Microbial Dynamics between Yeasts and Acetic Acid Bacteria in Kombucha: Impacts on the Chemical Composition of the Beverage
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Microbial Species
2.2. Growth Conditions
2.3. Chemical Analyses
2.4. Statistical Analyses
3. Results and Discussion
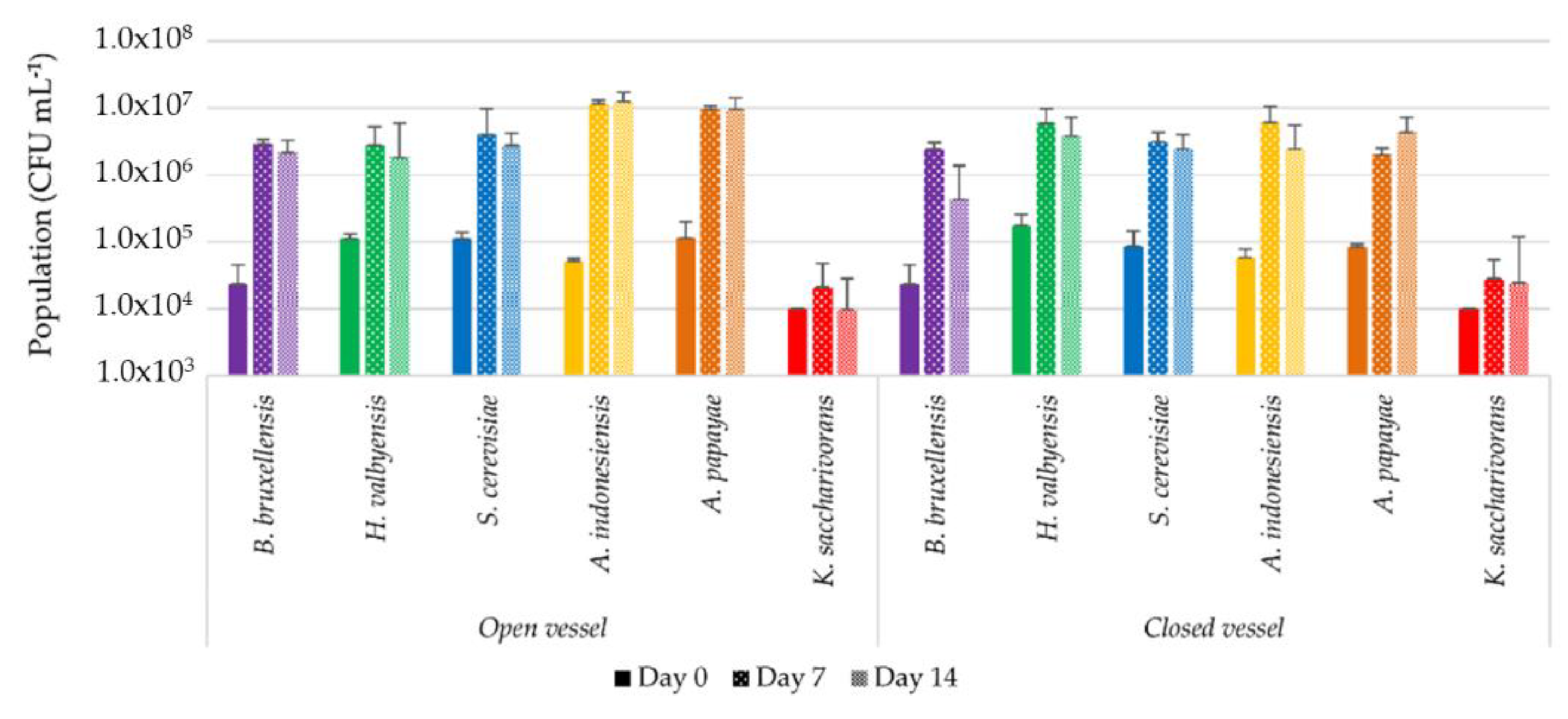
3.1. Isolation and Identification of Yeast and Bacterial Strains
3.2. Characterization of Pure Cultures in Sugared Black Tea
3.3. Comparison of Yeast-Acetic Acid Bacteria Cocultures with Original Kombucha Fermentation
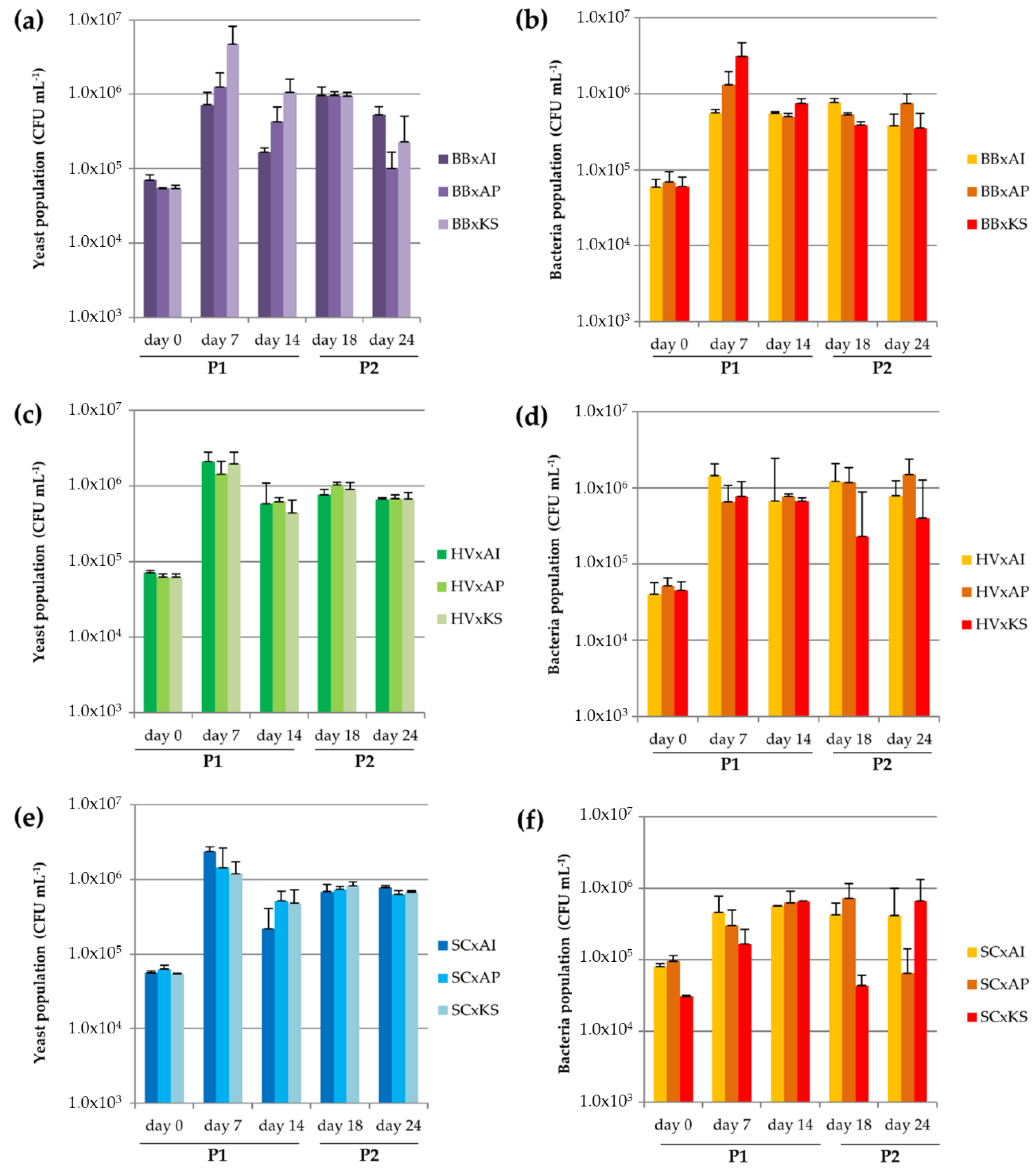
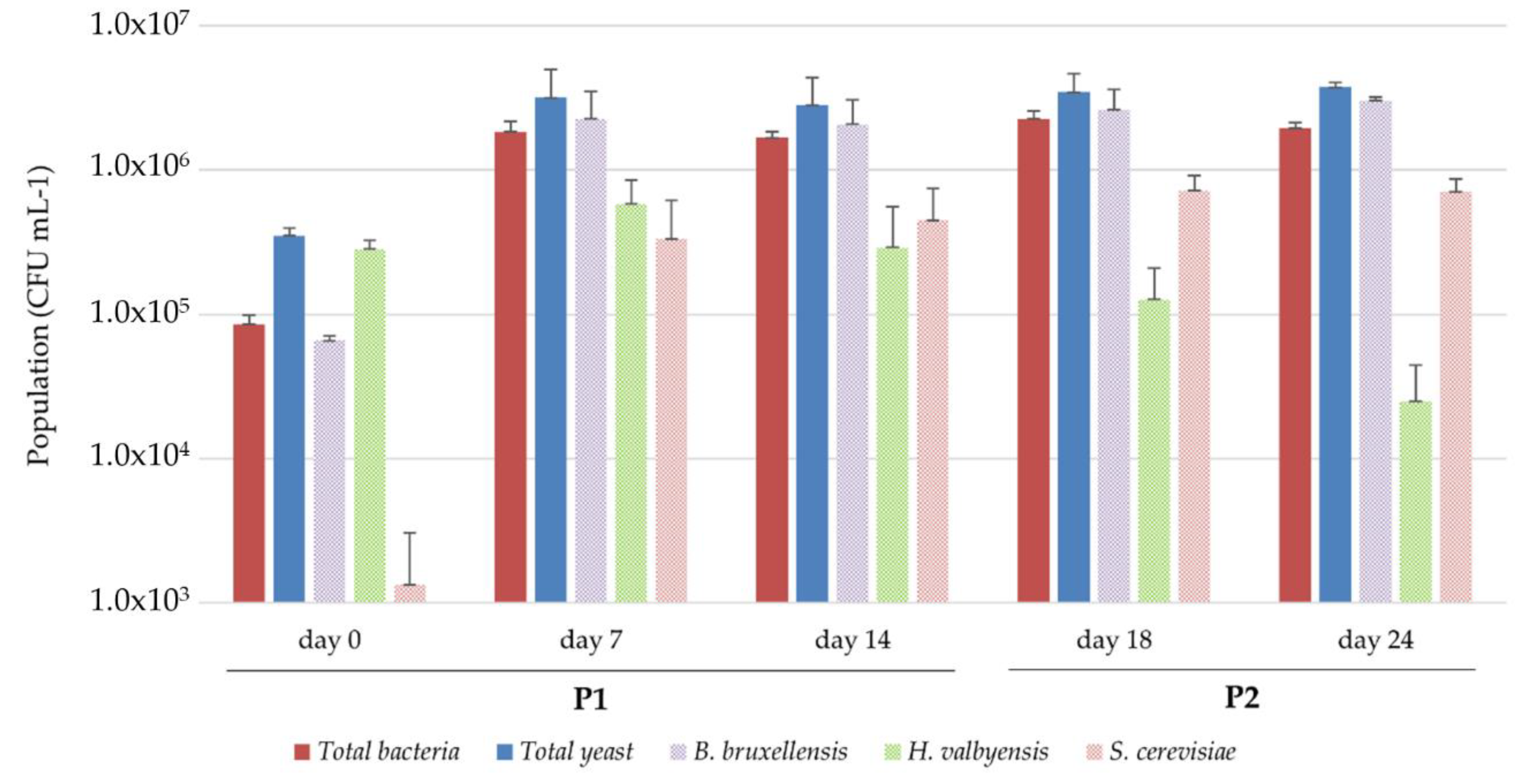
3.3.1. Microbial Dynamics
3.3.2. Utilization of Carbohydrates
3.3.3. Variations in Ethanol Content
3.3.4. Variations in the Free Amino Nitrogen Content
3.3.5. Acidification by the Production of Organic Acids
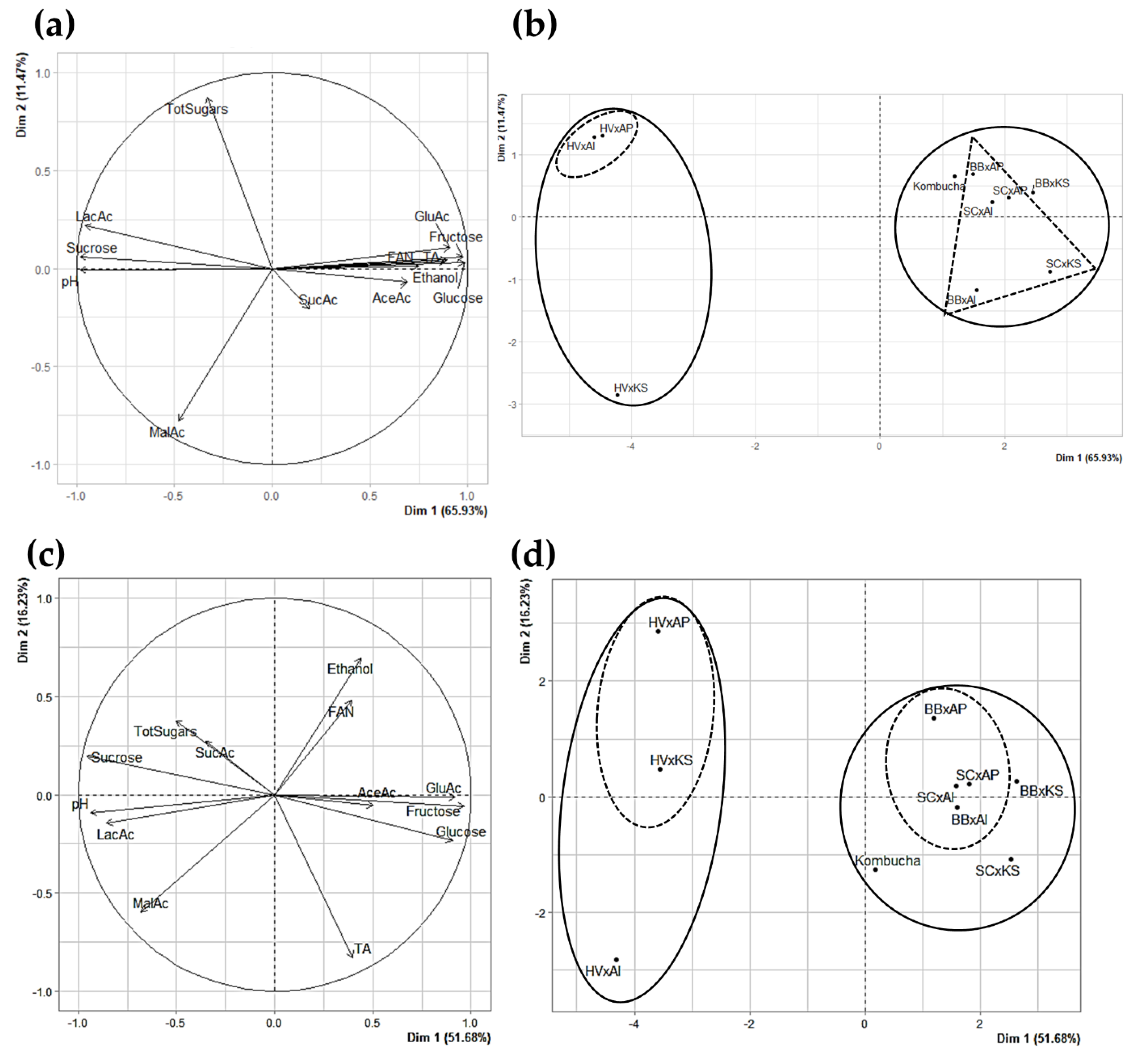
3.3.6. Principle Component Analysis as a Visual Tool for Understanding Complex Microbial Interactions
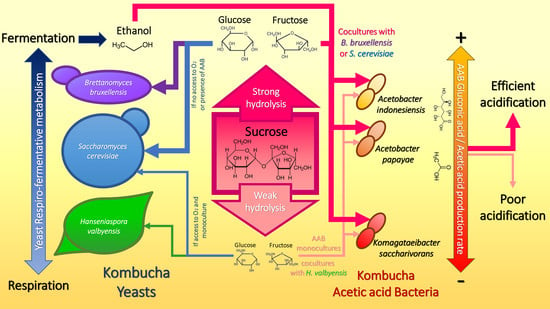
3.4. Sucrose Utilization Strategies as a Basis of Microbial Interactions in Kombucha
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ivey, M.; Massel, M.; Phister, T.G. Microbial Interactions in Food Fermentations. Annu. Rev. Food Sci. Technol. 2013, 4, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, S.; de Bok, F.A.M.; Hugenholtz, J.; van Hylckama Vlieg, J.E.T. Unraveling Microbial Interactions in Food Fermentations: From Classical to Genomics Approaches. Appl. Environ. Microbiol. 2008, 74, 4997–5007. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, B.E.; Dutton, R.J. Fermented Foods as Experimentally Tractable Microbial Ecosystems. Cell 2015, 161, 49–55. [Google Scholar] [CrossRef]
- Villarreal-Soto, S.A.; Beaufort, S.; Bouajila, J.; Souchard, J.-P.; Taillandier, P. Understanding Kombucha Tea Fermentation: A Review. J. Food Sci. 2018, 83, 580–588. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbaša, R.V.; Lončar, E.S.; Vitas, J.S.; Sathishkumar, M. A Review on Kombucha Tea-Microbiology, Composition, Fermentation, Beneficial Effects, Toxicity, and Tea Fungus. Compr. Rev. Food Sci. Food Saf. 2014, 13, 538–550. [Google Scholar] [CrossRef]
- Dufresne, C.; Farnworth, E. Tea, Kombucha, and health: A review. Food Res. Int. 2000, 33, 409–421. [Google Scholar] [CrossRef]
- Lynch, K.M.; Zannini, E.; Wilkinson, S.; Daenen, L.; Arendt, E.K. Physiology of Acetic Acid Bacteria and Their Role in Vinegar and Fermented Beverages. Compr. Rev. Food Sci. Food Saf. 2019, 18, 587–623. [Google Scholar] [CrossRef]
- La China, S.; Zanichelli, G.; De Vero, L.; Gullo, M. Oxidative fermentations and exopolysaccharides production by acetic acid bacteria: A mini review. Biotechnol. Lett. 2018, 40, 1289–1302. [Google Scholar] [CrossRef]
- Tran, T.; Grandvalet, C.; Verdier, F.; Martin, A.; Alexandre, H.; Tourdot-Maréchal, R. Microbiological and technological parameters impacting the chemical composition and sensory quality of kombucha. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2050–2070. [Google Scholar] [CrossRef]
- Carlier, A.; Pessi, G.; Eberl, L. Microbial Biofilms and Quorum Sensing. In Principles of Plant-Microbe Interactions; Lugtenberg, B., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 45–52. ISBN 978-3-319-08574-6. [Google Scholar]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Gullo, M.; La China, S.; Falcone, P.M.; Giudici, P. Biotechnological production of cellulose by acetic acid bacteria: Current state and perspectives. Appl. Microbiol. Biotechnol. 2018, 102, 6885–6898. [Google Scholar] [CrossRef]
- May, A.; Narayanan, S.; Alcock, J.; Varsani, A.; Maley, C.; Aktipis, A. Kombucha: A novel model system for cooperation and conflict in a complex multi-species microbial ecosystem. PeerJ 2019, 7, e7565. [Google Scholar] [CrossRef]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Teoh, A.L.; Heard, G.; Cox, J. Yeast ecology of Kombucha fermentation. Int. J. Food Microbiol. 2004, 95, 119–126. [Google Scholar] [CrossRef]
- Liu, C.-H.; Hsu, W.-H.; Lee, F.-L.; Liao, C.-C. The isolation and identification of microbes from a fermented tea beverage, Haipao, and their interactions during Haipao fermentation. Food Microbiol. 1996, 13, 407–415. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, L.; Qi, L.; Liang, H.; Lin, X.; Li, S.; Yu, C.; Ji, C. Effect of synthetic microbial community on nutraceutical and sensory qualities of kombucha. Int. J. Food Sci. Technol. 2020. [Google Scholar] [CrossRef]
- Lawson, C.E.; Harcombe, W.R.; Hatzenpichler, R.; Lindemann, S.R.; Löffler, F.E.; O’Malley, M.A.; García Martín, H.; Pfleger, B.F.; Raskin, L.; Venturelli, O.S.; et al. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 2019, 17, 725–741. [Google Scholar] [CrossRef]
- Green, S.R.; Gray, P.P. A differential procedure for bacteriological studies useful in the fermentation industry. Arch. Biochem. Biophys. 1951, 32, 59–69. [Google Scholar] [CrossRef]
- Hall, J.F. Detection of wild yeasts in the brewery. J. Inst. Brew. 1971, 77, 513–516. [Google Scholar] [CrossRef]
- Pallmann, C.L.; Brown, J.A.; Olineka, T.L.; Cocolin, L.; Mills, D.A.; Bisson, L.F. Use of WL Medium to Profile Native Flora Fermentations. Am. J. Enol. Vitic. 2001, 52, 198–203. [Google Scholar]
- Simonin, S.; Alexandre, H.; Nikolantonaki, M.; Coelho, C.; Tourdot-Maréchal, R. Inoculation of Torulaspora delbrueckii as a bio-protection agent in winemaking. Food Res. Int. 2018. [Google Scholar] [CrossRef] [PubMed]
- Salma, M.; Rousseaux, S.; Sequeira-Le Grand, A.; Alexandre, H. Cytofluorometric detection of wine lactic acid bacteria: Application of malolactic fermentation to the monitoring. J. Ind. Microbiol. Biotechnol. 2013, 40, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Maoura, N.; Mbaiguinam, M.; Nguyen, H.V.; Gaillardin, C.; Pourquie, J. Identification and typing of the yeast strains isolated from bili bili, a traditional sorghum beer of Chad. Afr. J. Biotechnol. 2005, 4, 646–656. [Google Scholar] [CrossRef]
- Esteve-Zarzoso, B. Identification of yeasts by RFLP analysis of the 5.85 rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 2019, 49, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qian, P.-Y. Conservative Fragments in Bacterial 16S rRNA Genes and Primer Design for 16S Ribosomal DNA Amplicons in Metagenomic Studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef]
- Cvetković, D.; Markov, S.; Djurić, M.; Savić, D.; Velićanski, A. Specific interfacial area as a key variable in scaling-up Kombucha fermentation. J. Food Eng. 2008, 85, 387–392. [Google Scholar] [CrossRef]
- OIV. Méthode OIV-MA-AS313-01: Acidité Totale; Recueil International des Méthodes d’Analyse-OIV: Paris, France, 2009. [Google Scholar]
- MEBAK 2.6.4.1.1 Ninhydrin Method (Photometric Method, EBC). In MEBAK Wort, Beer, Beer-Based Beverages: Collection of Brewing Analysis Methods of the Mitteleuropäische Brautechnische Analysenkommission (MEBAK); Freising-Weihenstephan: Munich, Germany, 2013.
- Zou, X.; Zhang, S.; Chen, L.; Hu, J.; Hong, F.F. Determination of live and dead Komagataeibacter xylinus cells and first attempt at precise control of inoculation in nanocellulose production. Microb. Biotechnol. 2020, 13, 458–469. [Google Scholar] [CrossRef]
- Millet, V.; Lonvaud-Funel, A. The viable but non-culturable state of wine micro-organisms during storage. Lett. Appl. Microbiol. 2000, 30, 136–141. [Google Scholar] [CrossRef]
- Lee, J.; Schreiner, R.P. Free amino acid profiles from ‘Pinot noir’ grapes are influenced by vine N-status and sample preparation method. Food Chem. 2010, 119, 484–489. [Google Scholar] [CrossRef]
- Aguilar Uscanga, M.G.; Délia, M.-L.; Strehaiano, P. Brettanomyces bruxellensis: Effect of oxygen on growth and acetic acid production. Appl. Microbiol. Biotechnol. 2003, 61, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Custer, M. Onderzoekingen over Het Gistgeslacht Brettanomyces. Ph.D. Thesis, University of Delft, Delft, The Netherlands, 1940. [Google Scholar]
- Nogueira, A.; le Quéré, J.M.; Drilleau, J.F.; Wosiacki, G. Fermentation Behavior of Apiculate Yeast Hanseniaspora valbyensis in Cider Processing Conditions; International Commission of Agricultural Engineering (CIGR), Institut fur Landtechnik: Bonn, Germany, 2008; p. Ref.20. [Google Scholar]
- Özcan, S.; Vallier, L.G.; Flick, J.S.; Carlson, M.; Johnson, M. Expression of the SUC2 Gene of Saccharomyces cerevisiae is Induced by Low Levels of Glucose. Yeast 1997, 13, 127–137. [Google Scholar] [CrossRef]
- Meijer, M.M.C.; Boonstra, J.; Verkleij, A.J.; Verrips, C.T. Glucose Repression in Saccharomyces cerevisiae Is Related to the Glucose Concentration Rather Than the Glucose Flux. J. Biol. Chem. 1998, 273, 24102–24107. [Google Scholar] [CrossRef] [PubMed]
- Marques, W.L.; Raghavendran, V.; Stambuk, B.U.; Gombert, A.K. Sucrose and Saccharomyces cerevisiae: A relationship most sweet. FEMS Yeast Res. 2016, 16, fov107. [Google Scholar] [CrossRef] [PubMed]
- Berthels, N.; Corderootero, R.; Bauer, F.; Thevelein, J.; Pretorius, I. Discrepancy in glucose and fructose utilisation during fermentation by wine yeast strains. FEMS Yeast Res. 2004, 4, 683–689. [Google Scholar] [CrossRef]
- Berthels, N.J.; Cordero Otero, R.R.; Bauer, F.F.; Pretorius, I.S.; Thevelein, J.M. Correlation between glucose/fructose discrepancy and hexokinase kinetic properties in different Saccharomyces cerevisiae wine yeast strains. Appl. Microbiol. Biotechnol. 2008, 77, 1083–1091. [Google Scholar] [CrossRef]
- Velasco-Bedrán, H.; López-Isunza, F. The unified metabolism of Gluconacetobacter entanii in continuous and batch processes. Process Biochem. 2007, 42, 1180–1190. [Google Scholar] [CrossRef]
- Sharma, C.; Bhardwaj, N.K. Biotransformation of fermented black tea into bacterial nanocellulose via symbiotic interplay of microorganisms. Int. J. Biol. Macromol. 2019, 132, 166–177. [Google Scholar] [CrossRef]
- De Ley, J. Comparative carbohydrate metabolism and a proposal for a phylogenetic relationship of the acetic acid bacteria. Microbiology 1961, 24, 31–50. [Google Scholar] [CrossRef][Green Version]
- De Ley, J.; Schell, J. Lactate and Pyruvate Catabolism in Acetic Acid Bacteria. J. Gen. Microbiol. 1962, 29, 589–601. [Google Scholar] [CrossRef]
- Prust, C.; Hoffmeister, M.; Liesegang, H.; Wiezer, A.; Fricke, W.F.; Ehrenreich, A.; Gottschalk, G.; Deppenmeier, U. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 2005, 23, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Hu, J.; Fan, L.; Chen, Q. RNA-Seq-based transcriptomic and metabolomic analysis reveal stress responses and programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Drysdale, G.S.; Fleet, G.H. The effect of acetic acid bacteria upon the growth and metabolism of yeasts during the fermentation of grape juice. J. Appl. Bacteriol. 1989, 67, 471–481. [Google Scholar] [CrossRef]
- Chen, C.; Liu, B.Y. Changes in major components of tea fungus metabolites during prolonged fermentation. J. Appl. Microbiol. 2000, 89, 834–839. [Google Scholar] [CrossRef]
- Goh, W.N.; Rosma, A.; Kaur, B.; Fazilah, A.; Karim, A.A.; Bhat, R. Fermentation of black tea broth (Kombucha): I. Effects of sucrose concentration and fermentation time on the yield of microbial cellulose. Int. Food Res. J. 2012, 19, 109–117. [Google Scholar]
- Blanc, P.J. Characterization of the tea fungus metabolites. Biotechnol. Lett. 1996, 18, 139–142. [Google Scholar] [CrossRef]
- Sievers, M.; Lanini, C.; Weber, A.; Schuler-Schmid, U.; Teuber, M. Microbiology and fermentation balance in a kombucha beverage obtained from a tea fungus fermentation. Syst. Appl. Microbiol. 1995, 18, 590–594. [Google Scholar] [CrossRef]
- Andjelković, U.; Milutinović-Nikolić, A.; Jović-Jovičić, N.; Banković, P.; Bajt, T.; Mojović, Z.; Vujčić, Z.; Jovanović, D. Efficient stabilization of Saccharomyces cerevisiae external invertase by immobilisation on modified beidellite nanoclays. Food Chem. 2015, 168, 262–269. [Google Scholar] [CrossRef]
- Beteta, P.; Gascon, S. Localization of invertase in yeast vacuoles. FEBS Lett. 1971, 13, 297–300. [Google Scholar] [CrossRef]
- Margetić, A.; Vujčić, Z. Comparative study of stability of soluble and cell wall invertase from Saccharomyces cerevisiae. Prep. Biochem. Biotechnol. 2017, 47, 305–311. [Google Scholar] [CrossRef]
- Neumann, N.P.; Lampen, J.O. Glycoprotein structure of yeast invertase. Biochemistry 1969, 8, 3552–3556. [Google Scholar] [CrossRef] [PubMed]
- Blondin, B.; Ratomahenina, R. Purification and Properties of the β-Glucosidase of a Yeast Capable of Fermenting Cellobiose to Ethanol: Dekkera intermedia Van der Walt. Eur. J. Microbiol. Biotechnol. 1983, 17, 1–6. [Google Scholar] [CrossRef]
- Conterno, L.; Joseph, C.M.L.; Arvik, T.J.; Henick-Kling, T.; Bisson, L.F. Genetic and Physiological Characterization of Brettanomyces bruxellensis Strains Isolated from Wines. Am. J. Enol. Vitic. 2006, 139–147. [Google Scholar]
- Fia, G.; Giovani, G.; Rosi, I. Study of beta-glucosidase production by wine-related yeasts during alcoholic fermentation. A new rapid fluorimetric method to determine enzymatic activity. J. Appl. Microbiol. 2005, 99, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Roach, M.J.; Borneman, A.R. New genome assemblies reveal patterns of domestication and adaptation across Brettanomyces (Dekkera) species. BMC Genom. 2020, 21, 14. [Google Scholar] [CrossRef]
- Steenwyk, J.L.; Opulente, D.A.; Kominek, J.; Shen, X.-X.; Zhou, X.; Labella, A.L.; Bradley, N.P.; Eichman, B.F.; Čadež, N.; Libkind, D.; et al. Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts. PLoS Biol. 2019, 17, e3000255. [Google Scholar] [CrossRef]
- Alexandre, H.; Heintz, D.; Chassagne, D.; Guilloux-Benatier, M.; Charpentier, C.; Feuillat, M. Protease A activity and nitrogen fractions released during alcoholic fermentation and autolysis in enological conditions. J. Ind. Microbiol. Biotechnol. 2001, 26, 235–240. [Google Scholar] [CrossRef]
- Guilloux-Benatier, M.; Chassagne, D.; Alexandre, H.; Charpentier, C.; Feuillat, M. Influence of yeast autolysis after alcoholic fermentation on the development of Brettanomyces/Dekkera in wine. OENO One 2001, 35, 157. [Google Scholar] [CrossRef]
- Charoenchai, C.; Fleet, G.H.; Henschke, P.A.; Todd, B.E.N. Screening of non-Saccharomyces wine yeasts for the presence of extracellular hydrolytic enzymes. Aust. J. Grape Wine Res. 1997, 3, 2–8. [Google Scholar] [CrossRef]
- Younes, B.; Cilindre, C.; Villaume, S.; Parmentier, M.; Jeandet, P.; Vasserot, Y. Evidence for an Extracellular Acid Proteolytic Activity Secreted by Living Cells of Saccharomyces cerevisiae PlR1: Impact on Grape Proteins. J. Agric. Food Chem. 2011, 59, 6239–6246. [Google Scholar] [CrossRef] [PubMed]
- Guilloux-Benatier, M.; Chassagne, D. Comparison of Components Released by Fermented or Active Dried Yeasts after Aging on Lees in a Model Wine. J. Agric. Food Chem. 2003, 51, 746–751. [Google Scholar] [CrossRef]
- Torres, A.P.; Oliveira, F.A.R.; Silva, C.L.M.; Fortuna, S.P. The influence of pH on the kinetics of acid hydrolysis of sucrose. J. Food Process Eng. 1994, 17, 191–208. [Google Scholar] [CrossRef]
- Jakob, F.; Quintero, Y.; Musacchio, A.; Estrada-de los Santos, P.; Hernández, L.; Vogel, R.F. Acetic acid bacteria encode two levansucrase types of different ecological relationship. Environ. Microbiol. 2019, 21, 4151–4165. [Google Scholar] [CrossRef]
- Balasubramaniam, K.; Kannangara, P.N. Sucrose Phosphorylase and Invertase Activities in Bacteria. J. Natl. Sci. Found. Sri Lanka 1982, 10, 169–180. [Google Scholar] [CrossRef]
- Albergaria, H.; Francisco, D.; Gori, K.; Arneborg, N.; Gírio, F. Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 2010, 86, 965–972. [Google Scholar] [CrossRef]
- Branco, P.; Viana, T.; Albergaria, H.; Arneborg, N. Antimicrobial peptides (AMPs) produced by Saccharomyces cerevisiae induce alterations in the intracellular pH, membrane permeability and culturability of Hanseniaspora guilliermondii cells. Int. J. Food Microbiol. 2015, 205, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Pereznevado, F.; Albergaria, H.; Hogg, T.; Girio, F. Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 2006, 108, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Mas, A.; Esteve-Zarzoso, B. The Interaction between Saccharomyces cerevisiae and Non-Saccharomyces Yeast during Alcoholic Fermentation Is Species and Strain Specific. Front. Microbiol. 2016, 7, Article 502. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Tourdot-Maréchal, R.; Morge, C.; Sparrow, C.; Liu, Y.; Quintanilla-Casas, B.; Vichi, S.; Alexandre, H. Non-Saccharomyces Yeasts Nitrogen Source Preferences: Impact on Sequential Fermentation and Wine Volatile Compounds Profile. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Renault, P.E.; Albertin, W.; Bely, M. An innovative tool reveals interaction mechanisms among yeast populations under oenological conditions. Appl. Microbiol. Biotechnol. 2013, 97, 4105–4119. [Google Scholar] [CrossRef]
- Soares, E.V. Flocculation in Saccharomyces cerevisiae: A review: Yeast flocculation: A review. J. Appl. Microbiol. 2011, 110, 1–18. [Google Scholar] [CrossRef]
- Greig, D.; Travisano, M. The Prisoner’s Dilemma and polymorphism in yeast SUC genes. Proc. R. Soc. Lond. B Biol. Sci. 2004, 271. [Google Scholar] [CrossRef]
- Celiker, H.; Gore, J. Competition between species can stabilize public-goods cooperation within a species. Mol. Syst. Biol. 2012, 8, 621. [Google Scholar] [CrossRef] [PubMed]
- Jayabalan, R.; Marimuthu, S.; Swaminathan, K. Changes in content of organic acids and tea polyphenols during kombucha tea fermentation. Food Chem. 2007, 102, 392–398. [Google Scholar] [CrossRef]
- Harrington, K.I.; Sanchez, A. Eco-evolutionary dynamics of complex social strategies in microbial communities. Commun. Integr. Biol. 2014, 7, e28230. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Gore, J. Feedback between Population and Evolutionary Dynamics Determines the Fate of Social Microbial Populations. PLoS Biol. 2013, 11, e1001547. [Google Scholar] [CrossRef] [PubMed]
- Langenberg, A.-K.; Bink, F.J.; Wolff, L.; Walter, S.; von Wallbrunn, C.; Grossmann, M.; Heinisch, J.J.; Schmitz, H.-P. Glycolytic Functions Are Conserved in the Genome of the Wine Yeast Hanseniaspora uvarum, and Pyruvate Kinase Limits Its Capacity for Alcoholic Fermentation. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Leistner, L. Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 2000, 55, 181–186. [Google Scholar] [CrossRef]
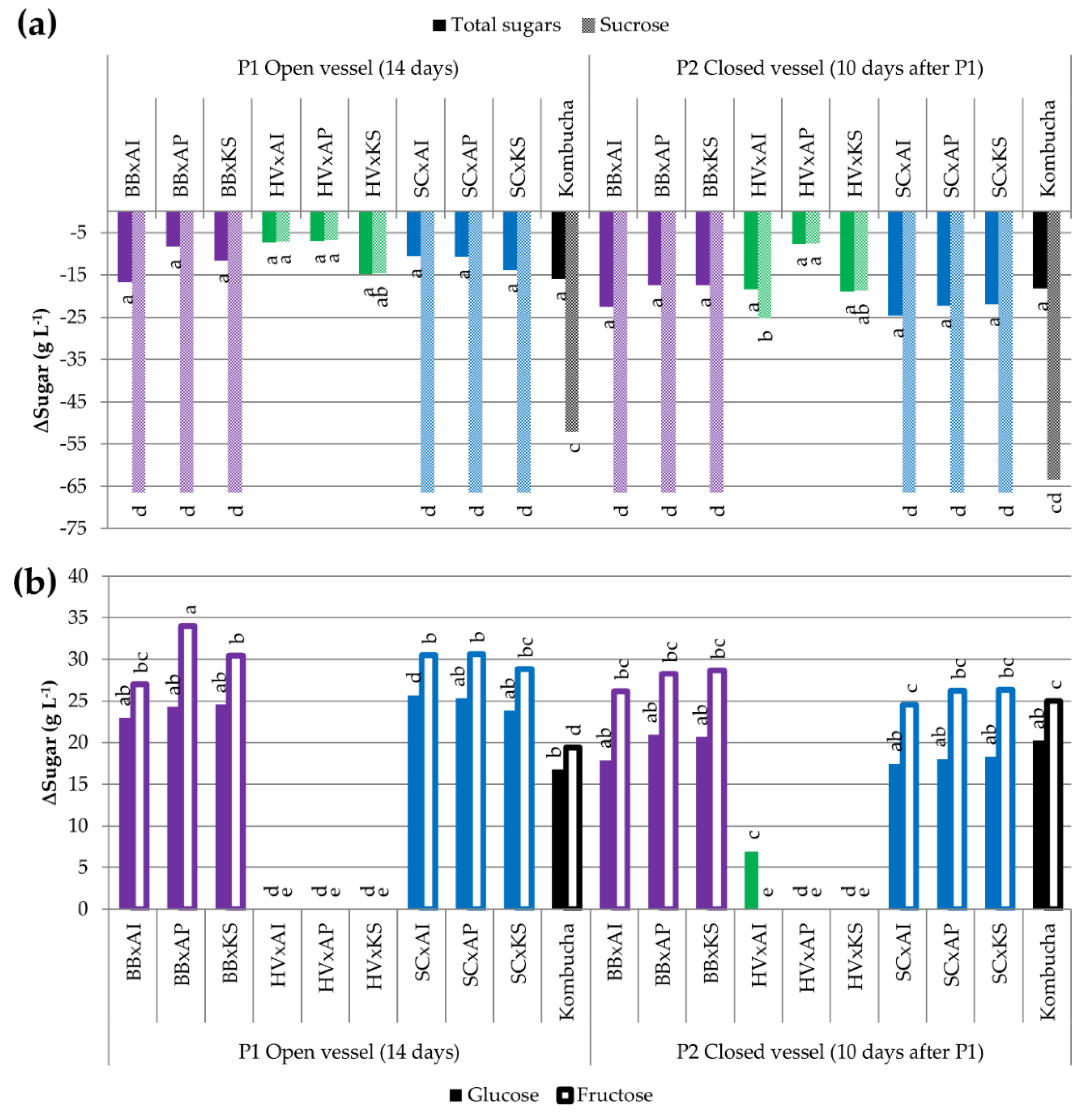
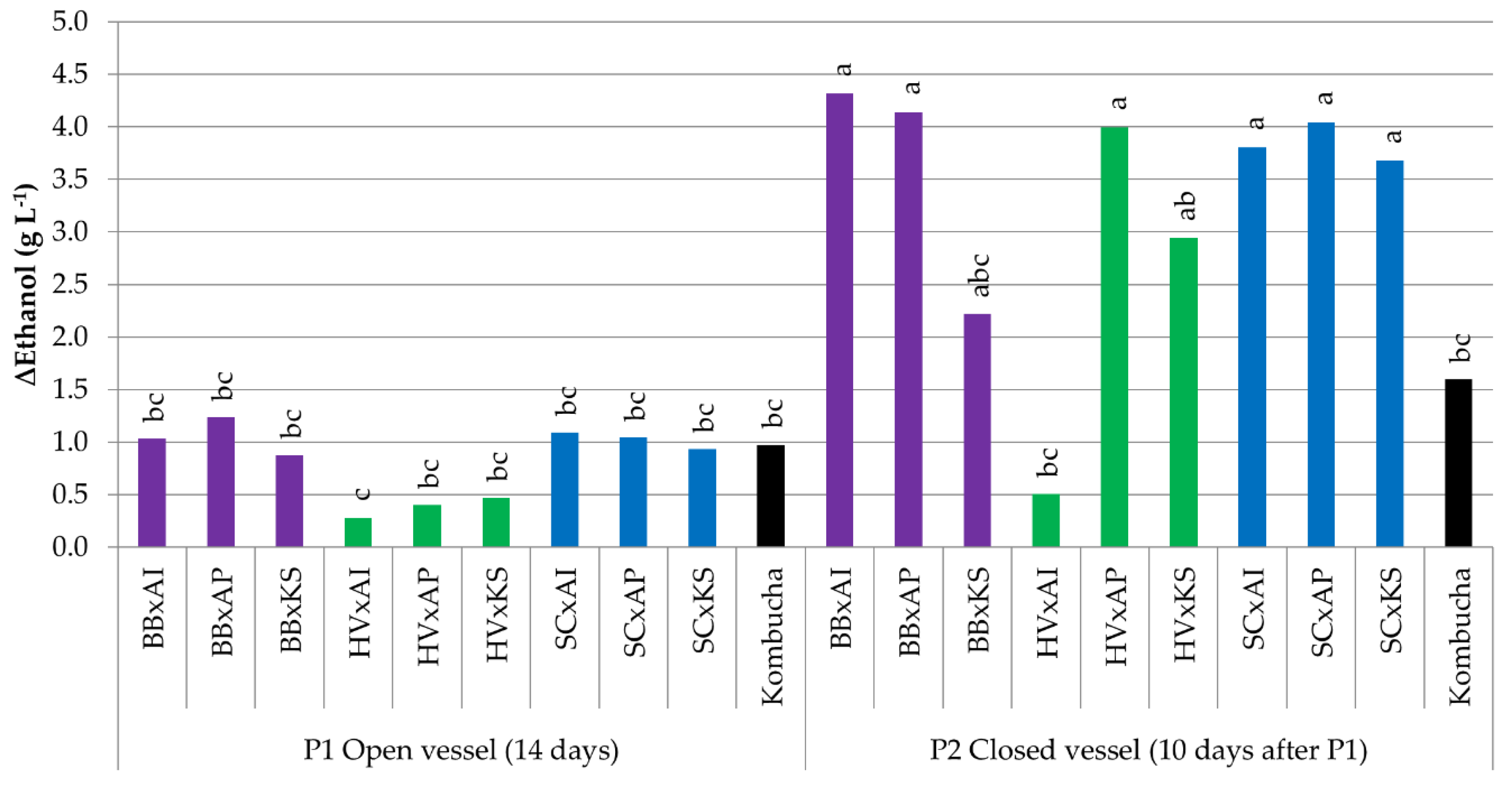
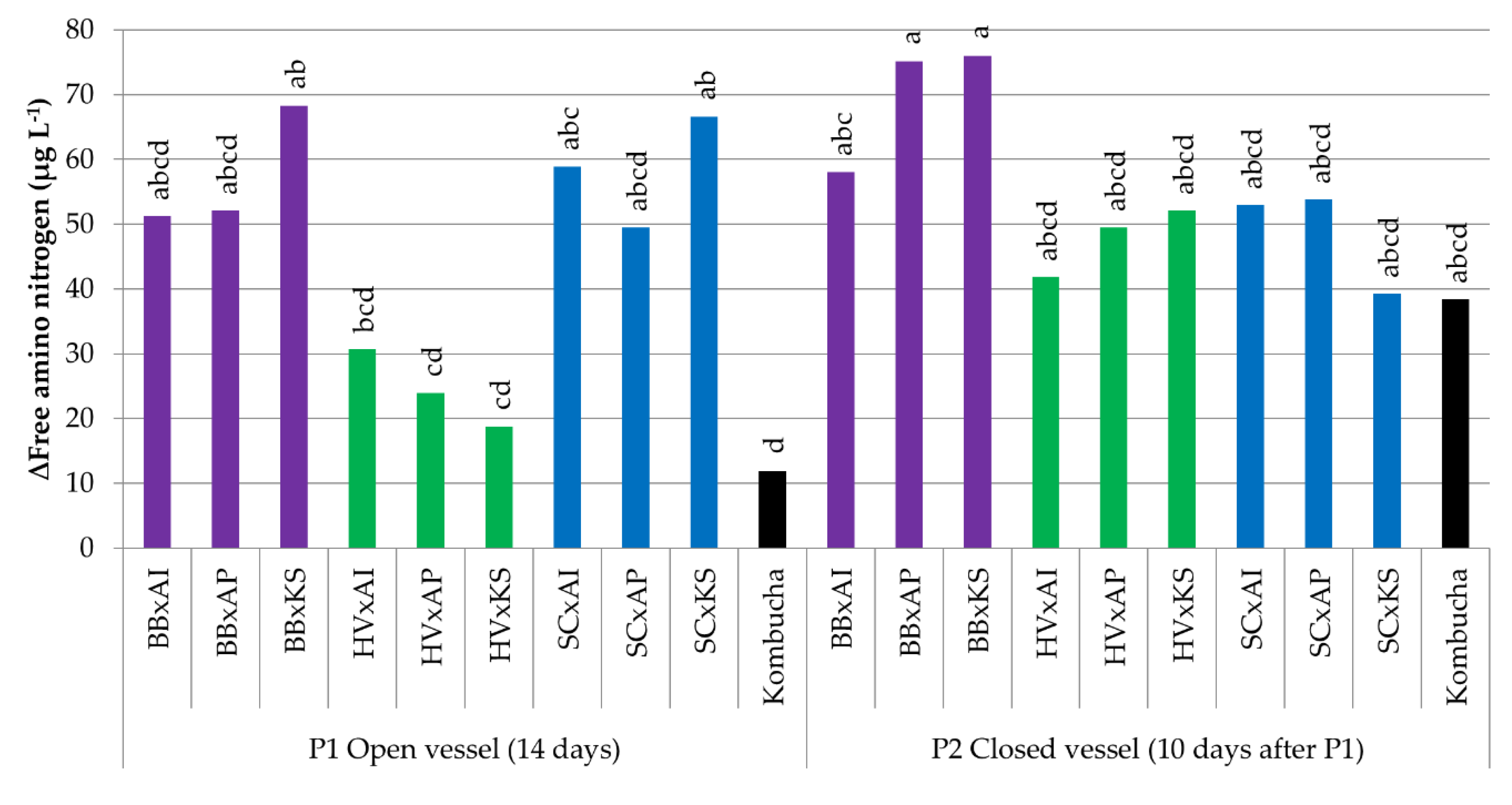
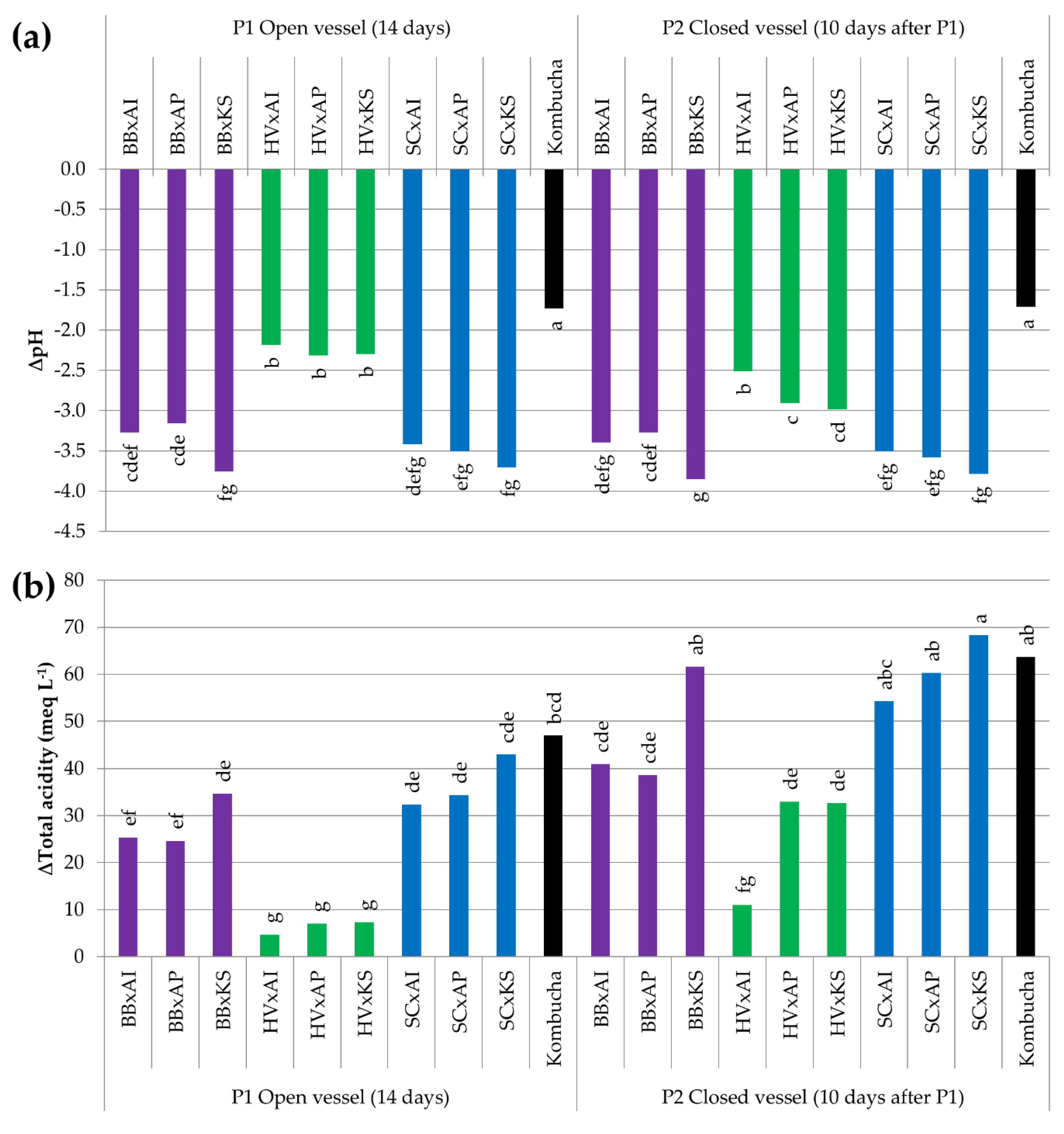
| Microorganism | B. bruxellensis | H. valbyensis | S. cerevisiae | A. indonesiensis | A. papayae | K. saccharivorans | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Incubation Condition | Open Vessel | Closed Vessel | Open Vessel | Closed Vessel | Open Vessel | Closed Vessel | Open Vessel | Closed Vessel | Open Vessel | Closed Vessel | Open Vessel | Closed Vessel |
| ∆Total sugars (g L−1) * | −10.7 a | −10.6 a | −2.2 a | −9.4 a | 0.0 a | −7.9 a | −3.6 a | −0.9 a | −0.7 a | 0.0 a | −6.2 a | −10.0 a |
| ∆Sucrose (g L−1) | −68.3 d | −68.3 d | −2.2 a | −15.4 ab | −36.1 c | −68.3 d | −6.9 ab | −0.9 a | −1.0 a | 0.0 a | −12.3 ab | −19.6 b |
| ∆Glucose (g L−1) | 27.0 a | 26.9 a | 0.0 d | 0.9 d | 18.9 b | 28.5 a | 2.1 d | nd | nd | nd | 4.2 d | 9.0 c |
| ∆Fructose (g L−1) | 29.8 a | 30.8 a | 0.0 d | 5.0 c | 17.2 b | 31.8 a | 1.1 cd | nd | 0.2 d | 0.4 d | 1.9 cd | 0.6 d |
| ∆Ethanol (g L−1) | 3.2 a | 2.5 b | 0.1 d | 1.5 c | 0.4 d | 2.4 b | nd | <0.1 | nd | nd | nd | <0.1 |
| ∆Free amino nitrogen (µg L−1) | 37 a | 52 a | 7 b | 31 ab | 3 b | 54 a | 3 b | 30 ab | 8 b | 29 ab | 8 b | 28 ab |
| ∆pH | −2.13 cd | −1.86 cd | −1.07 b | −2.26 cd | −0.38 a | −1.67 c | −2.64 d | −2.25 cd | −2.05 cd | −2.04 cd | −2.61 d | −2.50 d |
| ∆Total acidity (meq L−1) | 8.0 abc | 6.3 abc | 2.0 c | 6.7 abc | 1.0 c | 4.7 bc | 11.0 abc | 19.3 a | 7.0 abc | 5.7 bc | 19.3 a | 16.7 ab |
| ∆Acetic acid (g L−1) | 0.38 a | 0.25 a | nd | <0.1 | <0.1 | nd | 0.25 a | 0.49 a | 0.28 a | 0.38 a | 0.40 a | 0.56 a |
| ∆Gluconic acid (g L−1) | <0.05 | <0.05 | <0.05 | <0.05 | <0.05 | <0.05 | 0.91 a | 0.11 b | 0.56 ab | 0.18 b | 1.94 a | 0.12 b |
| ∆Lactic acid (g L−1) | nd | nd | 0.46 a | 0.20 abc | 0.32 ab | nd | nd | nd | nd | nd | <0.1 | nd |
| ∆Succinic acid (g L−1) | 1.27 a | 1.19 a | 0.11 c | 0.76 b | 0.34 c | 1.27 a | <0.1 | <0.1 | nd | nd | 0.14 c | 0.16 c |
| ∆Malic acid (g L−1) | 0.92 a | 1.4 a | nd | 1.00 a | nd | 1.24 a | nd | 0.22 b | <0.1 | 0.18 b | nd | <0.1 |
| Cocultures | Acetobacter indonesiensis | Acetobacter papayae | Komagataeibacter saccharivorans |
|---|---|---|---|
| Brettanomyces bruxellensis | BB x AI | BB x AP | BB x KS |
| Hanseniaspora valbyensis | HV x AI | HV x AP | HV x KS |
| Saccharomyces cerevisiae | SC x AI | SC x AP | SC x KS |
| Fermentation Phase | Coculture | ∆Acetic Acid (g L−1) | ∆Gluconic Acid (g L−1) | ∆Lactic Acid (g L−1) | ∆Succinic Acid (g L−1) | ∆Malic Acid (g L−1) |
|---|---|---|---|---|---|---|
| P1 Open vessel (14 days) | BB x AI | 0.7 bcd | 1.6 abc | nd | 0.3 abc | nd |
| BB x AP | 0.6 bcd | 1.4 abcd | nd | 0.3 abc | nd | |
| BB x KS | 2.7 bc | 1.7 ab | nd | nd | nd | |
| HV x AI | nd | 0.1 ef | 0.3 a | 0.2 bc | nd | |
| HV x AP | 0.2 cd | 0.1 f | 0.4 a | 0.2 abc | nd | |
| HV x KS | 0.4 c | <0.05 | 0.2 bc | 0.2 abc | 0.3 a | |
| SC x AI | 1.0 bcd | 1.2 abcde | nd | 0.3 abc | nd | |
| SC x AP | 0.8 c | 2.2 a | nd | 0.3 abc | nd | |
| SC x KS | 2.7 bcd | 1.3 abcde | nd | 0.4 abc | nd | |
| Kombucha | 1.6 bcd | 1.7 ab | 0.1 c | 0.2 abc | nd | |
| P2 Closed vessel (10 days after P1) | BB x AI | 2.0 bcd | 1.6 abc | nd | 0.6 abc | nd |
| BB x AP | 1.6 bcd | 1.2 abcde | nd | 0.7 ab | nd | |
| BB x KS | 12.7 a | 2.0 a | nd | nd | nd | |
| HV x AI | 0.7 cd | 0.3 def | 0.2 bc | 0.7 ab | 1.0 a | |
| HV x AP | 2.4 bcd | 0.5 cdef | 0.1 cd | 0.7 ab | nd | |
| HV x KS | 2.9 bcd | 0.4 def | 0.3 ab | 0.5 abc | 0.2 a | |
| SC x AI | 2.2 bcd | 2.3 a | nd | 0.8 a | nd | |
| SC x AP | 2.2 bcd | 2.4 a | nd | 0.8 a | nd | |
| SC x KS | 11.3 a | 2.2 a | nd | nd | nd | |
| Kombucha | 2.1 bcd | 0.8 bcdef | 0.1 cd | 0.3 abc | nd |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, T.; Grandvalet, C.; Verdier, F.; Martin, A.; Alexandre, H.; Tourdot-Maréchal, R. Microbial Dynamics between Yeasts and Acetic Acid Bacteria in Kombucha: Impacts on the Chemical Composition of the Beverage. Foods 2020, 9, 963. https://doi.org/10.3390/foods9070963
Tran T, Grandvalet C, Verdier F, Martin A, Alexandre H, Tourdot-Maréchal R. Microbial Dynamics between Yeasts and Acetic Acid Bacteria in Kombucha: Impacts on the Chemical Composition of the Beverage. Foods. 2020; 9(7):963. https://doi.org/10.3390/foods9070963
Chicago/Turabian StyleTran, Thierry, Cosette Grandvalet, François Verdier, Antoine Martin, Hervé Alexandre, and Raphaëlle Tourdot-Maréchal. 2020. "Microbial Dynamics between Yeasts and Acetic Acid Bacteria in Kombucha: Impacts on the Chemical Composition of the Beverage" Foods 9, no. 7: 963. https://doi.org/10.3390/foods9070963
APA StyleTran, T., Grandvalet, C., Verdier, F., Martin, A., Alexandre, H., & Tourdot-Maréchal, R. (2020). Microbial Dynamics between Yeasts and Acetic Acid Bacteria in Kombucha: Impacts on the Chemical Composition of the Beverage. Foods, 9(7), 963. https://doi.org/10.3390/foods9070963