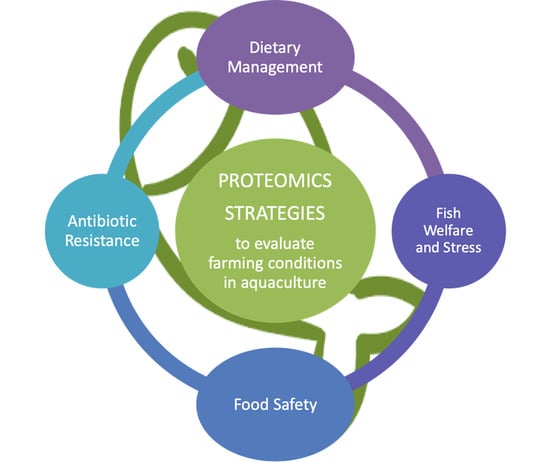
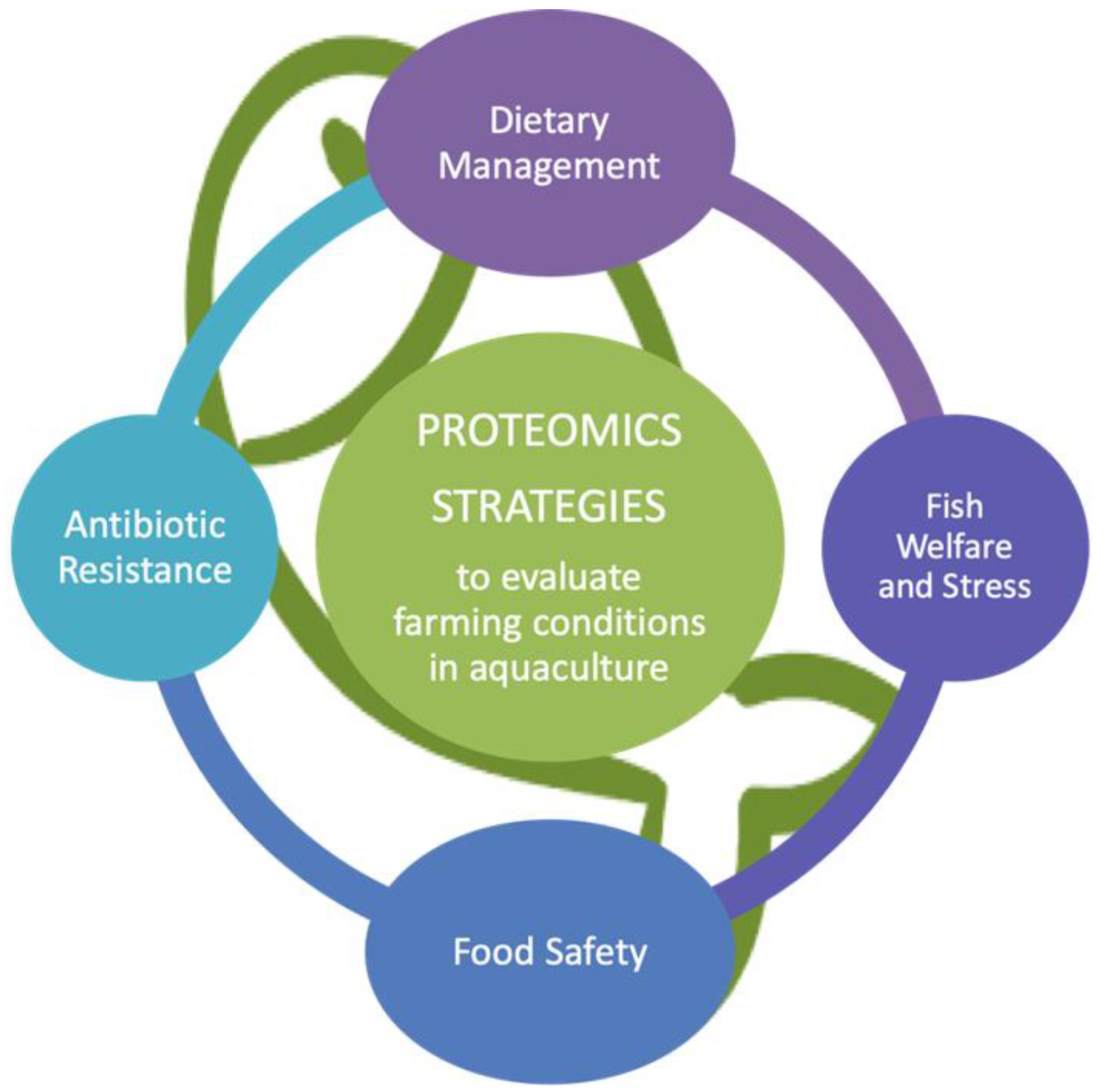
Proteomic Strategies to Evaluate the Impact of Farming Conditions on Food Quality and Safety in Aquaculture Products
Abstract
1. Introduction to Proteomics in Aquaculture
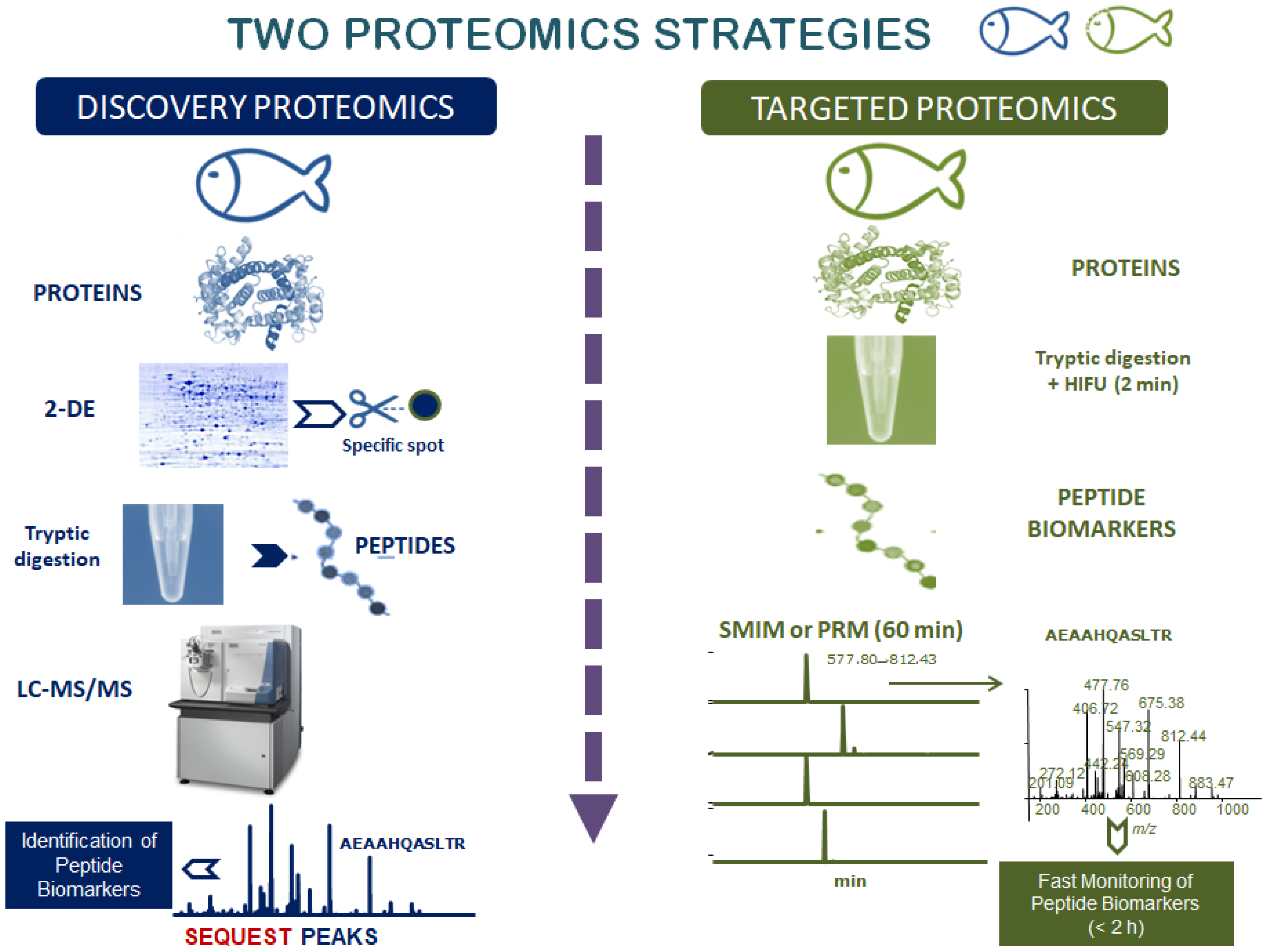
2. Workflow of Proteomics: Discovery and Targeted Proteomics
3. Application of Proteomics to Evaluate the Farming Conditions on Food Quality and Safety in Aquaculture Products
3.1. Dietary Management in Aquaculture
3.2. Fish Welfare and Stress Response in Aquaculture
3.3. Food Safety in Aquaculture
3.3.1. Food Safety in Aquaculture: Biotic Hazards
3.3.2. Food Safety in Aquaculture: Abiotic Hazards
3.4. Antibiotic Resistance in Aquaculture
4. Concluding Remarks and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). The State of World Fisheries and Aquaculture 2018: Meeting the Sustainable Development Goals; Food and Agriculture Organization of the United Nations: Rome, Italy, 2008. [Google Scholar]
- Chintagari, S.; Hazard, N.; Edwards, H.G.; Jadeja, R.; Janes, M. Risks associated with fish and seafood. Microbiol. Spectr. 2017, 5, 1–16. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Zolla, L. We are what we eat: Food safety and proteomics. J. Proteome Res. 2012, 11, 26–36. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Silva, T.S.; Dias, J.; Jessen, F. Proteomics in aquaculture: Applications and trends. J. Proteom. 2012, 75, 4325–4345. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Campos, A.; Kuruvilla, J.; Schrama, D.; Cristobal, S. Proteomics in Aquaculture: Quality and safety. In Proteomics in Food Science, from Farm to Fork; Colgrave, M.L., Ed.; Elsevier: London, UK, 2017; pp. 279–290. [Google Scholar]
- Pandey, A.; Mann, M. Proteomics to study genes and genomes. Nature 2000, 405, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.; Cañas, B.; Gallardo, J.M. Proteomics for the assessment of quality and safety of fishery products. Food Res. Int. 2013, 54, 972–979. [Google Scholar] [CrossRef]
- Piñeiro, C.; Carrera, M.; Cañas, B.; Lekube, X.; Martínez, I. Proteomics and food analysis: Principles, techniques and applications. In Handbook of Food Analysis-Two Volume Set; CRC Press: Boca Raton, FL, USA, 2015; pp. 393–416. [Google Scholar]
- Holton, T.A.; Vijayakumar, V.; Khaldi, N. Bioinformatics: Current perspectives and future directions for food and nutritional research facilitated by a Food-Wiki database. Trends Food Sci. Technol. 2013, 34, 5–17. [Google Scholar] [CrossRef]
- Gallardo, J.M.; Carrera, M.; Ortea, I. Proteomics in food science. In Foodomics: Advanced Mass Spectrometry in Modern Food Science and Nutrition; Cifuentes, A., Ed.; JohnWiley & Sons Inc.: Hoboken, NJ, USA, 2013; pp. 125–165. [Google Scholar]
- Carrera, M.; Mateos, J.; Gallardo, J.M. Data treatment in food proteomics. In Reference Module in Food Science; Elsevier: London, UK, 2019. [Google Scholar] [CrossRef]
- Rabilloud, T.; Lelong, C. Two-dimensional gel electrophoresis in proteomics: A tutorial. J. Proteom. 2011, 74, 1829–1841. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Piñeiro, C.; Vázquez, J.; Gallardo, J.M. De novo mass spectrometry sequencing and characterization of species-specific peptides from nucleoside diphosphate kinase B for the classification of commercial fish species belonging to the family Merlucciidae. J. Proteome Res. 2007, 6, 3070–3080. [Google Scholar] [CrossRef]
- Wolters, D.A.; Washburn, M.P.; Yates, J.R., 3rd. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001, 73, 5683–5690. [Google Scholar] [CrossRef]
- Carrera, M.; Ezquerra-Brauer, J.M.; Aubourg, S.P. Characterization of the jumbo squid (Dosidicus gigas) skin by-product by shotgun proteomics and protein-based bioinformatics. Mar. Drugs 2019, 18, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.C.; Yates, J.R., 3rd. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Eng, J.K.; McCormack, A.L.; Yates, J.R.I.I.I. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Kall, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Mann, M. Peptide sequencing by mass spectrometry for homology searches and cloning of genes. J. Protein Chem. 1997, 16, 481–490. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Scigelova, M.; Maroto, F.; Dufresne, C.; Vázquez, J. High Throughput de novo Sequencing. 2007. Available online: http://www.thermo.com/ (accessed on 23 July 2020).
- Carrera, M.; Cañas, B.; Vázquez, J.; Gallardo, J.M. Extensive de novo sequencing of new parvalbumin isoforms using a novel combination of bottom-up proteomics, accurate molecular mass measurement by FTICR-MS, and selected MS/MS ion monitoring. J. Proteome Res. 2010, 9, 4393–4406. [Google Scholar] [CrossRef]
- Ortea, I.; Cañas, B.; Gallardo, J.M. Mass spectrometry characterization of species-specific peptides from arginine kinase for the identification of commercially relevant shrimp species. J. Proteome Res. 2009, 8, 5356–5362. [Google Scholar] [CrossRef]
- Ong, S.E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labelling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef]
- Mateos, J.; Landeira-Abia, A.; Fafián-Labora, J.A.; Fernández-Pernas, P.; Lesende-Rodríguez, I.; Fernández-Puente, P.; Fernández-Moreno, M.; Delmiro, A.; Martín, M.A.; Blanco, F.J.; et al. iTRAQ-based analysis of progerin expression reveals mitocondrial dysfunction, reactive oxygen species accumulation and altered proteostasis. Stem Cell Res. Ther. 2015, 6, 119. [Google Scholar] [CrossRef]
- Robotti, E.; Marengo, E. 2D-DIGE and fluorescence image analysis. Methods Mol. Biol. 2018, 1664, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Stryiński, R.; Mateos, J.; Pascual, S.; González, A.F.; Gallardo, J.M.; Łopieńska-Biernat, E.; Medina, I.; Carrera, M. Proteome profiling of L3 and L4 Anisakis simplex development stages by TMT-based quantitative proteomics. J. Proteom. 2019, 201, 1–11. [Google Scholar] [CrossRef]
- López-Ferrer, D.; Ramos-Fernández, A.; Martínez-Bartolomé, S.; García-Ruiz, P.; Vázquez, J. Quantitative proteomics using 16O/18O labeling and linear ion trap mass spectrometry. Proteomics 2006, 6 (Suppl. S1), S4–S11. [Google Scholar] [CrossRef]
- Mueller, L.N.; Rinner, O.; Schmidt, A.; Letarte, S.; Bodenmiller, B.; Brusniak, M.Y.; Vitek, O.; Aebersold, R.; Müller, M. SuperHirn—A novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 2007, 7, 3470–3480. [Google Scholar] [CrossRef] [PubMed]
- Borràs, E.; Sabidó, E. What is targeted proteomics? A concise revision of targeted acquisition and targeted data analysis in mass spectrometry. Proteomics 2017, 17, 17–18. [Google Scholar] [CrossRef]
- Aebersold, R.; Bensimon, A.; Collins, B.C.; Ludwig, C.; Sabido, E. Applications and developments in targeted proteomics: From SRM to DIA/SWATH. Proteomics 2016, 16, 2065–2067. [Google Scholar] [CrossRef] [PubMed]
- Lange, V.; Picotti, P.; Domon, B.; Aebersold, R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jorge, I.; Casas, E.M.; Villar, M.; Ortega-Pérez, I.; López-Ferrer, D.; Martínez-Ruiz, A.; Carrera, M.; Marina, A.; Martínez, P.; Serrano, H.; et al. High-sensitivity analysis of specific peptides in complex samples by selected MS/MS ion monitoring and linear ion trap mass spectrometry: Application to biological studies. J. Mass Spectrom. 2007, 42, 1391–1403. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; López-Ferrer, D.; Piñeiro, C.; Vázquez, J.; Gallardo, J.M. Fast monitoring of species-specific peptide biomarkers using high-intensity-focused-ultrasound-assisted tryptic digestion and selected MS/MS ion monitoring. Anal. Chem. 2011, 83, 5688–5695. [Google Scholar] [CrossRef]
- Carrera, M.; Gallardo, J.M.; Pascual, S.; González, A.F.; Medina, I. Protein biomarker discovery and fast monitoring for the identification and detection of Anisakids by parallel reaction monitoring (PRM) mass spectrometry. J. Proteom. 2016, 142, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11, 016717. [Google Scholar] [CrossRef] [PubMed]
- Beynon, R.J.; Doherty, M.K.; Pratt, J.M.; Gaskell, S.J. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2005, 2, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Röst, H.; Malmström, L.; Aebersold, R. A computational tool to detect and avoid redundancy in selected reaction monitoring. Mol. Cell. Proteom. 2012, 11, 540–549. [Google Scholar] [CrossRef]
- Bereman, M.S.; MacLean, B.; Tomazela, D.M.; Liebler, D.C.; MacCoss, M.J. The development of selected reaction monitoring methods for targeted proteomics via empirical refinement. Proteomics 2012, 12, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Carpene, E.; Martin, B.; Dalla Libera, L. Biochemical differences in lateral muscle of wild and farmed gilthead sea bream (series Sparus aurata L.). Fish Physiol. Biochem. 1998, 19, 229–238. [Google Scholar] [CrossRef]
- Martinez, I.; Standal, I.B.; Aursand, M.; Yamashita, Y.; Yamashita, M. Analytical Methods to differentiate farmed from wild seafood. In Handbook of Seafood and Seafood Products Analysis; Nollet, L., Toldrá, F., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 215–232. [Google Scholar]
- Chiozzi, R.Z.; Capriotti, A.L.; Cavalieri, C.; La Barbera, G.; Montone, C.M.; Piovesana, S.; Lagana, A. Label-Free shotgun proteomics approach to characterize muscle tissue from farmed and wild European sea bass (Dicentrarchus labrax). Food Anal. Method. 2018, 11, 292–301. [Google Scholar] [CrossRef]
- Torstensen, B.E.; Espe, M.; Sanden, M.; Stubhaug, I.; Waagbø, R.; Hemre, G.I.; Fontanillas, F.; Nordgarden, U.; Hevrøy, E.M.; Olsvik, P.; et al. Novel production of Atlantic salmon (Salmo salar) protein based on combined replacement of fish meal and fish oil with plant meal and vegetable oil blends. Aquaculture 2008, 285, 193–200. [Google Scholar] [CrossRef]
- Dalmo, R.A.; Bøgwald, J. ß-glucans as conductors of immune symphonies. Fish Shellfish Immunol. 2008, 25, 384–396. [Google Scholar] [CrossRef]
- Ghaedi, G.; Keyvanshokooh, S.; Mohammadi Azarm, H.; Akhlaghi, M. Proteomic analysis of muscle tissue from rainbow trout (Oncorhynchus mykiss) fed dietary β-glucan. Iran. J. Vet. Res. 2016, 17, 184–189. [Google Scholar]
- Martin, S.A.M.; Vilhelmsson, O.; Médale, F.; Watt, P.; Kaushik, S.; Houlihan, D.F. Proteomic sensitivity to dietary manipulations in rainbow trout. Biochim. Biophys. Acta 2003, 1651, 17–29. [Google Scholar] [CrossRef]
- Estruch, G.; Martínez-Llorens, S.; Tomás-Vidal, A.; Monge-Ortiz, R.; Jover-Cerdá, M.; Brown, P.B.; Peñaranda, D.S. Impact of high dietary plant protein with or without marine ingredients in gut musosa proteome of gilthead seabream (Sparus aurata, L.). J. Proteom. 2020, 216, 103672. [Google Scholar] [CrossRef] [PubMed]
- Nasopoulou, C.; Zabetakis, I. Benefits of fish oil replacement by plant originated oils in compounded fish feeds. A review. LWT 2012, 47, 217–224. [Google Scholar] [CrossRef]
- Morais, S.; Silva, T.; Cordeiro, O.; Rodrigues, P.; Guy, D.R.; Bron, J.E.; Taggart, J.B.; Bell, J.G.; Tocher, D.R. Effects of genotype and dietary fish oil replacement with vegetable oil on the intestinal transcriptome and proteome of Atlantic salmon (Salmo salar). BMC Genom. 2012, 13, 448. [Google Scholar] [CrossRef]
- Monti, G.; De Napoli, L.; Mainolfi, P.; Barone, R.; Guida, M.; Marino, G.; Amoresano, A. Monitoring food quality by microfluidic electrophoresis, gas chromatography, and mass spectrometry techniques: Effects of aquaculture on the sea bass (Dicentrarchus labrax). Anal. Chem. 2005, 77, 2587–2594. [Google Scholar] [CrossRef]
- Belghit, I.; Lock, E.J.; Fumière, O.; Lecrenier, M.C.; Renard, P.; Dieu, M.; Berntssen, M.H.G.; Palmblad, M.; Rasinger, J.D. Species-specific discrimination of insect meals for aquafeeds by direct comparison of tandem mass spectra. Animals 2019, 9, 222. [Google Scholar] [CrossRef]
- Marco-Ramell, A.; de Almeida, A.M.; Cristobal, S.; Rodrigues, P.; Roncada, P.; Bassols, A. Proteomics and the search for welfare and stress biomarkers in animal production in the one-health context. Mol. Biosyst. 2016, 12, 2024–2035. [Google Scholar] [CrossRef]
- Raposo de Magalhães, C.; Schrama, D.; Farinha, A.P.; Revets, D.; Kuehn, A.; Planchon, S.; Rodrigues, P.M.; Marco Cerqueira, M.A. Protein changes as robust signatures of fish chronic stress: A proteomics approach in fish welfare research. BMC Genom. 2020, 21, 309. [Google Scholar] [CrossRef]
- Mommsen, T.P.; Vijayan, M.M.; Moon, T.W. Cortisol in teleosts: Dynamics, mechanisms of action, and metabolic regulation. Rev. Fish Biol. Fish. 1999, 9, 211–268. [Google Scholar] [CrossRef]
- Torres-Velarde, J.; Llera-Herrera, R.; García-Gasca, T.; García-Gasca, A. Mechanisms of stress-related muscle atrophy in fish: An ex vivo approach. Mech. Dev. 2018, 154, 162–169. [Google Scholar] [CrossRef]
- Christiansen, J.S.; Ringø, E.; Jobling, M. Effects of sustained exercise on growth and body composition of first-feeding fry of Arctic charr, Salvelinus alpinus (L.). Aquaculture 1989, 79, 329–335. [Google Scholar] [CrossRef]
- Christiansen, J.S.; Jobling, M. The behaviour and the relationship between food intake and growth of juvenile Arctic charr, Salvelinus alpinus L. subjected to sustained exercise. Can. J. Zool. 1990, 68, 2185–2191. [Google Scholar] [CrossRef]
- Jobling, M.; Baardvik, B.M.; Christiansen, J.S.; Jørgensen, E.H. The effects of prolonged exercise training on growth performance and production parameters in fish. Aquac. Int. 1993, 1, 95–111. [Google Scholar] [CrossRef]
- Eguiraun, H.; Casquero, O.; Sørensen, A.J.; Martinez, I. Reducing the number of individuals to monitor shoaling fish systems—Application of the Shannon entropy to construct a biological warning system model. Front. Physiol. 2018, 9, 493. [Google Scholar] [CrossRef]
- Christiansen, J.S.; Martinez, I.; Jobling, M.; Amin, A. Rapid somatic growth and muscle damage in a salmonid fish. Basic Appl. Myol. 1992, 2, 235–239. [Google Scholar]
- Stickland, N.C. Growth and development of muscle fibres in the rainbow trout (Salmo gairdneri). J. Anat. 1983, 137, 323–333. [Google Scholar] [PubMed]
- Rossi, G.; Messina, G. Comparative myogenesis in teleosts and mammals. Cell. Mol. Life Sci. 2014, 71, 3081–3099. [Google Scholar] [CrossRef] [PubMed]
- Nemova, N.N.; Lysenko, L.A.; Kantserova, N.P. Degradation of skeletal muscle protein during growth and development of salmonid fish. Russ. J. Dev. Biol. 2016, 47, 161–172. [Google Scholar] [CrossRef]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Perelló, M.; Salmerón, C.; Riera-Codina, M.; Ibarz, A.; Fernández-Borràs, J.; Blasco, J.; Capilla, E.; et al. Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture 2017, 467, 28–40. [Google Scholar] [CrossRef]
- Stockdale, F.E. Myogenic cell lineages. Dev. Biol. 1992, 154, 284–298. [Google Scholar] [CrossRef]
- Bigard, A.X.; Janmot, C.; Sanchez, H.; Serrurier, B.; Pollet, S.; d’Albis, A. Changes in myosin heavy chain profile of mature regenerated muscle with endurance training in rat. Acta Physiol. Scand. 1999, 165, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Cappuccinelli, R.; Tedde, V.; Pagnozzi, D.; Porcu, M.C.; Bonaglini, E.; Roggio, T.; Uzzau, S. Proteomic analysis of muscle tissue from gilthead sea bream (Sparus aurata, L.) farmed in offshore floating cages. Aquaculture 2010, 309, 245–252. [Google Scholar] [CrossRef]
- Silva, T.S.; Cordeiro, O.D.; Matos, E.D.; Wulff, T.; Dias, J.P.; Jessen, F.; Rodrigues, P.M. Effects of preslaughter stress levels on the post-mortem sarcoplasmic proteomic profile of gilthead seabream muscle. J. Agric. Food Chem. 2012, 60, 9443–9453. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.N.; Zheng, G.D.; Wu, C.B.; Jiang, X.Y.; Zou, S.M. Identification of proteins differentially expressed in the gills of grass carp (Ctenopharyngodon idella) after hypoxic stress by two-dimensional gel electrophoresis analysis. Fish Physiol. Biochem. 2019, 45, 743–752. [Google Scholar] [CrossRef]
- Timmins-Schiffman, E.B.; Crandall, G.A.; Vadopalas, B.; Riffle, M.E.; Nunn, B.L.; Roberts, S.B. Integrating discovery-driven proteomics and selected reaction monitoring to develop a noninvasive assay for geoduck reproductive maturation. J. Proteome Res. 2017, 16, 3298–3309. [Google Scholar] [CrossRef]
- Spencer, L.H.; Horwith, M.; Lowe, A.T.; Venkataraman, Y.R.; Timmins-Schiffman, E.; Nunn, B.L.; Roberts, S.B. Pacific geoduck (Panopea generosa) resilience to natural pH variation. Comp. Biochem. Physiol. Part D Genomics Proteom. 2019, 30, 91–101. [Google Scholar] [CrossRef]
- Freitas, J.; Vaz-Pires, P.; Câmara, J.S. From aquaculture production to consumption: Freshness, safety, traceability and authentication, the four pillars of quality. Aquaculture 2020, 518, 734857. [Google Scholar] [CrossRef]
- Teklemariam, A.D.; Tessema, F.; Abayneh, T. Review on evaluation of safety of fish and fish products. Int. J. Fish Aquat. Stud. 2015, 3, 111–117. [Google Scholar]
- Huss, H.H. Assurance of Seafood Quality. In FAO Fishery Technical Paper No. 334; FAO: Rome, Italy, 1994; p. 169. [Google Scholar]
- Jahncke, M.L.; Schwarz, M.H. Public, animal and environmental aquaculture health issues in industrialized countries. In Public, Animal and Environmental Aquaculture Health Issues; Jahncke, M., Garrett, E.S., Reilly, A., Martin, R.E., Cole, E., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 2002; pp. 67–102. [Google Scholar]
- Carrera, M.; Böhme, K.; Gallardo, J.M.; Barros-Velázquez, J.; Cañas, B.; Calo-Mata, P. Characterization of foodborne strains of Staphylococcus aureus by shotgun proteomics: Functional networks, virulence factors and species-specific peptide biomarkers. Front. Microbiol. 2017, 8, 2458. [Google Scholar] [CrossRef]
- Hazen, T.H.; Martinez, R.J.; Chen, Y.F.; Lafon, P.C.; Garrett, N.M.; Parsons, M.B.; Bopp, C.A.; Sullards, M.C.; Sobecky, P.A. Rapid identification of Vibrio parahaemolyticus by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2009, 75, 6745–6756. [Google Scholar] [CrossRef]
- Kazazić, S.P.; Topić Popović, N.; Strunjak-Perović, I.; Babić, S.; Florio, D.; Fioravanti, M.; Bojanić, K.; Čož-Rakovac, R. Matrix-assisted laser desorption/ionization time of flight mass spectrometry identification of Vibrio (Listonella) anguillarum isolated from sea bass and sea bream. PLoS ONE 2019, 14, e0225343. [Google Scholar] [CrossRef] [PubMed]
- Li, J.N.; Zhao, Y.T.; Cao, S.L.; Wang, H.; Zhang, J.J. Integrated transcriptomic and proteomic analyses of grass carp intestines after vaccination with a double-targeted DNA vaccine of Vibrio mimicus. Fish Shellfish Immunol. 2020, 98, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Böhme, K.; Fernández-No, I.C.; Barros-Velázquez, J.; Gallardo, J.M.; Calo-Mata, P.; Cañas, B. Species differentiation of seafood spoilage and pathogenic gram-negative bacteria by MALDI-TOF mass fingerprinting. J. Proteome Res. 2010, 9, 3169–3183. [Google Scholar] [CrossRef]
- Piamsomboon, P.; Jaresitthikunchai, J.; Hung, T.Q.; Roytrakul, S.; Wongtavatchai, J. Identification of bacterial pathogens in cultured fish with a custom peptide database constructed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). BMC Vet. Res. 2020, 16, 52. [Google Scholar] [CrossRef] [PubMed]
- Saleh, S.; Staes, A.; Deborggraeve, S.; Gevaert, K. Targeted proteomics for studying pathogenic bacteria. Proteomics 2019, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, B.; Tian, J.; Sun, W.; Wang, G.; Qian, A.; Wang, C.; Shan, X.; Kang, Y. An iTRAQ-based comparative proteomics analysis of the biofilm and planktonic states of Aeromonas veronii TH0426. Int. J. Mol. Sci. 2020, 21, 1450. [Google Scholar] [CrossRef]
- Saleh, M.; Kumar, G.; Abdel-Baki, A.S.; Dkhil, M.A.; El-Matbouli, M.; Al-Quraishy, S. Quantitative proteomic profiling of immune responses to Ichthyophthirius multifiliis in common carp skin mucus. Fish Shellfish Immunol. 2019, 84, 834–842. [Google Scholar] [CrossRef]
- Sangsuriya, P.; Huang, J.Y.; Chu, Y.F.; Phiwsaiya, K.; Leekitcharoenphon, P.; Meemetta, W.; Senapin, S.; Huang, W.P.; Withyachumnarnkul, B.; Flegel, T.W.; et al. Construction and application of a protein interaction map for white spot syndrome virus (WSSV). Mol. Cell. Proteom. 2014, 13, 269–282. [Google Scholar] [CrossRef]
- Chan, L.L.; Sit, W.H.; Lam, P.K.; Hsieh, D.P.; Hodgkiss, I.J.; Wan, J.M.; Ho, A.Y.; Choi, N.M.; Wang, D.Z.; Dudgeon, D. Identification and characterization of a “biomarker of toxicity” from the proteome of the paralytic shellfish toxin-producing dinoflagellate Alexandrium tamarense (Dinophyceae). Proteomics 2006, 6, 654–666. [Google Scholar] [CrossRef]
- Carrera, M.; Garet, E.; Barreiro, A.; Garcés, E.; Pérez, D.; Guisande, C.; González-Fernández, A. Generation of monoclonal antibodies for the specific immunodetection of the toxic dinoflagellate Alexandrium minutum Halim from Spanish waters. Harmful Algae 2010, 9, 272–280. [Google Scholar] [CrossRef]
- Hennon, G.M.M.; Dyhrman, S.T. Progress and promise of omics for predicting the impacts of climate change on harmful algal blooms. Harmful Algae 2020, 91, 101587. [Google Scholar] [CrossRef] [PubMed]
- Piñeiro, C.; Cañas, B.; Carrera, M. The role of proteomics in the study of the influence of climate change on seafood products. Food Res. Int. 2010, 43, 1791–1802. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Gallardo, J.M. Advanced proteomics and systems biology applied to study food allergy. Curr. Opin. Food Sci. 2018, 22, 9–16. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Gallardo, J.M. Rapid direct detection of the major fish allergen, parvalbumin, by selected MS/MS ion monitoring mass spectrometry. J. Proteom. 2012, 75, 3211–3220. [Google Scholar] [CrossRef]
- Abdel Rahman, A.M.; Kamath, S.; Lopata, A.L.; Helleur, R.J. Analysis of the allergenic proteins in black tiger prawn (Penaeus monodon) and characterization of the major allergen tropomyosin using mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 2462–2470. [Google Scholar] [CrossRef] [PubMed]
- De Magalhães, C.R.; Schrama, D.; Fonseca, F.; Kuehn, A.; Morisset, M.; Ferreira, S.R.; Gonçalves, A.; Rodrigues, P.M. Effect of EDTA enriched diets on farmed fish allergenicity and muscle quality; a proteomics approach. Food Chem. 2020, 305, 125508. [Google Scholar] [CrossRef]
- Mi, J.; Orbea, A.; Syme, N.; Ahmed, M.; Cajaraville, M.P.; Cristobal, S. Peroxisomal proteomics, a new tool for risk assessment of peroxisome proliferating pollutants in the marine environment. Proteomics 2005, 5, 3954–3965. [Google Scholar] [CrossRef]
- Apraiz, I.; Leoni, G.; Lindenstrand, D.; Persson, J.O.; Cristobal, S. Proteomic analysis of mussels exposed to fresh and weathered Prestige’s oil. J. Proteom. Bioinf. 2009, 2, 255–261. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Huang, L.; Zhang, Y.; Ke, C.H.; Huang, H.Q. Proteomic approach for identifying gonad differential proteins in the oyster (Crassostrea angulata) following food-chain contamination with HgCl2. J. Proteom. 2003, 94, 37–53. [Google Scholar] [CrossRef]
- Campos, A.; Danielsson, G.; Farinha, A.P.; Kuruvilla, J.; Warholm, P.; Cristobal, S. Shotgun proteomics to unravel marine mussel (Mytilus edulis) response to long-term exposure to low salinity and propranolol in a Baltic Sea microcosm. J. Proteom. 2016, 137, 97–106. [Google Scholar] [CrossRef]
- Gouveia, D.; Chaumot, A.; Charnot, A.; Almunia, C.; François, A.; Navarro, L.; Armengaud, J.; Salvador, A.; Geffard, O. Ecotoxico-proteomics for aquatic environmental monitoring: First in situ application of a new proteomics-based multibiomarker assay using caged amphipods. Environ. Sci. Technol. 2017, 51, 13417–13426. [Google Scholar] [CrossRef] [PubMed]
- Barros, D.; Pradhan, A.; Mendes, V.M.; Manadas, B.; Santos, P.M.; Pascoal, C.; Cássio, F. Proteomics and antioxidant enzymes reveal different mechanisms of toxicity induced by ionic and nanoparticulate silver in bacteria. Environ. Sci. Nano 2019, 6, 1207–1218. [Google Scholar] [CrossRef]
- Lulijwa, R.; Rupia, E.J.; Alfaro, A.C. Antibiotic use in aquaculture, policies and regulation, health and environmental risks: A review of the top 15 major producers. Rev. Aquac. 2020, 12, 640–663. [Google Scholar] [CrossRef]
- Yao, Z.; Li, W.; Lin, Y.; Wu, Q.; Yu, F.; Lin, W.; Lin, X. Proteomic analysis reveals that metabolic flows affect the susceptibility of Aeromonas hydrophila to antibiotics. Sci. Rep. 2016, 6, 39413. [Google Scholar] [CrossRef]
- Peng, B.; Li, H.; Peng, X. Proteomics approach to understand bacterial antibiotic resistance strategies. Expert Rev. Proteom. 2019, 16, 829–839. [Google Scholar] [CrossRef]
- Li, W.; Yao, Z.; Sun, L.; Hu, W.; Cao, J.; Lin, W.; Lin, X. Proteomics analysis reveals a potential antibiotic cocktail therapy strategy for Aeromonas hydrophila infection in biofilm. J. Proteome Res. 2016, 15, 1810–1820. [Google Scholar] [CrossRef]
- Li, W.; Ali, F.; Cai, Q.; Yao, Z.; Sun, L.; Lin, W.; Lin, X. Reprint of: Quantitative proteomic analysis reveals that chemotaxis is involved in chlortetracycline resistance of Aeromonas hydrophila. J. Proteom. 2018, 180, 138–146. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Abdeen, E.; Al-Dubaib, M.; Alsayeqh, A.; Ibrahem, M.; Hamada, M.; Alenzi, A.; Moussa, I.; Hemeg, H.A. Proteomic characterization and discrimination of Aeromonas species recovered from meat and water samples with a spotlight on the antimicrobial resistance of Aeromonas hydrophila. Microbiologyopen 2019, 8, e782. [Google Scholar] [CrossRef]
- Zhu, W.; Zhou, S.; Chu, W. Comparative proteomic analysis of sensitive and multi-drug resistant Aeromonas hydrophila isolated from diseased fish. Microb. Pathog. 2019, 139, 103930. [Google Scholar] [CrossRef]
- Sun, L.; Chen, H.; Lin, W.; Lin, X. Quantitative proteomic analysis of Edwardsiella tarda in response to oxytetracycline stress in biofilm. J. Proteom. 2017, 150, 141–148. [Google Scholar] [CrossRef]
- Su, Y.B.; Kuang, S.F.; Peng, X.X.; Li, H. The depressed P cycle contributes to the acquisition of ampicillin resistance in Edwardsiella piscicida. J. Proteom. 2020, 212, 103562. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrera, M.; Piñeiro, C.; Martinez, I. Proteomic Strategies to Evaluate the Impact of Farming Conditions on Food Quality and Safety in Aquaculture Products. Foods 2020, 9, 1050. https://doi.org/10.3390/foods9081050
Carrera M, Piñeiro C, Martinez I. Proteomic Strategies to Evaluate the Impact of Farming Conditions on Food Quality and Safety in Aquaculture Products. Foods. 2020; 9(8):1050. https://doi.org/10.3390/foods9081050
Chicago/Turabian StyleCarrera, Mónica, Carmen Piñeiro, and Iciar Martinez. 2020. "Proteomic Strategies to Evaluate the Impact of Farming Conditions on Food Quality and Safety in Aquaculture Products" Foods 9, no. 8: 1050. https://doi.org/10.3390/foods9081050
APA StyleCarrera, M., Piñeiro, C., & Martinez, I. (2020). Proteomic Strategies to Evaluate the Impact of Farming Conditions on Food Quality and Safety in Aquaculture Products. Foods, 9(8), 1050. https://doi.org/10.3390/foods9081050