Ancestral Wheat Types Release Fewer Celiac Disease Related T Cell Epitopes than Common Wheat upon Ex Vivo Human Gastrointestinal Digestion
Abstract
1. Introduction
2. Materials and Methods
2.1. Wheat Sample Collection
2.2. Wheat Characterization
2.3. Ex vivo Digestion of Wheat Porridge
2.4. Peptide Profile by HPLC-ESI MS/MS
2.5. MS Analysis Spectra Identification
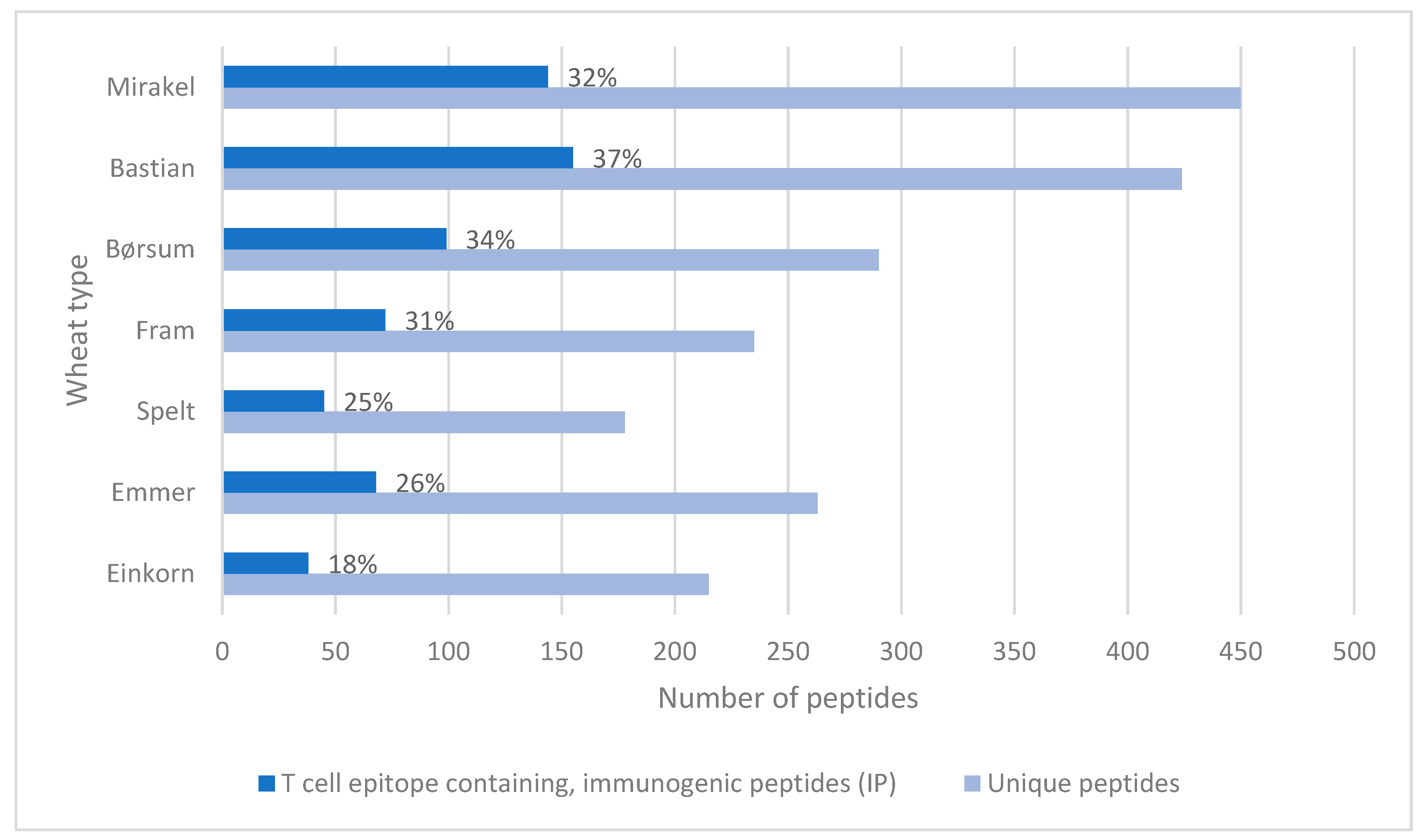
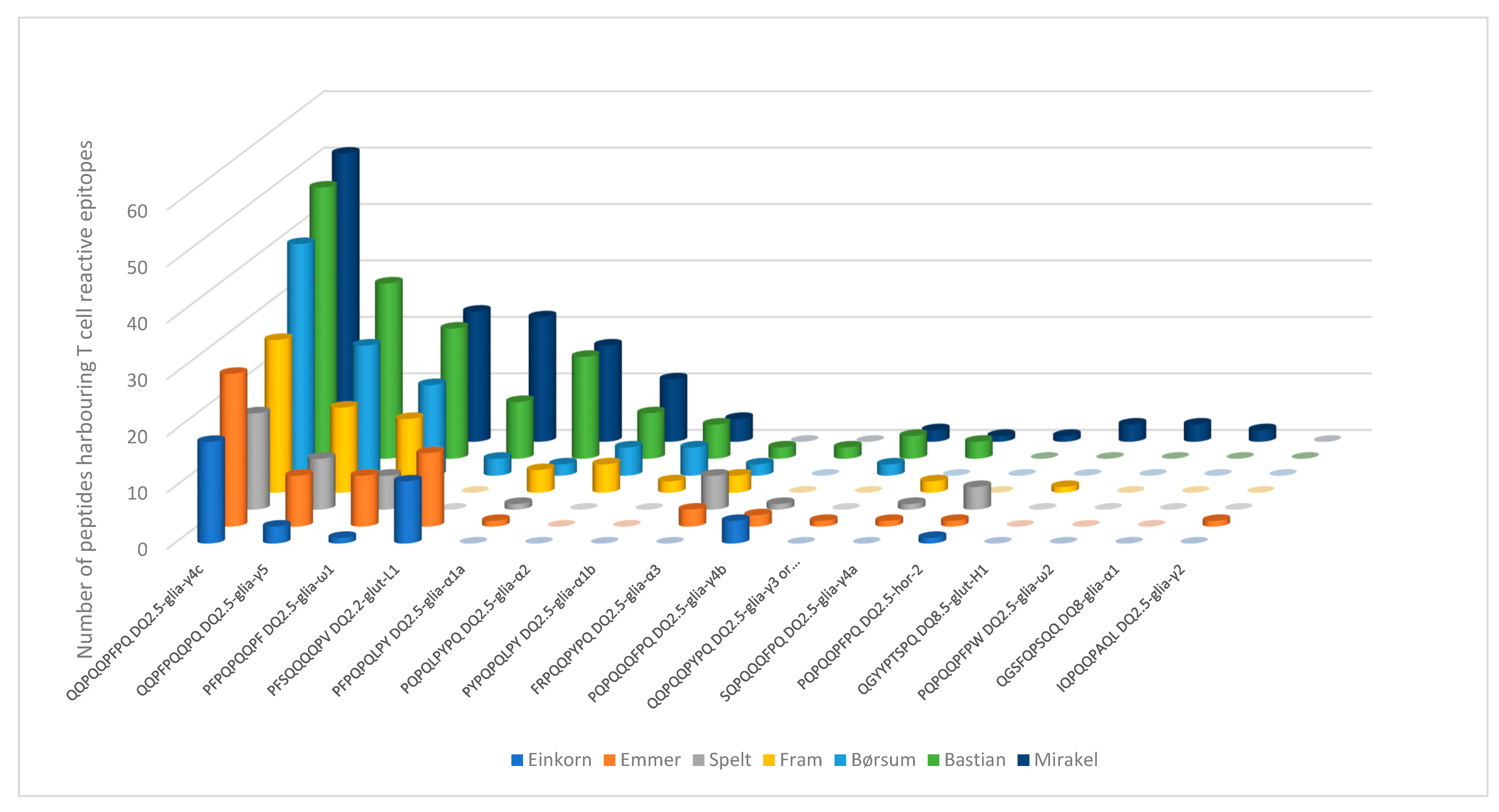
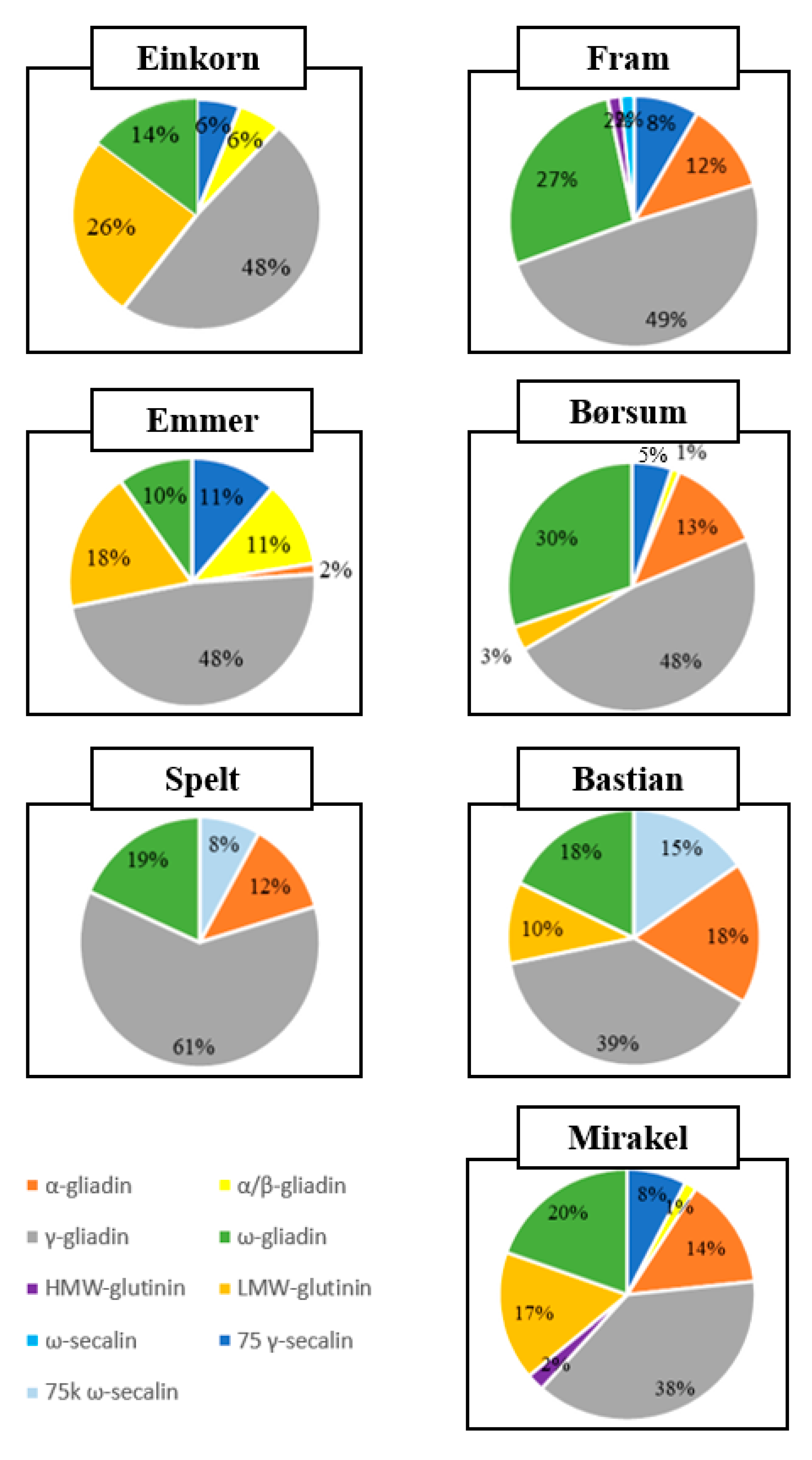
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lohi, S.; Mustalahti, K.; Kaukinen, K.; Laurila, K.; Collin, P.; Rissanen, H.; Lohi, O.; Bravi, E.; Gasparin, M.; Reunanen, A.; et al. Increasing prevalence of coeliac disease over time. Aliment. Pharmacol. Ther. 2007, 26, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Green, P.H.; Cellier, C. Celiac disease. New Engl. J. Med. 2007, 357, 1731–1743. [Google Scholar] [CrossRef] [PubMed]
- Katz, K.D.; Rashtak, S.; Lahr, B.D.; Melton, J.L.I.; Krause, P.K.; Maggi, K.; Talley, N.J.; Murray, J.A. Screening for celiac disease in a north american population: Sequential serology and gastrointestinal symptoms. Am. J. Gastroenterol. 2011, 106, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Schalk, K.; Lexhaller, B.; Koehler, P.; Scherf, K.A. Isolation and characterization of gluten protein types from wheat, rye, barley and oats for use as reference materials. PLoS ONE 2017, 12, e0172819. [Google Scholar] [CrossRef] [PubMed]
- Kagnoff, M.F. Celiac disease: Pathogenesis of a model immunogenetic disease. J. Clin. Investig. 2007, 117, 41–49. [Google Scholar] [CrossRef]
- Sollid, L.M. Coeliac disease: Dissecting a complex inflammatory disorder. Nat. Rev. Immunol. 2002, 2, 647. [Google Scholar] [CrossRef]
- Karell, K.; Louka, A.S.; Moodie, S.J.; Ascher, H.; Clot, F.; Greco, L.; Ciclitira, P.J.; Sollid, L.M.; Partanen, J.; Members of the European Genetics Cluster on Celiac Disease. HLA types in celiac disease patients not carrying the DQA1* 05-DQB1* 02 (DQ2) heterodimer: Results from the European Genetics Cluster on Celiac Disease. Hum. Immunol. 2003, 64, 469–477. [Google Scholar] [CrossRef]
- Trynka, G.; Hunt, K.A.; Bockett, N.A.; Romanos, J.; Mistry, V.; Szperl, A.; Bakker, S.F.; Bardella, M.T.; Bhaw-Rosun, L.; Castillejo, G.; et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011, 43, 1193. [Google Scholar] [CrossRef]
- Shewry, P.R.; Halford, N.G.; Lafiandra, D. Genetics of wheat gluten proteins. Adv. Genet. 2003, 49, 111–184. [Google Scholar]
- Shan, L.; Molberg, Ø.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef]
- Stepniak, D.; Vader, L.W.; Kooy, Y.; van Veelen, P.A.; Moustakas, A.; Papandreou, N.A.; Eliopoulos, E.; Drijfhout, J.W.; Papadopoulos, G.K.; Koning, F. T-cell recognition of HLA-DQ2-bound gluten peptides can be influenced by an N-terminal proline at p-1. Immunogenetics 2005, 57, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Arentz-Hansen, H.; Körner, R.; Molberg, Ø.; Quarsten, H.; Vader, W.; Kooy, Y.M.; Lundin, K.E.A.; Koning, F.; Roepstorff, P.; Sollid, L.M.; et al. The intestinal T cell response to α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J. Exp. Med. 2000, 191, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Sjostrom, H.; Lundin, K.E.; Molberg, O.; Korner, R.; McAdam, S.N.; Anthonsen, D.; Quarsten, H.; Norén, O.; Roepstorff, P.; Thorsby, E.; et al. Identification of a gliadin T-cell epitope in coeliac disease: General importance of gliadin deamidation for intestinal T-cell recognition. Scand. J. Immunol. 1998, 48, 111–115. [Google Scholar] [CrossRef]
- Gianfrani, C.; Camarca, A.; Mazzarella, G.; Di Stasio, L.; Giardullo, N.; Ferranti, P.; Picariello, G.; Aufiero, V.R.; Picascia, S.; Troncone, R.; et al. Extensive in vitro gastrointestinal digestion markedly reduces the immune-toxicity of triticum monococcum wheat: Implication for celiac disease. Mol. Nutr. Food Res. 2015, 59, 1844–1854. [Google Scholar] [CrossRef]
- Huang, S.; Sirikhachornkit, A.; Su, X.; Faris, J.; Gill, B.; Haselkorn, R.; Gornicki, P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 2002, 99, 8133–8138. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Liu, D.; Yang, W.; Kishii, M.; Mao, L. Synthetic hexaploid wheat: Yesterday, today, and tomorrow. Engineering 2018, 4, 552–558. [Google Scholar] [CrossRef]
- Wieser, H. Comparative investigations of gluten proteins from different wheat species. III. N-terminal amino acid sequences of α-gliadins potentially toxic for coeliac patients. Eur. Food Res. Technol. 2001, 213, 183–186. [Google Scholar]
- Deng, Y.; Gruppen, H.; Wierenga, P.A. Comparison of protein hydrolysis catalyzed by bovine, porcine, and human trypsins. J. Agric. Food Chem. 2018, 66, 4219–4232. [Google Scholar] [CrossRef]
- Sollid, L.M.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 2012, 64, 455–460. [Google Scholar] [CrossRef]
- McCleary, B.; Gibson, T.; Solah, V.; Mugford, D. Total starch measurement in cereal products: Interlaboratory evaluation of a rapid enzymic test procedure. Cereal Chem. 1994, 71, 501–504. [Google Scholar]
- Ulleberg, E.K.; Comi, I.; Holm, H.; Herud, E.B.; Jacobsen, M.; Vegarud, G.E. Human gastrointestinal juices intended for use in in vitro digestion models. Food Dig. 2011, 2, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Molberg, Ø.; Mcadam, S.N.; Körner, R.; Quarsten, H.; Kristiansen, C.; Madsen, L.; Fugger, L.; Scott, H.; Norén, O.; Roepstorff, P.; et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 1998, 4, 713. [Google Scholar] [CrossRef] [PubMed]
- van de Wal, Y.; Kooy, Y.; van Veelen, P.; Peña, S.; Mearin, L.; Papadopoulos, G.; Koning, F. Cutting edge: Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J. Immunol. 1998, 161, 1585–1588. [Google Scholar]
- Ciccocioppo, R.; Di Sabatino, A.; Corazza, G.R. The immune recognition of gluten in coeliac disease. Clin. Exp. Immunol. 2005, 140, 408–416. [Google Scholar] [CrossRef]
- Mamone, G.; Ferranti, P.; Rossi, M.; Roepstorff, P.; Fierro, O.; Malorni, A.; Addeo, F. Identification of a peptide from α-gliadin resistant to digestive enzymes: Implications for celiac disease. J. Chromatogr. B 2007, 855, 236–241. [Google Scholar] [CrossRef]
- Gianfrani, C.; Siciliano, R.A.; Facchiano, A.M.; Camarca, A.; Mazzeo, M.F.; Costantini, S.; Salvati, V.M.; Maurano, F.; Mazzarella, G.; Iaquinto, G.; et al. Transamidation of wheat flour inhibits the response to gliadin of intestinal T cells in celiac disease. Gastroenterology 2007, 133, 780–789. [Google Scholar] [CrossRef]
- Asledottir, T.; Picariello, G.; Mamone, G.; Ferranti, P.; Roseth, A.; Devold, T.G.; Vegarud, G.E. Degradation of beta-casomorphin-7 through in vitro gastrointestinal and jejunal brush border membrane digestion. J. Dairy Sci. 2019, 102, 8622–8629. [Google Scholar] [CrossRef]
- Prandi, B.; Tedeschi, T.; Folloni, S.; Galaverna, G.; Sforza, S. Peptides from gluten digestion: A comparison between old and modern wheat varieties. Food Res. Int. 2017, 91, 92–102. [Google Scholar] [CrossRef]
- Malalgoda, M.; Meinhardt, S.W.; Simsek, S. Detection and quantitation of immunogenic epitopes related to celiac disease in historical and modern hard red spring wheat cultivars. Food Chem. 2018, 264, 101–107. [Google Scholar] [CrossRef]
- Molberg, Ø.; Uhlen, A.K.; Sollid, L.M.; Jensen, T.; Flæte, N.S.; Arntz-Hansen, H.; Raki, M.; Lundin, K.E.A.; Sollid, L.M. Mapping of gluten T-cell epitopes in the bread wheat ancestors: Implications on celiac disease. Gastoenterology 2005, 128, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Dorum, S.; Steinsbo, O.; Bergseng, E.; Arntzen, M.O.; de Souza, G.A.; Sollid, L.M. Gluten-specific antibodies of celiac disease gut plasma cells recognize long proteolytic fragments that typically harbor T-cell epitopes. Sci Rep. 2016, 6, 25565. [Google Scholar] [CrossRef] [PubMed]
- Hüe, S.; Mention, J.-J.; Monteiro, R.C.; Zhang, S.; Cellier, C.; Schmitz, J.; Verkarre, V.; Fodil, N.; Bahram, S.; Cerf-Bensussan, N.; et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity 2004, 21, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, L.; Ciacci, C.; Ricciardelli, I.; Vacca, L.; Raia, V.; Auricchio, S.; Picard, J.; Osman, M.; Quaratino, S.; Londei, P.M. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 2003, 362, 30–37. [Google Scholar] [CrossRef]
- Barone, M.V.; Zanzi, D.; Maglio, M.; Nanayakkara, M.; Santagata, S.; Lania, G.; Miele, E.; Ribecco, M.T.S.; Maurano, F.; Auricchio, R.; et al. Gliadin-mediated proliferation and innate immune activation in celiac disease are due to alterations in vesicular trafficking. PLoS ONE 2011, 6, e17039. [Google Scholar] [CrossRef]
- Aronsson, C.A.; Lee, H.S.; Koletzko, S.; Uusitalo, U.; Yang, J.; Virtanen, S.M.; Liu, E.; Lernmark, Å.; Norris, J.M.; Agardh, D. Effects of gluten intake on risk of celiac disease: A case-control study on a Swedish birth cohort. Clin. Gastroenterol. Hepatol. 2016, 14, 403–409. [Google Scholar] [CrossRef]

| Wheat Type | Species | Genome | Variety | Breeding Company/Origin | Marked Release |
|---|---|---|---|---|---|
| Einkorn | T. monococcum | AA | Unknown | ||
| Emmer | T. dicoccon | AABB | Gotland | ||
| Spelt | T. aestivum var. spelta | AABBDD | Vit Gotland | ||
| Common wheat | T. aestivum var. aestivum | AABBDD | Fram | Norwegian landrace | Before 1900 |
| AABBDD | Børsum | Norwegian Agricultural University (NLH) | 1936 | ||
| AABBDD | Bastian | Graminor, Norway | 1989 | ||
| AABBDD | Mirakel | Graminor, Norway | 2012 |
| Wheat Sample | TKW (g) | Protein (%) | Starch (%) |
|---|---|---|---|
| Einkorn | 30.5 | 10.3 | 66.5 |
| Emmer | 31.7 | 11.0 | 50.0 |
| Spelt | 41.1 | 10.6 | 60.0 |
| Fram | 32.3 | 8.2 | 63.1 |
| Børsum | 31.9 | 9.2 | 55.6 |
| Bastian | 33.1 | 10.2 | 58.8 |
| Mirakel | 38.5 | 9.1 | 63.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asledottir, T.; Rehman, R.; Mamone, G.; Picariello, G.; Devold, T.G.; Vegarud, G.E.; Røseth, A.; Lea, T.E.; Halstensen, T.S.; Ferranti, P.; et al. Ancestral Wheat Types Release Fewer Celiac Disease Related T Cell Epitopes than Common Wheat upon Ex Vivo Human Gastrointestinal Digestion. Foods 2020, 9, 1173. https://doi.org/10.3390/foods9091173
Asledottir T, Rehman R, Mamone G, Picariello G, Devold TG, Vegarud GE, Røseth A, Lea TE, Halstensen TS, Ferranti P, et al. Ancestral Wheat Types Release Fewer Celiac Disease Related T Cell Epitopes than Common Wheat upon Ex Vivo Human Gastrointestinal Digestion. Foods. 2020; 9(9):1173. https://doi.org/10.3390/foods9091173
Chicago/Turabian StyleAsledottir, Tora, Rashida Rehman, Gianfranco Mamone, Gianluca Picariello, Tove Gulbrandsen Devold, Gerd Elisabeth Vegarud, Arne Røseth, Tor Erling Lea, Trond S. Halstensen, Pasquale Ferranti, and et al. 2020. "Ancestral Wheat Types Release Fewer Celiac Disease Related T Cell Epitopes than Common Wheat upon Ex Vivo Human Gastrointestinal Digestion" Foods 9, no. 9: 1173. https://doi.org/10.3390/foods9091173
APA StyleAsledottir, T., Rehman, R., Mamone, G., Picariello, G., Devold, T. G., Vegarud, G. E., Røseth, A., Lea, T. E., Halstensen, T. S., Ferranti, P., & Uhlen, A. K. (2020). Ancestral Wheat Types Release Fewer Celiac Disease Related T Cell Epitopes than Common Wheat upon Ex Vivo Human Gastrointestinal Digestion. Foods, 9(9), 1173. https://doi.org/10.3390/foods9091173






