020617 (accessed on 15 December 2021).
- EPA. Sodium Dodecyl Sulfate. 151-21-3 | DTXSID1026031. 2021. Available online: https://comptox.epa.gov/dashboard/chemical/details/DTXSID1026031 (accessed on 15 December 2021).
- Madrigal-Santillán, E.; Madrigal-Bujaidar, E.; Álvarez-González, I.; Sumaya-Martínez, M.T.; Gutiérrez-Salinas, J.; Bautista, M.; Morales-González, Á.; García-Luna, Y.; González-Rubio, M.; Aguilar-Faisal, J.L.; et al. Review of natural products with hepatoprotective effects. World, J. Gastroenterol. 2014, 20, 14787–14804. [Google Scholar] [CrossRef] [PubMed]
- Khanal, T.; Kim, H.G.; Jin, S.W.; Shim, E.; Han, H.J.; Noh, K.; Park, S.; Lee, D.H.; Kang, W.; Yeo, H.K.; et al. Protective role of metabolism by intestinal microflora in butyl paraben-induced toxicity in HepG2 cell cultures. Toxicol. Lett. 2012, 213, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Rial, S.A.; Ravaut, G.; Malaret, T.B.; Bergeron, K.-F.; Mounier, C. Hexanoic, Octanoic and Decanoic Acids Promote Basal and Insulin-Induced Phosphorylation of the Akt-mTOR Axis and a Balanced Lipid Metabolism in the HepG2 Hepatoma Cell Line. Molecules 2018, 23, 2315. [Google Scholar] [CrossRef]
- Ulanowska, M.; Olas, B. Biological Properties and Prospects for the Application of Eugenol—A Review. Int. J. Mol. Sci. 2021, 22, 3671. [Google Scholar] [CrossRef]
- Shah, K.H.; Verma, R.J. Butyl p-hydroxybenzoic acid induces oxidative stress in mice liver—An in vivo study. Acta Pol. Pharm. 2011, 68, 875–879. [Google Scholar]
- Bondi, C.A.M.; Marks, J.L.; Wroblewski, L.B.; Raatikainen, H.S.; Lenox, S.R.; Gebhardt, K.E. Human and Environmental Toxicity of Sodium Lauryl Sulfate (SLS): Evidence for Safe Use in Household Cleaning Products. Environ. Health Insights 2015, 9, 27–32. [Google Scholar] [CrossRef]
- Niraula, T.P.; Bhattarai, A.; Chatterjee, S.K. Sodium dodecylsulphate: A very useful surfactant for scientific investigation. J. Knowl. Innov. 2014, 2, 111–113. [Google Scholar]
- OECD. SIDS Initial Assessment Report for SIAM 5; Sodium Dodecyl Sulphate (CAS No: 151-21-3). 2005, p. 17. Available online: https://hpvchemicals.oecd.org/ui/handler.axd?id=7ffa18c7-5c4c-4766-91d7-5967b67aeba3 (accessed on 10 December 2021).
- NTP. Chemical Effects in Biological Systems. 2021. Available online: https://cebs.niehs.nih.gov/cebs/ (accessed on 10 December 2021).
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2020, 49, D1138–D1143. [Google Scholar] [CrossRef]
- Watford, S.; Pham, L.; Wignall, J.; Shin, R.; Martin, M.T.; Friedman, K.P. ToxRefDB version 2.0: Improved utility for predictive and retrospective toxicology analyses. Reprod. Toxicol. 2019, 89, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, Z.; Addington, C.K.; Dionisio, K.L.; Lyons, D.; Tornero-Velez, R.; Phillips, K.A.; Buckley, T.J.; Isaacs, K.K. Mining of Consumer Product Ingredient and Purchasing Data to Identify Potential Chemical Coexposures. Environ. Health Perspect. 2021, 129, 067006. [Google Scholar] [CrossRef] [PubMed]
- CDC. Fourth National Report on Human Exposure to Environmental Chemicals. 2021. Available online: https://www.cdc.gov/biomonitoring/pdf/fourthreport_updatedtables_feb2015.pdf (accessed on 25 March 2022).
- EPA. Air Quality System (AQS). 2021. Available online: https://www.epa.gov/aqs (accessed on 10 December 2021).
- Ring, C.L.; Arnot, J.A.; Bennett, D.H.; Egeghy, P.P.; Fantke, P.; Huang, L.; Isaacs, K.K.; Jolliet, O.; Phillips, K.A.; Price, P.S.; et al. Consensus Modeling of Median Chemical Intake for the U.S. Population Based on Predictions of Exposure Pathways. Environ. Sci. Technol. 2018, 53, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Ragerlab. Ragerlab Github. 2022. Available online: https://github.com/Ragerlab (accessed on 25 March 2022).
- Carberry, C.K.; Turla, T.; Koval, L.E.; Hartwell, H.; Fry, R.C.; Rager, J.E. Dataset for Chemical Mixtures in Household Environments: In Silico Predictions and In Vitro Testing of Potential Joint Action on PPARg in Human Liver Cells. UNC Dataverse, V1. 2022. Available online: https://dataverse.unc.edu/dataset.xhtml?persistentId=10.15139/S3/31A4OD (accessed on 18 April 2022).
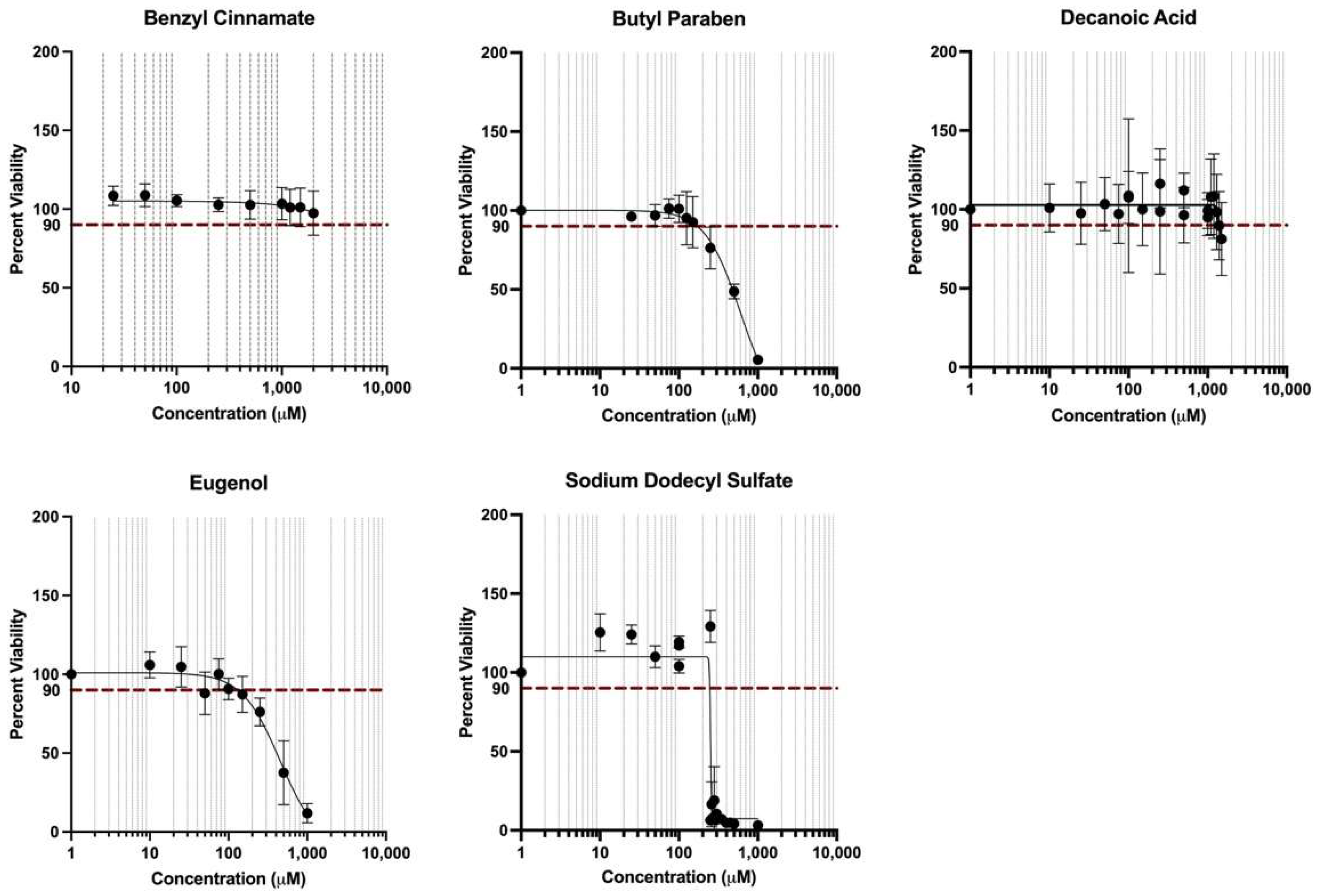
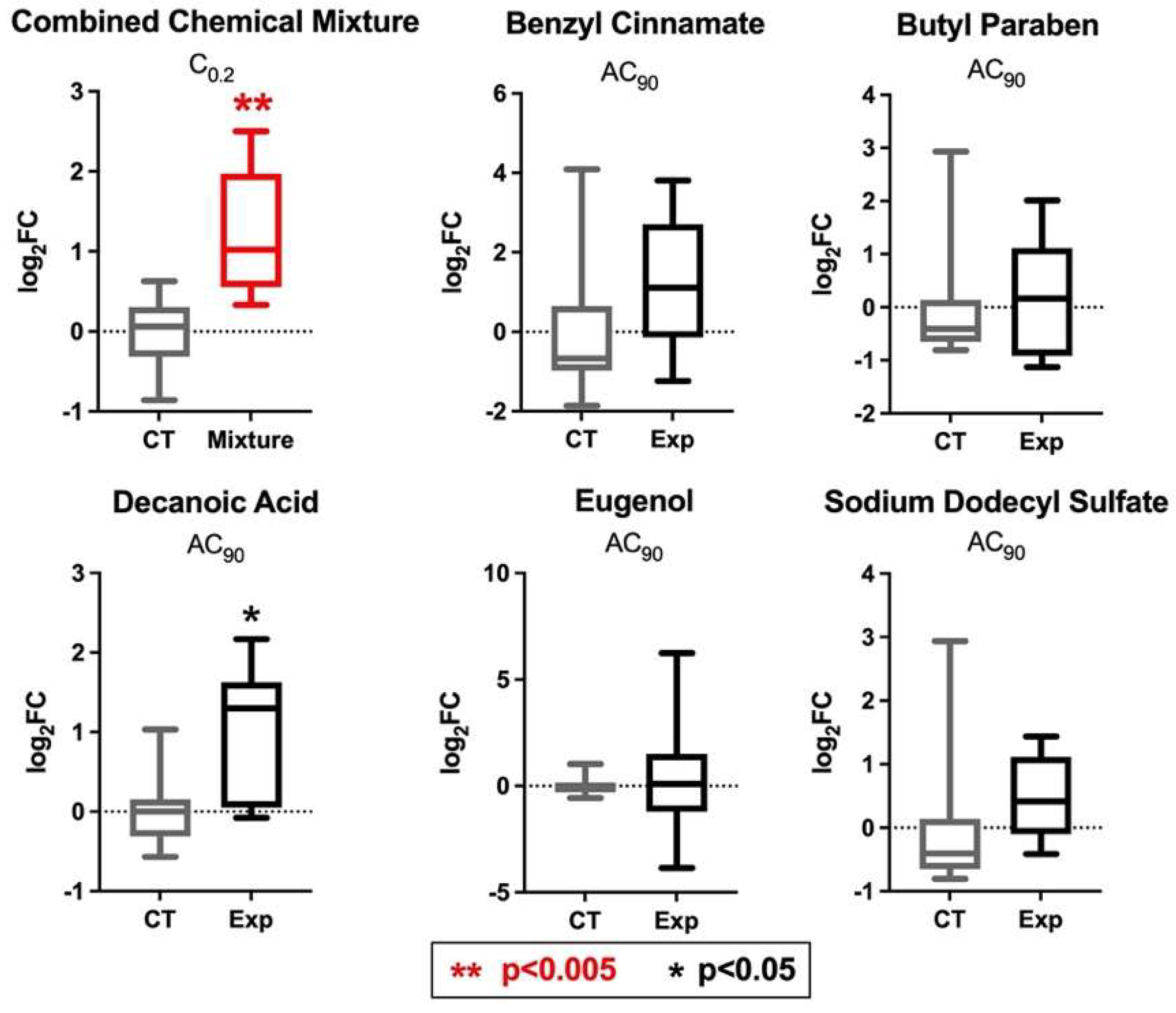
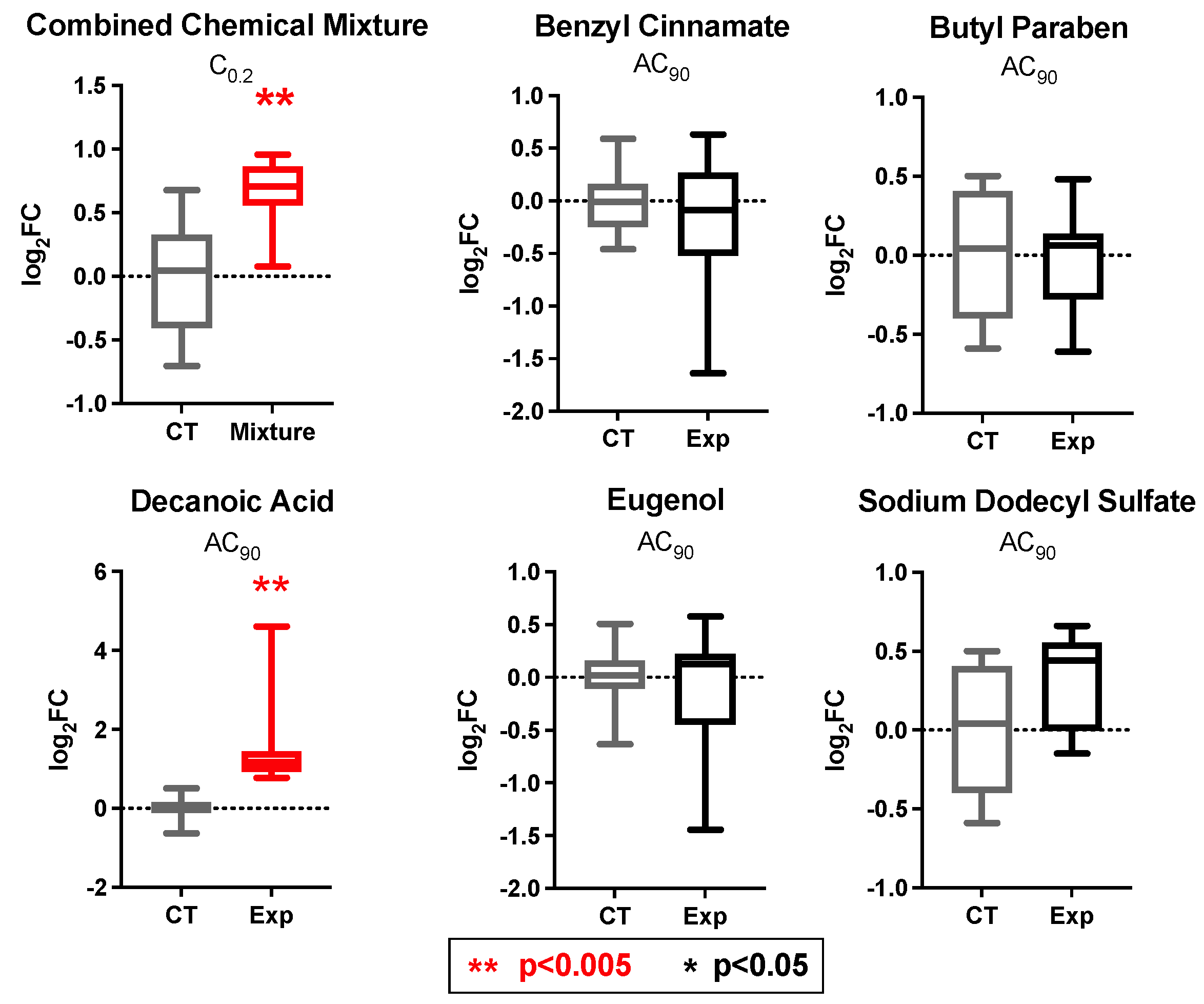
| Chemical Name | CASRN | Individual Chemical Concentrations Tested (AC90) (μM) | Concentrations of Chemicals Tested as a Mixture (at C0.2) 1 (μM) |
|---|---|---|---|
| Benzyl cinnamate | 103-41-3 | 1000 | 200 |
| Butylparaben | 94-26-8 | 150 | 30 |
| Decanoic acid | 334-48-5 | 1300 | 260 |
| Eugenol | 97-53-0 | 150 | 30 |
| Sodium Dodecyl Sulfate | 151-21-3 | 250 | 50 |
| % Viability of each treatment 2 | 90% | 136% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carberry, C.K.; Turla, T.; Koval, L.E.; Hartwell, H.; Fry, R.C.; Rager, J.E. Chemical Mixtures in Household Environments: In Silico Predictions and In Vitro Testing of Potential Joint Action on PPARγ in Human Liver Cells. Toxics 2022, 10, 199. https://doi.org/10.3390/toxics10050199
Carberry CK, Turla T, Koval LE, Hartwell H, Fry RC, Rager JE. Chemical Mixtures in Household Environments: In Silico Predictions and In Vitro Testing of Potential Joint Action on PPARγ in Human Liver Cells. Toxics. 2022; 10(5):199. https://doi.org/10.3390/toxics10050199
Chicago/Turabian StyleCarberry, Celeste K., Toby Turla, Lauren E. Koval, Hadley Hartwell, Rebecca C. Fry, and Julia E. Rager. 2022. "Chemical Mixtures in Household Environments: In Silico Predictions and In Vitro Testing of Potential Joint Action on PPARγ in Human Liver Cells" Toxics 10, no. 5: 199. https://doi.org/10.3390/toxics10050199
APA StyleCarberry, C. K., Turla, T., Koval, L. E., Hartwell, H., Fry, R. C., & Rager, J. E. (2022). Chemical Mixtures in Household Environments: In Silico Predictions and In Vitro Testing of Potential Joint Action on PPARγ in Human Liver Cells. Toxics, 10(5), 199. https://doi.org/10.3390/toxics10050199






