Integration of Genotoxic Biomarkers in Environmental Biomonitoring Analysis Using a Multi-Biomarker Approach in Three-Spined Stickleback (Gasterosteus aculeatus Linnaeus, 1758)
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sampling Sites
2.2. Morphometric Indices
2.3. Biomarkers Analysis
2.3.1. Peripherical Erythrocyte Counts, Mortality and Genotoxic Biomarkers
2.3.2. Innate Immune Capacities and Biochemical Parameters
2.4. Statistical Analysis
3. Results
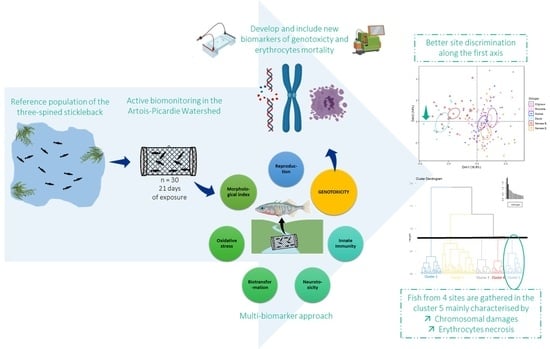
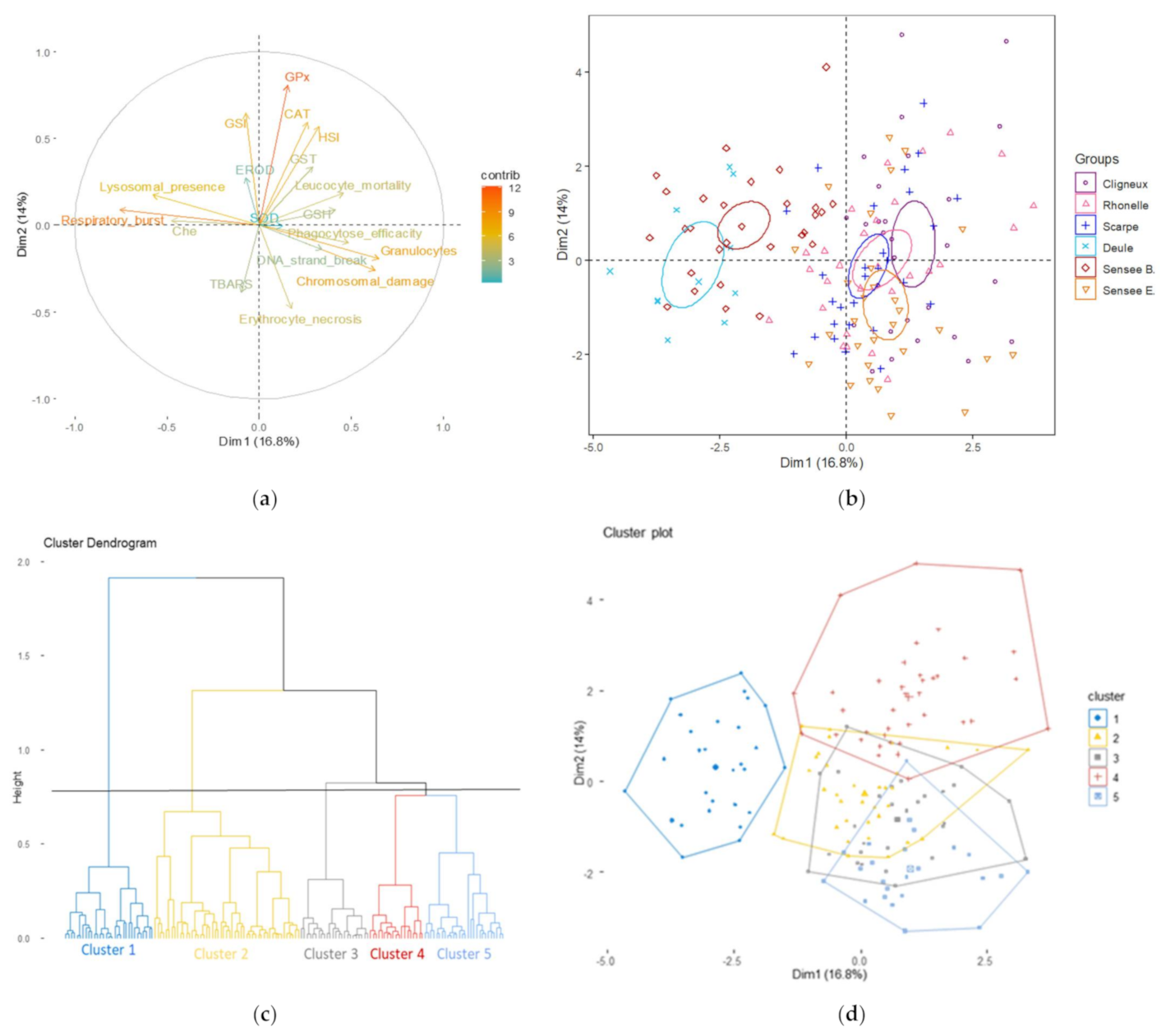
3.1. Effect of Current Biomarkers on Site Discrimination
3.2. Evolution of Site Discrimination by Adding Genotoxic Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brack, W.; Ait-Aissa, S.; Altenburger, R.; Cousins, I.; Dulio, V.; Escher, B.; Focks, A.; Ginebreda, A.; Hering, D.; Hilscherová, K.; et al. Let Us Empower the WFD to Prevent Risks of Chemical Pollution in European Rivers and Lakes. Environ. Sci. Eur. 2019, 31, 47. [Google Scholar] [CrossRef]
- Brack, W.; Dulio, V.; Ågerstrand, M.; Allan, I.; Altenburger, R.; Brinkmann, M.; Bunke, D.; Burgess, R.M.; Cousins, I.; Escher, B.I.; et al. Towards the Review of the European Union Water Framework Directive: Recommendations for More Efficient Assessment and Management of Chemical Contamination in European Surface Water Resources. Sci. Total Environ. 2017, 576, 720–737. [Google Scholar] [CrossRef] [PubMed]
- Amiard-Triquet, C.; Amiard, J.-C.; Mouneyrac, C. Aquatic Ecotoxicology: Advancing Tools for Dealing with Emerging Risks; Academic Press: Cambridge, MA, USA, 2015; ISBN 978-0-12-801176-8. [Google Scholar]
- Milinkovitch, T.; Geffard, O.; Geffard, A.; Mouneyrac, C.; Chaumot, A.; Xuereb, B.; Fisson, C.; Minier, C.; Auffret, M.; Perceval, O.; et al. Biomarkers as Tools for Monitoring within the Water Framework Directive Context: Concept, Opinions and Advancement of Expertise. Environ. Sci. Pollut. Res. 2019, 26, 32759–32763. [Google Scholar] [CrossRef] [PubMed]
- Brack, W.; Escher, B.I.; Müller, E.; Schmitt-Jansen, M.; Schulze, T.; Slobodnik, J.; Hollert, H. Towards a Holistic and Solution-Oriented Monitoring of Chemical Status of European Water Bodies: How to Support the EU Strategy for a Non-Toxic Environment? Environ. Sci. Eur. 2018, 30, 33. [Google Scholar] [CrossRef]
- Sanchez, W.; Porcher, J.-M. Utilisation des biomarqueurs pour la caractérisation de l’état écotoxicologique des masses d’eau. TSM 2009, 29–38. [Google Scholar] [CrossRef]
- Vethaak, A.; Dick, H.K.; Davies, I.M.; Thain, J.E.; Gubbins, M.J.; Martinez-Gomez, C.; Robinson, C.D.; Moffat, C.F.; Thierry, B.; Thomas, M.; et al. Integrated Indicator Framework and Methodology for Monitoring and Assessment of Hazardous Substances and Their Effects in the Marine Environment. Mar. Environ. Res. 2017, 124, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Ifremer Evaluation Du Descripteur 8 “Contaminants Dans Le Milieu” En France Métropolitainre—Rapport Scientifique Pour l’évaluation 2018 Au Titre de La DCSMM 2018. Available online: https://www.documentation.eauetbiodiversite.fr/notice/evaluation-du-descripteur-8-contaminants-dans-le-milieu-en-france-metropolitaine0 (accessed on 4 January 2022).
- Sanchez, W.; Katsiadaki, I.; Piccini, B.; Ditche, J.-M.; Porcher, J.-M. Biomarker Responses in Wild Three-Spined Stickleback (Gasterosteus aculeatus L.) as a Useful Tool for Freshwater Biomonitoring: A Multiparametric Approach. Environ. Int. 2008, 34, 490–498. [Google Scholar] [CrossRef]
- Ossana, N.A.; Eissa, B.L.; Baudou, F.G.; Castañé, P.M.; Soloneski, S.; Ferrari, L. Multibiomarker Response in Ten Spotted Live-Bearer Fish Cnesterodon decemmaculatus (Jenyns, 1842) Exposed to Reconquista River Water. Ecotoxicol. Environ. Saf. 2016, 133, 73–81. [Google Scholar] [CrossRef]
- Catteau, A.; Bado-Nilles, A.; Beaudouin, R.; Tebby, C.; Joachim, S.; Palluel, O.; Turiès, C.; Chrétien, N.; Nott, K.; Ronkart, S.; et al. Water Quality of the Meuse Watershed: Assessment Using a Multi-Biomarker Approach with Caged Three-Spined Stickleback (Gasterosteus aculeatus L.). Ecotoxicol. Environ. Saf. 2021, 208, 111407. [Google Scholar] [CrossRef]
- Carvalho, R.N.; Arukwe, A.; Ait-Aissa, S.; Bado-Nilles, A.; Balzamo, S.; Baun, A.; Belkin, S.; Blaha, L.; Brion, F.; Conti, D.; et al. Mixtures of Chemical Pollutants at European Legislation Safety Concentrations: How Safe Are They? Toxicol. Sci. 2014, 141, 218–233. [Google Scholar] [CrossRef]
- Wootton, R.J. A Functional Biology of Sticklebacks; University of California Press: Berkeley, CA, USA, 1984; ISBN 978-0-520-05381-6. [Google Scholar]
- Pottinger, T.G.; Carrick, T.R.; Yeomans, W.E. The Three-Spined Stickleback as an Environmental Sentinel: Effects of Stressors on Whole-Body Physiological Indices. J. Fish Biol. 2002, 61, 207–229. [Google Scholar] [CrossRef]
- Catteau, A.; Bado-Nilles, A.; Beaudouin, R.; Joachim, S.; Palluel, O.; Turiès, C.; Galet, C.; Geffard, A.; Porcher, J.-M. An Active Biomonitoring Approach Using Three-Spined Stickleback (Gasterosteus aculeatus, L.) to Assess the Efficiency of a Constructed Wetland as Tertiary Treatment of Wastewater. Ecol. Indic. 2020, 114, 106238. [Google Scholar] [CrossRef]
- Hani, Y.M.I.; Marchand, A.; Turies, C.; Kerambrun, E.; Palluel, O.; Bado-Nilles, A.; Beaudouin, R.; Porcher, J.-M.; Geffard, A.; Dedourge-Geffard, O. Digestive Enzymes and Gut Morphometric Parameters of Threespine Stickleback (Gasterosteus aculeatus): Influence of Body Size and Temperature. PLoS ONE 2018, 13, e0194932. [Google Scholar] [CrossRef]
- Sanchez, W.; Burgeot, T.; Porcher, J.-M. A Novel “Integrated Biomarker Response” Calculation Based on Reference Deviation Concept. Environ. Sci. Pollut. Res. 2013, 20, 2721–2725. [Google Scholar] [CrossRef] [PubMed]
- Le Guernic, A.; Sanchez, W.; Bado-Nilles, A.; Palluel, O.; Turies, C.; Chadili, E.; Cavalié, I.; Delahaut, L.; Adam-Guillermin, C.; Porcher, J.-M.; et al. In Situ Effects of Metal Contamination from Former Uranium Mining Sites on the Health of the Three-Spined Stickleback (Gasterosteus aculeatus, L.). Ecotoxicology 2016, 25, 1234–1259. [Google Scholar] [CrossRef]
- Oikari, A. Caging Techniques for Field Exposures of Fish to Chemical Contaminants. Aquat. Toxicol. 2006, 78, 370–381. [Google Scholar] [CrossRef]
- Le Guernic, A.; Sanchez, W.; Palluel, O.; Bado-Nilles, A.; Turies, C.; Chadili, E.; Cavalié, I.; Adam-Guillermin, C.; Porcher, J.-M.; Geffard, A.; et al. In Situ Experiments to Assess Effects of Constraints Linked to Caging on Ecotoxicity Biomarkers of the Three-Spined Stickleback (Gasterosteus aculeatus L.). Fish Physiol. Biochem. 2016, 42, 643–657. [Google Scholar] [CrossRef]
- Catteau, A.; Le Guernic, A.; Marchand, A.; Hani, Y.M.I.; Palluel, O.; Turiès, C.; Bado-Nilles, A.; Dedourge-Geffard, O.; Geffard, A.; Porcher, J.-M. Impact of Confinement and Food Access Restriction on the Three-Spined Stickleback (Gasterosteus aculeatus, L.) during Caging: A Multi-Biomarker Approach. Fish Physiol. Biochem. 2019, 45, 1261–1276. [Google Scholar] [CrossRef]
- Claxton, L.D.; Houk, V.S.; Hughes, T.J. Genotoxicity of Industrial Wastes and Effluents. Mutat. Res. 1998, 410, 237–243. [Google Scholar] [CrossRef]
- Kaur, R.; Shah, T.K.; Kaur, S.; Rani, S. A Review on Genotoxicity in Aquatic Organisms and Environment. J. Pharm. Innov. 2018, 7, 353–359. [Google Scholar]
- Bolognesi, C.; Cirillo, S. Genotoxicity Biomarkers in Aquatic Bioindicators. Curr. Zool. 2014, 60, 273–284. [Google Scholar] [CrossRef]
- Jha, A.N. Ecotoxicological Applications and Significance of the Comet Assay. Mutagenesis 2008, 23, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Osman, A.G.M.; Reheem, A.-E.-B.M.A.E.; Moustafa, M.A.; Mahmoud, U.M.; Abuel-Fadl, K.Y.; Kloas, W. In Situ Evaluation of the Genotoxic Potential of the River Nile: I. Micronucleus and Nuclear Lesion Tests of Erythrocytes of Oreochromis niloticus niloticus (Linnaeus, 1758) and Clarias gariepinus (Burchell, 1822). Toxicol. Environ. Chem. 2011, 93, 1002–1017. [Google Scholar] [CrossRef]
- Hussain, B.; Sultana, T.; Sultana, S.; Masoud, M.S.; Ahmed, Z.; Mahboob, S. Fish Eco-Genotoxicology: Comet and Micronucleus Assay in Fish Erythrocytes as in Situ Biomarker of Freshwater Pollution. Saudi J. Biol. Sci. 2018, 25, 393–398. [Google Scholar] [CrossRef]
- Orsière, T. De La Génotoxicologie à La Biosurveillance. J. Natl. Santé Trav. BTP Ann. 2005, 28, 25. [Google Scholar]
- Collins, A.R.; Dobson, V.L.; Dušinská, M.; Kennedy, G.; Štětina, R. The Comet Assay: What Can It Really Tell Us? Mutat. Res. /Fundam. Mol. Mech. Mutagenesis 1997, 375, 183–193. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A Simple Technique for Quantitation of Low Levels of DNA Damage in Individual Cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef]
- Tice, R.R.; Agurell, E.; Anderson, D.; Burlinson, B.; Hartmann, A.; Kobayashi, H.; Miyamae, Y.; Rojas, E.; Ryu, J.-C.; Sasaki, Y.F. Single cell gel/comet assay: Guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagenesis 2000, 35, 206–221. [Google Scholar] [CrossRef]
- de Lapuente, J.; Lourenço, J.; Mendo, S.A.; Borràs, M.; Martins, M.G.; Costa, P.M.; Pacheco, M. The Comet Assay and Its Applications in the Field of Ecotoxicology: A Mature Tool That Continues to Expand Its Perspectives. Front. Genet. 2015, 6. [Google Scholar] [CrossRef]
- Gajski, G.; Žegura, B.; Ladeira, C.; Novak, M.; Sramkova, M.; Pourrut, B.; Del Bo’, C.; Milić, M.; Gutzkow, K.B.; Costa, S.; et al. The Comet Assay in Animal Models: From Bugs to Whales – (Part 2 Vertebrates). Mutat. Res./Rev. Mutat. Res. 2019, 781, 130–164. [Google Scholar] [CrossRef]
- Osman, A.G.M. Genotoxicity Tests and Their Contributions in Aquatic Environmental Research. J. Environ. Prot. 2014, 05, 1391. [Google Scholar] [CrossRef]
- Santos, R.; Aude, J.; Olivier, P.; Mélissa, P.-L.; Aurélien, B.; Christophe, B.; Marc, P.J.; Sylvie, B.; Alain, D.; Wilfried, S. Characterization of a Genotoxicity Biomarker in Three-Spined Stickleback (Gasterosteus aculeatus L.): Biotic Variability and Integration in a Battery of Biomarkers for Environmental Monitoring. Environ. Toxicol. 2016, 31, 415–426. [Google Scholar] [CrossRef]
- Wirzinger, G.; Weltje, L.; Gercken, J.; Sordyl, H. Genotoxic Damage in Field-Collected Three-Spined Sticklebacks (Gasterosteus aculeatus L.): A Suitable Biomonitoring Tool? Mutat. Res./Genet. Toxicol. Environ. Mutagenesis 2007, 628, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Theodorakis, C.W.; D’Surney, S.J.; Shugart, L.R. Detection of Genotoxic Insult as DNA Strand Breaks in Fish Blood Cells by Agarose Gel Electrophoresis. Environ. Toxicol. Chem. 1994, 13, 1023–1031. [Google Scholar] [CrossRef]
- Otto, F.J.; Oldiges, H. Flow Cytogenetic Studies in Chromosomes and Whole Cells for the Detection of Clastogenic Effects. Cytometry 1980, 1, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Vindeløv, L.L.; Christensen, I.J. A Review of Techniques and Results Obtained in One Laboratory by an Integrated System of Methods Designed for Routine Clinical Flow Cytometric DNA Analysis. Cytometry 1990, 11, 753–770. [Google Scholar] [CrossRef]
- Goanvec, C.; Theron, M.; Lacoue-Labarthe, T.; Poirier, E.; Guyomarch, J.; Le-Floch, S.; Laroche, J.; Nonnotte, L.; Nonnotte, G. Flow Cytometry for the Evaluation of Chromosomal Damage in Turbot Psetta maxima (L.) Exposed to the Dissolved Fraction of Heavy Fuel Oil in Sea Water: A Comparison with Classical Biomarkers. J. Fish Biol. 2008, 73, 395–413. [Google Scholar] [CrossRef]
- Marchand, A.; Porcher, J.-M.; Turies, C.; Chadili, E.; Palluel, O.; Baudoin, P.; Betoulle, S.; Bado-Nilles, A. Evaluation of Chlorpyrifos Effects, Alone and Combined with Lipopolysaccharide Stress, on DNA Integrity and Immune Responses of the Three-Spined Stickleback, Gasterosteus aculeatus. Ecotoxicol. Environ. Saf. 2017, 145, 333–339. [Google Scholar] [CrossRef]
- Theodorakis, C.W.; Bickham, J.W.; Donnelly, K.C.; McDonald, T.J.; Willink, P.W. DNA Damage in Cichlids from an Oil Production Facility in Guatemala. Ecotoxicology 2012, 21, 496–511. [Google Scholar] [CrossRef]
- Sanchez, W.; Aït-Aïssa, S.; Palluel, O.; Ditche, J.-M.; Porcher, J.-M. Preliminary Investigation of Multi-Biomarker Responses in Three-Spined Stickleback (Gasterosteus aculeatus L.) Sampled in Contaminated Streams. Ecotoxicology 2007, 16, 279–287. [Google Scholar] [CrossRef]
- Consulter Les Données de Qualité Des Rivières | Agence de l’Eau Artois-Picardie. Available online: https://www.eau-artois-picardie.fr/donnees-sur-leau/visualiser-et-telecharger-les-donnees (accessed on 10 January 2022).
- Catteau, A.; Porcher, J.-M.; Bado-Nilles, A.; Bonnard, I.; Bonnard, M.; Chaumot, A.; David, E.; Dedourge-Geffard, O.; Delahaut, L.; Delorme, N.; et al. Interest of a Multispecies Approach in Active Biomonitoring: Application in the Meuse Watershed. Sci. Total Environ. 2022, 808, 152148. [Google Scholar] [CrossRef]
- Bado-Nilles, A.; Jolly, S.; Porcher, J.-M.; Palluel, O.; Geffard, A.; Gagnaire, B.; Betoulle, S.; Sanchez, W. Applications in Environmental Risk Assessment of Leucocyte Apoptosis, Necrosis and Respiratory Burst Analysis on the European Bullhead, Cottus sp. Environ. Pollut. 2014, 184, 9–17. [Google Scholar] [CrossRef]
- Fulton, T.W. Rate of Growth of Sea Fishes; Neill, Company: London, UK, 1902. [Google Scholar]
- Slooff, W.; Kreijl, C.F.V.; Baars, A.J. Relative Liver Weights and Xenobiotic-Metabolizing Enzymes of Fish from Polluted Surface Waters in the Netherlands. Aquat. Toxicol. 1983, 4, 1–14. [Google Scholar] [CrossRef]
- Lofts, B.; Pickford, G.E.; Atz, J.W. Effects of Methyl Testosterone on the Testes of a Hypophysectomized Cyprinodont Fish, Fundulus heteroclitus. Gen. Comp. Endocrinol. 1966, 6, 74–88. [Google Scholar] [CrossRef]
- Goanvec, C.; Theron, M.; Poirier, E.; Floch, S.L.; Laroche, J.; Nonnotte, L.; Nonnotte, G. Evaluation of Chromosomal Damage by Flow Cytometry in Turbot (Scophthalmus maximus L.) Exposed to Fuel Oil. Biomarkers 2004, 9, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Mélissa, P.-L.; Aurélien, B.; Emmanuelle, V.; Marc, P.J.; Sylvie, B.; Alain, D.; Wilfried, S. Kinetic Response of a Genotoxicity Biomarker in the Three-Spined Stickleback and Implication for Environmental Monitoring. Ecotoxicol. Environ. Saf. 2014, 102, 6–11. [Google Scholar] [CrossRef]
- Langie, S.A.S.; Azqueta, A.; Collins, A.R. The Comet Assay: Past, Present, and Future. Front. Genet. 2015, 6. [Google Scholar] [CrossRef]
- Bado-Nilles, A.; Betoulle, S.; Geffard, A.; Porcher, J.-M.; Gagnaire, B.; Sanchez, W. Flow Cytometry Detection of Lysosomal Presence and Lysosomal Membrane Integrity in the Three-Spined Stickleback (Gasterosteus aculeatus L.) Immune Cells: Applications in Environmental Aquatic Immunotoxicology. Environ. Sci. Pollut. Res. 2013, 20, 2692–2704. [Google Scholar] [CrossRef]
- Gagnaire, B.; Bado-Nilles, A.; Betoulle, S.; Amara, R.; Camilleri, V.; Cavalié, I.; Chadili, E.; Delahaut, L.; Kerambrun, E.; Orjollet, D.; et al. Former Uranium Mine-Induced Effects in Caged Roach: A Multiparametric Approach for the Evaluation of in Situ Metal Toxicity. Ecotoxicology 2015, 24, 215–231. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for Lipid Peroxides in Animal Tissues by Thiobarbituric Acid Reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Paoletti, F.; Aldinucci, D.; Mocali, A.; Caparrini, A. A Sensitive Spectrophotometric Method for the Determination of Superoxide Dismutase Activity in Tissue Extracts. Anal. Biochem. 1986, 154, 536–541. [Google Scholar] [CrossRef]
- Vandeputte, C.; Guizon, I.; Genestie-Denis, I.; Vannier, B.; Lorenzon, G. A Microtiter Plate Assay for Total Glutathione and Glutathione Disulfide Contents in Cultured/Isolated Cells: Performance Study of a New Miniaturized Protocol. Cell Biol. Toxicol. 1994, 10, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Paglia, D.E.; Valentine, W.N. Studies on the Quantitative and Qualitative Characterization of Erythrocyte Glutathione Peroxidase. J. Lab. Clin. Med. 1967, 70, 158–169. [Google Scholar] [CrossRef]
- Babo, S.; Vasseur, P. In Vitro Effects of Thiram on Liver Antioxidant Enzyme Activities of Rainbow Trout (Oncorhynchus mykiss). Aquat. Toxicol. 1992, 22, 61–68. [Google Scholar] [CrossRef]
- Flammarion, P.; Devaux, A.; Nehls, S.; Migeon, B.; Noury, P.; Garric, J. Multibiomarker Responses in Fish from the Moselle River (France). Ecotoxicol. Environ. Saf. 2002, 51, 145–153. [Google Scholar] [CrossRef]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-Transferases the First Enzymatic Step in Mercapturic Acid Formation. J. Biol. Chem. 1974, 7130–7139. [Google Scholar] [CrossRef]
- Sanchez, W.; Palluel, O.; Meunier, L.; Coquery, M.; Porcher, J.-M.; Ait-Aissa, S. Copper-Induced Oxidative Stress in Three-Spined Stickleback: Relationship with Hepatic Metal Levels. Environ. Toxicol. Pharmacol. 2005, 19, 177–183. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A New and Rapid Colorimetric Determination of Acetylcholinesterase Activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Catteau, A. Pertinence d’une Approche Multibiomarqueurs En Biosurveillance Active Avec l’épinoche à Trois Épines (Gasterosteus aculeatus). Ph.D. Thesis, University of Reims Champagne-Ardenne, Reims, France, 2019. [Google Scholar]
- Pereira, P.; de Pablo, H.; Vale, C.; Pacheco, M. Combined Use of Environmental Data and Biomarkers in Fish (Liza aurata) Inhabiting a Eutrophic and Metal-Contaminated Coastal System—Gills Reflect Environmental Contamination. Mar. Environ. Res. 2010, 69, 53–62. [Google Scholar] [CrossRef]
- Sanchez, W.; Piccini, B.; Maillot-Maréchal, E.; Porcher, J.-M. Comparison of Two Reference Systems for Biomarker Data Analysis in a Freshwater Biomonitoring Context. Environ. Int. 2010, 36, 377–382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jolly, S.; Bado-Nilles, A.; Lamand, F.; Turies, C.; Chadili, E.; Porcher, J.-M.; Betoulle, S.; Sanchez, W. Multi-Biomarker Approach in Wild European Bullhead, Cottus sp., Exposed to Agricultural and Urban Environmental Pressures: Practical Recommendations for Experimental Design. Chemosphere 2012, 87, 675–683. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marchand, A.; Tebby, C.; Beaudouin, R.; Catteau, A.; Porcher, J.-M.; Turiès, C.; Bado-Nilles, A. Reliability Evaluation of Biomarker Reference Ranges for Mesocosm and Field Conditions: Cellular Innate Immunomarkers in Gasterosteus aculeatus. Sci. Total Environ. 2020, 698, 134333. [Google Scholar] [CrossRef] [PubMed]
- Soldatov, A.A. Peculiarities of Organization and Functioning of the Fish Red Blood System. J. Evol. Biochem. Phys. 2005, 41, 272–281. [Google Scholar] [CrossRef]
- Avery, E.H.; Lee, B.L.; Freedland, R.A.; Cornelius, C.E. Bile Pigments in Gallbladder and Freshly-Secreted Hepatic Duct Bile from Fed and Fasted Rainbow Trout, Oncorhynchus mykiss. Comp. Biochem. Physiol. Part A Physiol. 1992, 101, 857–861. [Google Scholar] [CrossRef]
- Dinnen, R.D.; Tomlinson, S.M.; Hart, D.; Chopra, C.; Heddle, J.A. Application of a Micronucleus Assay to the Peripheral Blood Cells of Rainbow Trout, Salmo gairdneri. Can. Tech. Rep. Fish. Aquat. Sci 1988, 1607, 69–78. [Google Scholar]
- Nikinmaa, M. Environmental Regulation of the Function of Circulating Erythrocytes via Changes in Age Distribution in Teleost Fish: Possible Mechanisms and Significance. Mar. Genom. 2020, 49, 100717. [Google Scholar] [CrossRef]
- Bickham, J.W. Flow Cytometry as a Technique to Monitor the Effects of Environmental Genotoxins on Wildlife Populations. In In Situ Evaluation of Biological Hazards of Environmental Pollutants; Sandhu, S.S., Lower, W.R., de Serres, F.J., Suk, W.A., Tice, R.R., Eds.; Environmental Science Research; Springer: Boston, MA, USA, 1990; pp. 97–108. ISBN 978-1-4684-5808-4. [Google Scholar]
- Otto, F.J.; Oldiges, H.; Göhde, W.; Jain, V.K. Flow Cytometric Measurement of Nuclear DNA Content Variations as a Potential in Vivo Mutagenicity Test. Cytometry 1981, 2, 189–191. [Google Scholar] [CrossRef]
- Galloway, S.M. Chromosome Aberrations Induced in Vitro: Mechanisms, Delayed Expression, and Intriguing Questions. Environ. Mol. Mutagenesis 1994, 23, 44–53. [Google Scholar] [CrossRef]
- Benabdelmouna, A.; Ledu, C. The Mass Mortality of Blue Mussels (Mytilus spp.) from the Atlantic Coast of France Is Associated with Heavy Genomic Abnormalities as Evidenced by Flow Cytometry. J. Invertebr. Pathol. 2016, 138, 30–38. [Google Scholar] [CrossRef]
- Peixoto, M.S.; de Oliveira Galvão, M.F.; Batistuzzo de Medeiros, S.R. Cell Death Pathways of Particulate Matter Toxicity. Chemosphere 2017, 188, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.R.; Alagawany, M. Erythrocytes as a Biological Model for Screening of Xenobiotics Toxicity. Chem. -Biol. Interact. 2018, 279, 73–83. [Google Scholar] [CrossRef]
- Marigomez, I.; Garmendia, L.; Soto, M.; Orbea, A.; Izagirre, U.; Cajaraville, M.P. Marine Ecosystem Health Status Assessment through Integrative Biomarker Indices: A Comparative Study after the Prestige Oil Spill “Mussel Watch”. Ecotoxicology 2013, 22, 486–505. [Google Scholar] [CrossRef] [PubMed]
- Davico, C.E.; Loteste, A.; Parma, M.J.; Poletta, G.; Simoniello, M.F. Stress Oxidative and Genotoxicity in Prochilodus lineatus (Valenciennes, 1836) Exposed to Commercial Formulation of Insecticide Cypermethrin. Drug Chem. Toxicol. 2020, 43, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilbeigi, M.; Kalbassi, M.R.; Seyedi, J.; Tayemeh, M.B.; Moghaddam, J.A. Intra and Extracellular Effects of Benzo [α] Pyrene on Liver, Gill and Blood of Caspian White Fish (Rutilus frissi kutum): Cyto-Genotoxicity and Histopathology Approach. Mar. Pollut. Bull. 2021, 163, 111942. [Google Scholar] [CrossRef]
- Fanali, L.Z.; De Oliveira, C.; Sturve, J. Enzymatic, Morphological, and Genotoxic Effects of Benzo[a]Pyrene in Rainbow Trout (Oncorhynchus mykiss). Environ. Sci. Pollut. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.; Ali, R.; Shahid, M.; Khan, M.A.; Zubair, M.; Ali, T.; Khan, Q.M. Genotoxic and Hematological Effects of Chlorpyrifos Exposure on Freshwater Fish Labeo rohita. Drug Chem. Toxicol. 2018, 41, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Mitkovska, V.; Chassovnikarova, T. Chlorpyrifos Levels within Permitted Limits Induce Nuclear Abnormalities and DNA Damage in the Erythrocytes of the Common Carp. Environ. Sci. Pollut. Res. 2020, 27, 7166–7176. [Google Scholar] [CrossRef] [PubMed]
- Paravani, E.V.; Simoniello, M.F.; Poletta, G.L.; Casco, V.H. Cypermethrin Induction of DNA Damage and Oxidative Stress in Zebrafish Gill Cells. Ecotoxicol. Environ. Saf. 2019, 173, 1–7. [Google Scholar] [CrossRef]
- Paravani, E.V.; Simoniello, M.F.; Poletta, G.L.; Zolessi, F.R.; Casco, V.H. Cypermethrin: Oxidative Stress and Genotoxicity in Retinal Cells of the Adult Zebrafish. Mutat. Res. /Genet. Toxicol. Environ. Mutagenesis 2018, 826, 25–32. [Google Scholar] [CrossRef]
- Yazdani, M. Comparative Toxicity of Selected PAHs in Rainbow Trout Hepatocytes: Genotoxicity, Oxidative Stress and Cytotoxicity. Drug Chem. Toxicol. 2020, 43, 71–78. [Google Scholar] [CrossRef] [PubMed]


| Cligneux | Rhonelle | Scarpe | Deule | Sensée | ||
|---|---|---|---|---|---|---|
| St Rémy du Nord | Artres | Biache-Saint-Vaast | Courrières | Bouchain | Etaing | |
| GPS coordinates | N 50°14′20.7″ E 3°53′39.0″ | N 50°17′50.4″ E 3°32′40.2″ | N 50°18′17.3″ E 2°56′38.1″ | N 50°28′08.2″ E 2°56′43.0″ | N 50°16′45.4″ E 3°18′18.6″ | N 50°16′43.3″ E 2°59′47.8″ |
| Sandre code | 01001452 | 01029000 | 01037000 | 01078000 | 01024000 | 01000274 |
| WFD data | Yes | Yes | Yes | Yes | Yes | No |
| Ecological status (2016–2018) | Bad | Mediocre | Mediocre | Mediocre | Mediocre | / |
| Chemical status (2016–2018) | Bad | Bad | Bad | Bad | Bad | / |
| Pressure | Poorly urbanised Agricultural practices | Poorly urbanised Agricultural practices | Polluted site Artificial canal | Modified river Waterway transport Urban | Natural stream Down stream | Agricultural practices Up stream |
| Temperatures (°C) | 8.5 ± 2.1 | 7.0 ± 1.6 | 10.6 ± 1.5 | 10.6 ± 2.8 | 10.2 ± 1.8 | 10.7 ± 0.9 |
| pH | 8.3 ± 0.1 | 8.3 ± 0.0 | 8.0 ± 0.1 | 8.1 ± 0.2 | 8.0 ± 0.1 | 7.8 ± 0.0 |
| Conductivity (µS/cm) | 625.0 ± 5.6 | 749.5 ± 4.9 | 694.5 ± 7.8 | 719.0 ± 25.5 | 582.5 ± 47.4 | 710.0 ± 1.4 |
| Oxygen rate (mg/L) | 10.7 ± 0.3 | 10.8 | 8.7 ± 1.5 | 7.5 ± 2.3 | 6.3 ± 0.9 | 9.4 ± 0.1 |
| Cligneux | Rhonelle | Scarpe | Deule | Sensée | ||||
|---|---|---|---|---|---|---|---|---|
| St Rémy du Nord | Artres | Biache-Saint-Vaast | Courrières | Bouchain | Etaing | |||
| Biometric index | n | M | 14 | 15 | 16 | 5 | 12 | 17 |
| F | 14 | 13 | 12 | 6 | 17 | 12 | ||
| Standard length (mm) | M/F | 49.68 c ±3.53 | 49.43 bc ±3.63 | 46.21 a ±4.60 | 44.36 a ±3.32 | 46.38 ab ±3.76 | 46.21 a ±3.77 | |
| Weight (g) | M/F | 1.87 c ±0.60 | 1.62 bc ±0.34 | 1.37 a ±0.42 | 1.15 a ±0.22 | 1.41 a ±0.61 | 1.40 ab ±0.36 | |
| Fulton’s condition | M | 1.6 a ±0.29 | 1.35 a ±0.13 | 1.38 a ±0.15 | 1.36 a ±0.15 | 1.30 a ±0.07 | 1.44 a ±0.14 | |
| F | 1.40 ±0.12 | 1.29 ±0.10 | 1.33 ±0.14 | 1.27 ±0.10 | 1.51 ±1.03 | 1.34 ±0.10 | ||
| Hepatosomatic index | M | 3.55 ±0.67 | 3.66 ±0.81 | 3.38 ±0.98 | 3.15 ±0.53 | 3.96 ±0.83 | 3.30 ±1.00 | |
| F | 4.27 ±1.08 | 5.10 ±1.43 | 4.41 ±1.26 | 4.08 ±1.16 | 3.62 ±1.15 | 3.89 ±0.79 | ||
| Reproductive system | Gonadosomatic index | M | 0.88 a ±0.31 | 1.09 ab ±0.24 | 1.15 b ±0.30 | 1.03 ab ±0.08 | 0.98 ab ±0.16 | 0.99 ab ±0.27 |
| F | 2.36 ±0.54 | 2.08 ±0.54 | 2.09 ±0.21 | 2.21 ±0.45 | 2.04 ±0.54 | 2.21 ±0.62 | ||
| Innate immune response | Leucocyte necrosis (%) | M/F | 10.46 bc ±3.77 | 12.33 c ±3.54 | 10.29 bc ±4.24 | 6.34 a ±1.23 | 8.71 ab ±4.24 | 6.99 b ±2.32 |
| Leucocyte apoptosis (%) | M/F | 8.70 c ±3.75 | 7.14 bc ±3.30 | 5.27 ab ±2.53 | 3.74 a ±0.99 | 4.74 a ±6.31 | 4.51 a ±1.84 | |
| Granulocytes–macrophages (%) | M/F | 53.47 c ±7.46 | 47.84 bc ±8.06 | 45.62 ab ±7.29 | 40.36 ab ±7.42 | 39.52 a ±9.39 | 48.30 bc ±8.77 | |
| Phagocytosis capacity (%) | M | 61.11 b ±6.12 | 54.80 a ±4.42 | 62.15 b ±4.52 | 49.01 a ±3.05 | 53.68 a ±4.52 | 60.11 b ±5.53 | |
| F | 54.96 ab ±5.56 | 57.11 ab ±9.90 | 63.16 c ±6.15 | 51.66 a ±2.77 | 51.53 a ±5.29 | 56.79 bc ±2.54 | ||
| Phagocytosis efficiency (%) | M/F | 16.55 b ±4.61 | 19.08 b ±6.81 | 25.76 c ±6.70 | 10.95 a ±2.22 | 13.01 a ±4.09 | 18.65 b ±7.26 | |
| Lysosomal presence (%) | M | 173.17 a ±31.48 | 234.04 ab ±38.86 | 338.26 d ±73.92 | 334.09 cde ±52.36 | 409.31 e ±90.56 | 268.97 bc ±69.05 | |
| F | 214.56 a ±39.17 | 227.16 bc ±44.74 | 405.29 c ±149.55 | 378.98 c ±66.64 | 462.36 c ±78.41 | 279.43 b ±77.37 | ||
| Respiratory burst index (U) | M/F | 1.05 bc ±0.53 | 1.15 c ±0.38 | 0.93 ab ±0.30 | 6.95 d ±2.38 | 3.87 d ±1.94 | 0.88 a ±0.50 | |
| Neurotoxicity | Che activity (U/g prot) | M/F | 38.61 b ±16.27 | 37.08 b ±16.67 | 27.23 a ±13.23 | 65.34 c ±24.12 | 43.03 b ±21.66 | 25.25 a ±14.46 |
| Metabolic detoxification | EROD activity (pmol/min/mg prot) | M | 5.35 bc ±4.28 | 6.11 c ±3.59 | 2.73 b ±1.77 | 6.56 c ±1.49 | 5.80 c ±3.22 | 1.11 a ±0.60 |
| F | 4.04 c ±1.68 | 4.38 c ±1.91 | 1.85 ab ±1.19 | 4.84 bc ±3.48 | 4.34 c ±3.11 | 1.15 a ±0.82 | ||
| GST activity (U/g prot) | M/F | 5083 ±1768 | 3915 ±1045 | 4466 ±1710 | 4038 ±1606 | 3813 ±1002 | 4113 ±1085 | |
| Antioxidant system | GSH content (µmol/g prot) | M | 7.79 b ±3.57 | 5.26 ab ±2.47 | 6.58 ab ±3.46 | 2.44 a ±3.19 | 5.55 ab ±2.68 | 7.13 b ±2.80 |
| F | 8.04 c ±5.80 | 3.12 ab ±2.41 | 6.04 bc ±3.11 | 1.60 a ±1.71 | 5.19 bc ±2.37 | 4.60 abc ±2.86 | ||
| GPx activity (U/g prot) | M | 17.49 ±4.76 | 14.74 ±5.21 | 17.63 ±11.01 | 12.53 ±6.64 | 18.57 ±4.66 | 16.14 ±5.52 | |
| F | 50.22 a ±25.77 | 32.49 a ±15.15 | 50.58 a ±21.29 | 29.55 a ±13.58 | 41.23 a ±17.00 | 34.57 a ±16.66 | ||
| SOD activity (U/g prot × 103) | M/F | 3.14 ±0.78 | 3.06 ±0.66 | 2.82 ±1.09 | 3.06 ±1.13 | 3.01 ±1.06 | 3.42 ±0.56 | |
| Catalase activity (U/g prot × 105) | M | 4.59 b ±2.42 | 2.99 ab ±1.81 | 2.01 a ±1.70 | 2.27 ab ±1.52 | 3.08 ab ±1.84 | 4.97 b ±2.47 | |
| F | 7.84 ab ±2.52 | 7.97 ab ±4.40 | 5.46 ab ±2.25 | 3.57 a ±2.22 | 6.32 ab ±2.32 | 8.94 b ±4.79 | ||
| Cell integrity | TBARS (nmol/g prot) | M | 131.87 ±37.25 | 130.14 ±32.40 | 153.09 ±33.30 | 142.59 ±27.82 | 122.70 ±29.66 | 153.37 ±56.04 |
| F | 82.87 a ±29.87 | 123.58 ab ±48.95 | 143.19 b ±23.78 | 134.90 ab ±55.11 | 118.51 ab ±48.65 | 143.49 b ±37.68 | ||
| Erythrocyte density | Fresh erythrocyte (cell/mm3 × 105) | M/F | 2.89 bc ±1.14 | 3.21 c ±0.88 | 2.42 b ±0.47 | 1.56 a ±0.67 | 2.01 a ±0.23 | 1.99 a ±0.79 |
| Erythrocyte mortality | Erythrocyte necrosis (%) | M | 9.21 bc ±7.50 | 2.08 a ±2.28 | 1.65 a ±0.75 | 7.19 bc ±7.17 | 2.66 ab ±1.39 | 11.99 c ±7.09 |
| F | 1.56 a ±0.67 | 1.34 a ±0.57 | 1.39 a ±0.80 | 2.13 a ±1.25 | 2.10 a ±0.69 | 6.11 b ±3.58 | ||
| Genotoxicity | DNA strand breaks (tail intensity %) | M/F | 28.74 ab ±16.92 | 38.78 b ±19.69 | 25.28 ab ±13.69 | 22.19 ab ±10.53 | 18.94 a ±7.85 | 22.36 a ±12.70 |
| Chromosomal damage | M/F | 2.41 b ±2.35 | 2.41 b ±2.19 | 2.31 b ±2.78 | −0.60 a ±2.18 | −2.34 a ±1.35 | 3.87 b ±4.72 |
| Cligneux | Rhonelle | Scarpe | Deule | Sensée | |||
|---|---|---|---|---|---|---|---|
| St Rémy du Nord | Artres | Biache-St-Vaast | Courrières | Bouchain | Etaing | ||
| Biometric, biochemical and immune biomarkers | Cluster 1 | - | - | - | 100 | 58.6 | - |
| Cluster 2 | - | 7.1 | 67.8 | - | 13.8 | 44.8 | |
| Cluster 3 | 53.6 | 57.1 | 17.8 | - | 20.7 | 37.9 | |
| Cluster 4 | 46.4 | 37.7 | 14.3 | - | 6.9 | 17.2 | |
| Genotoxic, biometric, biochemical and immune biomarkers | Cluster 1 | - | - | - | 100 | 62.1 | - |
| Cluster 2 | 7.1 | 25 | 32.1 | - | 24.1 | 31.0 | |
| Cluster 3 | 21.4 | 39.3 | 35.7 | - | 3.5 | - | |
| Cluster 4 | 46.4 | 32.1 | 28.6 | - | 10.3 | 17.2 | |
| Cluster 5 | 25 | 3.6 | 3.6 | - | - | 51.7 | |
| Biometric, Biochemical and Immune Biomarkers | Genotoxic, Biometric, Biochemical and Immune Biomarkers | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | ||
| Biometric index | HSI | - | −3.2 | 5.7 | −2.0 | - | −2.5 | 5.6 | −3.5 | |
| Reproductive system | GSI | - | −6.4 | 6.1 | - | - | −3.1 | 6.5 | −3.9 | |
| Innate immune responses | Leucocyte mortality | −3.9 | −3.4 | 3.7 | 2.9 | −4.1 | - | 5.7 | - | −2.2 |
| Granulocyte–macrophage | −5.6 | - | 5.3 | - | −5.5 | - | 3.8 | - | 2.1 | |
| Phagocytosis efficiency | −5.3 | 5.8 | - | - | −5.1 | 2.7 | 2.6 | - | - | |
| Lysosomal presence | 4.7 | 3.9 | −4.8 | −3.0 | 5.4 | - | - | - | −2.7 | |
| Respiratory burst index | 9.9 | −3.2 | −2.9 | −2.6 | 9.8 | - | −2.1 | −2.8 | −2.8 | |
| Neurotoxicity | Che activity | 5.8 | −2.3 | - | - | 5.9 | −3.7 | - | - | - |
| Metabolic detoxification | EROD activity | 2.4 | −4.9 | 2.3 | - | 2.1 | - | 2.0 | - | −2.3 |
| GST activity | - | −2.8 | - | 3.7 | - | −2.8 | - | 3.8 | - | |
| Antioxidant system | GSH content | −3.4 | - | - | - | −3.6 | - | - | - | 3.4 |
| GPx activity | - | −2.2 | −4.7 | 6.8 | - | −2.2 | −2.8 | 7.8 | −3.1 | |
| SOD activity | - | 5.1 | 2.5 | - | - | 5.1 | −4.1 | - | - | |
| CAT activity | - | −2.9 | −4.4 | 7.2 | - | −2.9 | −2.4 | 7.2 | - | |
| Cell integrity | TBARS content | - | 5.8 | - | −4.1 | - | −2.6 | 2.2 | −3.2 | 4.1 |
| Erythrocyte mortality | Erythrocyte necrosis | - | - | - | - | - | - | - | −2.7 | 8.6 |
| Genotoxicity | DNA strand breaks | / | / | / | / | −2.7 | −3.6 | 4.6 | - | - |
| Chromosomal damage | / | / | / | / | −5.9 | - | - | - | 5.4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cant, A.; Bonnard, M.; Porcher, J.-M.; Prygiel, J.; Catteau, A.; Delahaut, L.; Palluel, O.; Turiès, C.; Geffard, A.; Bado-Nilles, A. Integration of Genotoxic Biomarkers in Environmental Biomonitoring Analysis Using a Multi-Biomarker Approach in Three-Spined Stickleback (Gasterosteus aculeatus Linnaeus, 1758). Toxics 2022, 10, 101. https://doi.org/10.3390/toxics10030101
Cant A, Bonnard M, Porcher J-M, Prygiel J, Catteau A, Delahaut L, Palluel O, Turiès C, Geffard A, Bado-Nilles A. Integration of Genotoxic Biomarkers in Environmental Biomonitoring Analysis Using a Multi-Biomarker Approach in Three-Spined Stickleback (Gasterosteus aculeatus Linnaeus, 1758). Toxics. 2022; 10(3):101. https://doi.org/10.3390/toxics10030101
Chicago/Turabian StyleCant, Amélie, Marc Bonnard, Jean-Marc Porcher, Jean Prygiel, Audrey Catteau, Laurence Delahaut, Olivier Palluel, Cyril Turiès, Alain Geffard, and Anne Bado-Nilles. 2022. "Integration of Genotoxic Biomarkers in Environmental Biomonitoring Analysis Using a Multi-Biomarker Approach in Three-Spined Stickleback (Gasterosteus aculeatus Linnaeus, 1758)" Toxics 10, no. 3: 101. https://doi.org/10.3390/toxics10030101
APA StyleCant, A., Bonnard, M., Porcher, J.-M., Prygiel, J., Catteau, A., Delahaut, L., Palluel, O., Turiès, C., Geffard, A., & Bado-Nilles, A. (2022). Integration of Genotoxic Biomarkers in Environmental Biomonitoring Analysis Using a Multi-Biomarker Approach in Three-Spined Stickleback (Gasterosteus aculeatus Linnaeus, 1758). Toxics, 10(3), 101. https://doi.org/10.3390/toxics10030101