Identification of Arctic Food Fish Species for Anthropogenic Contaminant Testing Using Geography and Genetics
Abstract
1. Introduction
2. Materials and Methods
2.1. Indigenous Knowledge, Mapping of Fishing Locations, and Sampling Methodology
2.2. Contaminant Analysis
2.3. Statistical Methods for Contaminants
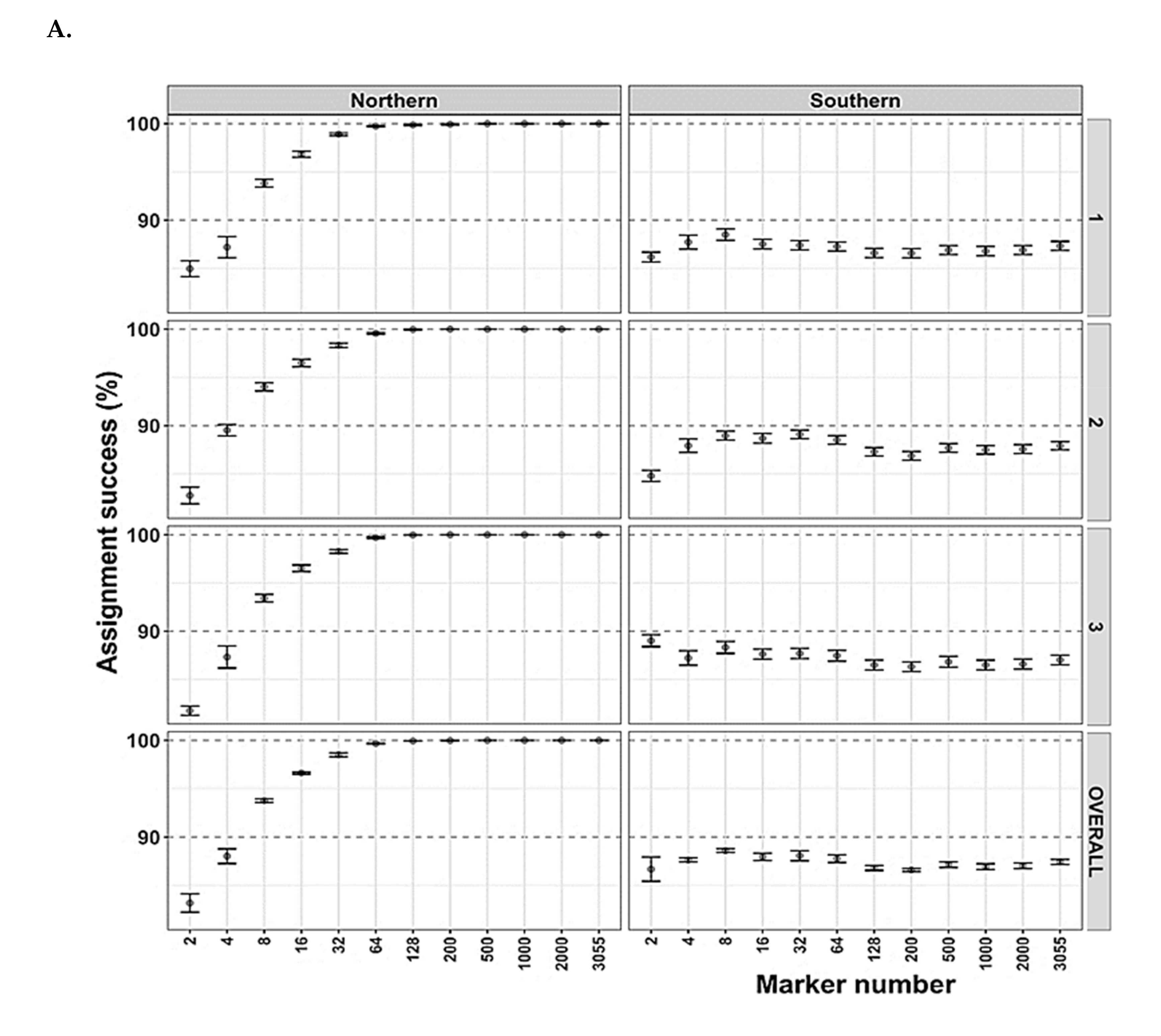
2.4. Genetic and Bioinformatic Analysis
3. Results
3.1. Mapping Subsistence Fishing Locations and Sampling
3.2. Variation of Hg, As, and PCB Concentrations by Fish Species
3.3. Geographical Analysis of Contaminant Levels
3.4. Arctic Char Geographical and Population Assignments as Revealed by Genetics
4. Discussion
4.1. Distribution of Contaminants by Species and Geography
4.2. Genetic Tools for Geographical or Population Assignments and the Future of Contaminant Monitoring
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Minamata Convention on Mercury. UN Environmental Program. 2013. Available online: http://www.mercuryconvention.org (accessed on 13 July 2020).
- EIP Associates. Polychlorinated biphenyls (PCBs) source identification, prepared for: Palo Alto Regional Water Quality Control Plant. 1997. Available online: http://www.cityofpaloalto.org/civicax/filebank/documents/3772 (accessed on 27 June 2020).
- Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological Profile for Polychlorinated Biphenyls (PCBs); Department of Health and Human Services; Public Health Service: Atlanta, GA, USA, 2000. Available online: https://www.atsdr.cdc.gov/ToxProfiles/tp.asp?id=142&tid=26 (accessed on 13 July 2020).
- Das, P.; McDonald, J.A.K.; Petrof, E.O.; Allen-Vercoe, E.; Walker, V.K. Nanosilver-Mediated Change in Human Intestinal Microbiota. J. Nanomed. Nanotechnol. 2014, 5, 235. [Google Scholar] [CrossRef]
- Carbery, M.; O’Connor, W.; Palanisami, T. Trophic transfer of microplastics and mixed contaminants in the marine food web and implications for human health. Environ. Int. 2018, 115, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, W.H.; Anlauf, K.G.; Barrie, L.A.; Lu, J.Y.; Steffan, A.; Schneeberger, D.R.; Berg, T. Arctic springtime depletion of mercury. Nature 1998, 394, 331–332. [Google Scholar] [CrossRef]
- Steffen, A.; Douglas, T.; Amyot, M.; Ariya, P.; Aspmo, K.; Berg, T.; Bottenheim, J.; Brooks, S.; Cobbett, F.; Dastoor, A.; et al. A synthesis of atmospheric mercury depletion event chemistry in the atmosphere and snow. Atmos. Chem. Phys. 2008, 8, 1445–1482. [Google Scholar] [CrossRef]
- Dietz, R.; Outridge, P.M.; Hobson, K.A. Anthropogenic contributions to mercury levels in present-day Arctic animals—A review. Sci Total Environ. 2009, 407, 6120–6131. [Google Scholar] [CrossRef]
- Durnford, D.; Dastoor, A.; Figueras-Nieto, D.; Ryjkov, A. Long-range transport of mercury to the Arctic and across Canada. Atmos. Chem. Phys. 2010, 10, 6063–6086. [Google Scholar] [CrossRef]
- Zhang, Y.; Jocob, D.J.; Horowitz, H.M.; Chen, L.; Amos, H.M.; Krabbenhoft, D.P.; Slemr, F.; St. Louis, V.L.; Sunderland, M. Observed decrease in atmospheric mercury explained by global decline in anthropogenic emissions. Proc. Natl. Acad. Sci. USA 2016, 113, 526–531. [Google Scholar] [CrossRef]
- Mackenzie, F.T.; Lantzy, R.J.; Paterson, V. Global trace metal cycles and predictions. J. Int. Assoc. Math Geol. 1979, 11, 99–142. [Google Scholar] [CrossRef]
- Wang, S.; Mulligan, C.N. Occurrence of arsenic contamination in Canada: Sources, behavior and distribution. Sci. Total Environ. 2006, 366, 701–721. [Google Scholar] [CrossRef]
- Danse, I.R.; Jaeger, R.J.; Kava, R.; Kroger, M.; London, W.M.; Lu, F.C. Position paper of the American Council on Science and Health: Public health concerns about environmental polychlorinated biphenyls (PCBs). Ecotoxicol. Environ. Saf. 1997, 38, 71–84. [Google Scholar] [CrossRef][Green Version]
- Eckhardt, S.; Breivik, K.; Manø, S.; Stohl, A. Record high peaks in PCB concentrations in the Arctic atmosphere due to long-range transport of biomass burning emissions. Atmos. Chem. Phys. 2007, 7, 4527–4536. [Google Scholar] [CrossRef]
- Boucher, O.; Muckle, G.; Ayotte, P.; Dewailly, E.; Jacobson, S.W.; Jacobson, J.L. Altered fine motor function at school age in Inuit children exposed to PCBs, methylmercury, and lead. Environ. Int. 2016, 95, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Creaser, C.S.; Wood, M.D.; Alcock, R.; Copplestone, D.; Crook, P.J. UK Soil and Herbage Pollutant Survey (UK SHS Report No. 8) Environmental Concentrations of Polychlorinated Biphenyls in UK Soil and Herbage; Environment Agency: Bristol, UK, 2007; ISBN 978-1-84432-7737. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/291162/scho0607bmtb-e-e.pdf (accessed on 13 July 2020).
- Thomas, D.J.; Tracey, B.; Marshal, H.; Norstrom, R.J. Arctic Terrestrial Ecosystem Contamination. Sci. Total Environ. 1992, 122, 135–164. [Google Scholar] [CrossRef]
- Muir, D.; Braune, B.; DeMarch, B.; Norstrom, R.; Wagemann, R.; Gamberg, M.; Poole, K.; Addison, R.; Bright, D.; Dodd, M.; et al. Ecosystem Uptake and Effects. Canadian Arctic Contaminants Assessment Report; Indian and Northern Affairs Canada: Ottawa, ON, Canada, 1997; Chapter 3; pp. 191–294. [Google Scholar]
- Brown, T.M.; Iverson, S.J.; Fisk, A.T.; Macdonald, R.W.; Helbing, C.C.; Reimer, K.J. Local contamination, and not feeding preferences, explains elevated PCB concentrations in Labrador ringed seals (Pusa hispida). Sci. Total Environ. 2015, 515, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Kidd, K.A.; Hesslein, R.H.; Fudge, R.J.P.; Hallard, K.A. The Influence of Trophic Level as Measured by δ15N on Mercury Concentrations in Freshwater Organisms. In Mercury as a Global Pollutant; Porcella, D.B., Huckabee, J.W., Wheatley, B., Eds.; Springer: Dordrecht, The Netherlands, 1995. [Google Scholar] [CrossRef]
- Trudel, M.; Rasmussen, J.B. Bioenergetics and mercury dynamics in fish: A modelling perspective. Can. J. Fish. Aquat. Sci. 2006, 63, 1890–1902. [Google Scholar] [CrossRef]
- Stern, G.; Loseto, L.; MacDonald, R.W.; Wang, F.; Zdanowicz, C.; Outridge, P.M.; Cole, A.; Hintelmann, H.; Steffen, A. How does climate change influence Arctic mercury. Sci. Total Environ. 2012, 42, 414. [Google Scholar] [CrossRef] [PubMed]
- Schuster, P.F.; Striegl, R.G.; Aiken, G.R.; Krabbenhoft, D.P.; Dewild, J.F.; Butler, K.; Kamark, B.; Dornblaser, M. Mercury export from the Yukon River Basin and potential response to a changing climate. Environ. Sci. Technol. 2011, 45, 9262–9267. [Google Scholar] [CrossRef] [PubMed]
- Chiasson-Gould, S.A.; Blais, J.M.; Poulain, A.J. Dissolved organic matter kinetically controls mercury bioavailability to bacteria. Environ. Sci. Technol. 2014, 48, 3153–3161. [Google Scholar] [CrossRef]
- Carlsson, P.; Breivik, K.; Brorström-Lundén, E. Polychlorinated biphenyls (PCBs) as sentinels for the elucidation of Arctic environmental change processes: A comprehensive review combined with ArcRisk project results. Environ. Sci. Pollut. Res. Int. 2018, 25, 22499–22528. [Google Scholar] [CrossRef]
- Schuster, P.F.; Schaefer, K.M.; Aiken, G.R.; Antweiler, R.C.; Dewild, J.F.; Gryziec, J.D.; Gusmeroli, A.; Hugelius, G.; Jafarov, E.; Krabbenhoft, D.P.; et al. Permafrost stores a globally significant amount of mercury. Geophys. Res. Lett. 2018, 45, 1463–1471. [Google Scholar] [CrossRef]
- Canadian Arctic Contaminants Assessment Report III (2012): Mercury in Canada’s North Government of Canada. Available online: http://science.gc.ca/eic/site/063.nsf/eng/h_7FE5B2F8.html (accessed on 13 July 2020).
- Helyar, S.J.; Hemmer-Hansen, J.; Bekkevold, D.; Taylor, M.I.; Ogden, R.; Limborg, M.T.; Cariani, A.; Maes, G.E.; Diopere, E.; Carvalho, G.R.; et al. Application of SNPs for population genetics of nonmodel organisms: New opportunities and challenges. Mol. Ecol. Resour. 2011, 11, 123–136. [Google Scholar] [CrossRef]
- Hess, J.E.; Matala, A.P.; Narum, S.R. Comparison of SNPs and microsatellites for fine-scale application of genetic stock identification of Chinook salmon in the Columbia river Basin. Mol. Ecol. Resour. 2011, 11, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Bernatchez, L.; Dempson, J.B.; Martin, S. Microsatellite gene diversity analysis in anadromous arctic char, Salvelinus alpinus, from Labrador, Canada. Can. J. Fish. Aquat. Sci. 1998, 55, 1264–1272. [Google Scholar] [CrossRef]
- Moore, J.-S.; Harris, L.N.; le Luyer, J.; Sutherland, B.J.G.; Rougemont, Q.; Tallman, R.F.; Fisk, A.T.; Bernatchez, L. Genomics and telemetry suggest a role for migration harshness in determining overwintering habitat choice, but not gene flow, in anadromous Arctic Char. Mol. Ecol. 2017, 26, 6784–6800. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; VC de Groot, P.; Clemente-Carvalho, R.; Lougheed, S.C. ddRAD genotyping reveals fine-scale genetic structure and a deeper genetic division in anadromous Arctic char (Salvelinus alpinus) in the Lower Northwest Passage, Nunavut. Can. J. Fish. Aquat. Sci. 2020, in press. [Google Scholar] [CrossRef]
- Narum, S.R.; Banks, M.; Beacham, T.D.; Bellinger, M.R.; Campbell, M.R.; Dekoning, J.; Elz, A.; Guthrie Iii, C.M.; Kozfkay, C.; Miller, K.M.; et al. Differentiating salmon populations at broad and fine geographical scales with microsatellites and single nucleotide polymorphisms. Mol. Ecol. 2008, 17, 3464–3477. [Google Scholar] [CrossRef]
- Smith, C.T.; Seeb, L.W. Number of alleles as a predictor of the relative assignment accuracy of short tandem repeat (STR) and single-nucleotide-polymorphism (SNP) baselines for chum salmon. Trans. Am. Fish. Soc. 2008, 137, 751–762. [Google Scholar] [CrossRef]
- Element, G.; Engel, K.; Neufeld, J.D.; Casselman, J.M.; van Coeverden de Groot, P.; Greer, C.W.; Walker, V.K. Seasonal habitat drives intestinal microbiome composition in anadromous Arctic char (Salvelinus alpinus). Environ. Microbiol. 2020, 22, 3112–3125. [Google Scholar] [CrossRef]
- Schott, S.; Qitsualik, J.; van Coeverden de Groot, P.; Okpakok, S.; Chapman, J.M.; Lougheed, S.; Walker, V.K. Operationalizing knowledge coevolution: Towards a sustainable fishery for Nunavummiut. Arctic Sci. 2020, in press. [Google Scholar] [CrossRef]
- Hamilton, E.F.; Element, G.; van Coeverden de Groot, P.; Engel, K.; Neufeld, J.D.; Shah, V.; Walker, V.K. Anadromous Arctic char microbiomes: Bioprospecting in the high Arctic. Front. Bioeng. Biotechnol. 2019, 7, 1–32. [Google Scholar] [CrossRef]
- Wu, Y.; Lougheed, D.R.; Lougheed, S.C.; Moniz, K.; Walker, V.K.; Colautti, R.I. baRcodeR: An open-source R package for sample labelling. Methods Ecol. Evol. 2020, 1–6. [Google Scholar] [CrossRef]
- Casselman, J.M. A Techniques Manual Outlining General Procedures for Preparing thin Sections of Fish Otoliths for Age and Growth Interpretation; Special publication of AFishESci. Inc.: Mallorytown, ON, USA, 2015; 30p. Available online: https://www.fisheries.noaa.gov/alaska/science-data/age-and-growth-fish-otolith-thin-sectioning-procedures (accessed on 13 July 2020).
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high-quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Christensen, K.A.; Rondeau, E.B.; Minkley, D.R.; Leong, J.S.; Nugent, C.M.; Danzmann, R.G.; Ferguson, M.M.; Stadnik, A.; Devlin, R.H.; Muzzerall, R.; et al. The Arctic charr (Salvelinus alpinus) genome and transcriptome assembly. PLoS ONE 2018. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Genome Project Data Processing S: The Sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Tang, F.; Ishwaran, H. Random Forest Missing Data Algorithms. Stat. Anal. Data Min. 2017, 10, 363–377. [Google Scholar] [CrossRef]
- Gosselin, T.; Archer, E. Grur: An R Package Tailored for RADseq Data Imputations. R Package Version 0.1.1. 2019. Available online: https://github.com/thierrygosselin/grur (accessed on 13 July 2020).
- Anderson, E.C. Assessing the power of informative subsets of loci for population assignment: Standard methods are upwardly biased. Mol. Ecol. Resour. 2010, 10, 701–710. [Google Scholar] [CrossRef]
- Anderson, E.; Waples, R.; Kalinowski, S. An improved method for predicting the accuracy of genetic stock identification. Can. J. Fish. Aquat. Sci. 2008, 65, 1475–1486. [Google Scholar] [CrossRef]
- Gosselin, T.; Anderson, E.C.; Bradbury, I. Assigner: Assignment Analysis with GBS/RAD Data Using R. R Package Version 0.5.6. 2019. Available online: http://thierrygosselin.github.io/assigner/ (accessed on 13 July 2020).
- Chapman, J.M.; Schott, S. Knowledge coevolution: Generating new understanding through bridging and strengthening distinct knowledge systems and empowering local knowledge holders. Sustain. Sci. 2020, 15, 931–943. [Google Scholar] [CrossRef]
- Scott, W.B.; Crossman, E.J. Freshwater Fishes of Canada; Fisheries Research Board of Canada: Ottawa, ON, Canada, 1973; p. 966. [Google Scholar] [CrossRef]
- Priest, H.; Usher, P.J. The Nunavut Wildlife Harvest Study, Nunavut. Wildlife Management Board, Nunavut Canada. 2004. Available online: http://www.nwmb.com/inu/publications/harvest-study/1824−156-nwhs-report−2004−156−0003/file (accessed on 21 November 2020).
- Weihs, F.; Okalik, P. Strategy for the Inuit Sealing Economy; Baffin Region Report; A report for the Baffin Regional Hunters and Trappers Committee of the Baffin Regional Council: Ottawa, ON, Canada, 2000; 160p. [Google Scholar]
- Usher, P.J. Standard Edible Weights of Harvested Species in the Inuvialuit Settlement Region; Report to the Northern Contaminants Program, Department of Indian Affairs and Northern Development; Department of Indian Affairs and Northern Development: Ottawa, ON, Canada, 2000. [Google Scholar]
- Statistics Canada Population Census Data for 1996 and 2001. Available online: www12.satcan.gc.ca (accessed on 21 November 2020).
- Health Canada. Health Canada’s Maximum Levels for Chemical Contaminants in Foods. Government of Canada. 2000. Available online: https://www.canada.ca/en/health-canada/services/food-nutrition/food-safety/chemical-contaminants/maximum-levels-chemical-contaminants-foods.html (accessed on 8 December 2020).
- McLaughlin, J.; Gessner, B.D. Fish Consumption Advice for Alaskans: A Risk Management Strategy to Optimize the Public’s Health. 2007. Available online: http://www.epi.hss.state.ak.us/bulletins/docs/rr2007_04.pdf (accessed on 1 December 2020).
- USEPA. Method 8082A, Revision 1, February 2007, Third Edition of the Test Methods for Evaluating Solid Waste, Physical/Chemical Methods, EPA Publication SW−846. Available online: https://www.epa.gov/sites/production/files/2015−12/documents/8082a.pdf (accessed on 1 December 2020).
- NEIWPCC. Mercury (in the New England States). 2007. Available online: https://neiwpcc.org/our-programs/nps/mercury/NEIWPCC (accessed on 6 March 2020).
- Government of Alberta. Fish Consumption Guidance: Mercury in Fish. Environmental Public Health Science Unit, Health Protection Branch, Public Health and Compliance Division, Alberta Health. 2019. Available online: https://open.alberta.ca/dataset/270e8456-7b8b-46c9-b87d-3508b22c319d/resource/bbf031cd-f28a-4e3e-b9b6-230164fda44c/download/fish-consumption-guidance-mercury-in-fish-2019.pdf (accessed on 6 March 2020).
- Sobolev, N.; Aksenov, A.; Sorokina, T.; Chashchin, V.; Ellingsen, D.G.; Nieboer, E.; Varakina, Y.; Veselkina, E.; Kotsur, D.; Thomassen, Y. Essential and non-essential trace elements in fish consumed by indigenous peoples of the European Russian Arctic. Environ. Pollut. 2019, 253, 966–973. [Google Scholar] [CrossRef]
- Meador, J.P.; Ernest, D.W.; Kagley, A. Bioaccumulation of arsenic in marine fish and invertebrates from Alaska and California. Arch. Environ. Contam. Toxicol. 2004, 47, 223–233. [Google Scholar] [CrossRef]
- Justice Canada. Food and Drug Regulations (C.R.C., c. 870). (Regulations are current to 2020−09−09). 2020. Available online: https://laws-lois.justice.gc.ca/eng/regulations/C.R.C.%2C_c._870/page-74.html#docCont (accessed on 9 September 2020).
- Bohn, A.; Fallis, B.W. Metal concentrations (As, Cd, Cu, Pb and Zn) in shorthorn sculpins, Myoxocephalus scorpius (Linnaeus), and arctic char, Salvelinus alpinus (Linnaeus), from the vicinity of Strathcona Sound, Northwest Territories. Water Res. 1978, 12, 659–663. [Google Scholar] [CrossRef]
- Muir, D.; Wang, X.; Bright, D.; Lockhart, L.; Köck, G. Spatial and temporal trends of mercury and other metals in landlocked char from lakes in the Canadian Arctic archipelago. Sci. Total Environ. 2005, 351, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.G. Trace element loadings to sediments of fourteen Ontario lakes and correlations with concentrations in fish. Can. J. Fish. Aquat. Sci. 1987, 44, 3–13. [Google Scholar] [CrossRef]
- de Rosemond, S.; Xie, Q.; Liber, K. Arsenic concentration and speciation in five freshwater fish species from Back Bay near Yellowknife, NT, CANADA. Environ. Monit. Assess. 2008, 147, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, C.J.; Brumbaugh, W.G. National contaminant biomonitoring program: Concentrations of arsenic, cadmium, copper, lead, mercury, selenium, and zinc in US freshwater fish, 1976–1984. Arch. Environ. Contam. Toxicol. 1990, 19, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Cott, P.A.; Zajdlik, B.A.; Palmer, M.J.; McPherson, M.D. Arsenic and mercury in lake whitefish and burbot near the abandoned Giant Mine on Great Slave Lake. J. Great Lakes Res. 2016, 42, 223–232. [Google Scholar] [CrossRef]
- Nicholson, L.; Rengan, K. Determination of arsenic in some Lake Michigan fish using neutron activation analysis. J. Radioanal. Nucl. Chem. 1979, 54, 355–360. [Google Scholar] [CrossRef]
- Bohn, A. Arsenic in marine organisms from West Greenland. Mar. Pollut. Bull. 1975, 6, 87–89. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority) Results of the monitoring of non dioxin-like PCBs in food and feed. EFSA J. 2010, 8, 1701. [CrossRef]
- Caumette, G.; Koch, I.; Reimer, K.J. Arsenobetaine formation in plankton: A review of studies at the base of the aquatic food chain. J. Environ. Monit. 2012, 14, 2841–2853. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, Z.A.; Stow, J.P.; Burgess, N.M.; Solomon, S.M.; Reimer, K.J. PCBs in sediments and the coastal food web near a local contaminant source in Saglek Bay, Labrador. Sci. Total Environ. 2005, 351, 264–284. [Google Scholar] [CrossRef] [PubMed]
- Hardell, S.; Tilander, H.; Welfinger-Smith, G.; Burger, J.; Carpenter, D.O. Levels of polychlorinated biphenyls (PCBs) and three organochlorine pesticides in fish from the Aleutian Islands of Alaska. PLoS ONE 2010, 5, e12396. [Google Scholar] [CrossRef] [PubMed]
- McKinney, M.A.; McMeans, B.C.; Tomy, G.T.; Rosenberg, B.; Ferguson, S.H.; Morris, A.; Muir, D.C.; Fisk, A.T. Trophic transfer of contaminants in a changing arctic marine food web: Cumberland Sound, Nunavut, Canada. Environ. Sci. Technol. 2012, 46, 9914–9922. [Google Scholar] [CrossRef] [PubMed]
- NCP (Northern Contaminants Program). Chapter 4 Annex Tables from Canadian Arctic Contaminants Assessment Report on Persistent Organic Pollutants; Annex Tables Are Available Only on Request from NCP; Muir, D., Kurt-Karakus, P., Stow, J., Eds.; Northern Contaminants Program, Aboriginal Affairs and Northern Development (now Crown-Indigenous Relations and Northern Affairs Canada): Ottawa, ON, Canada, 2013. [Google Scholar]
- Stantec. Human Health Risk Assessment for the Consumption of Country Foods in the Town of Hopedale, NL. Prepared for: Newfoundland and Labrador Department of Environment and Conservation. File No. 121413099 Final Report October 31, 2014. 47 pages + 398 pages appendices. 2014. Available online: https://www.gov.nl.ca/mae/files/env-protection-impactedsites-hopedale-hopedale-country-foods-hhera.-oct2014.pdf (accessed on 9 September 2020).
- Cabrerizo, A.; Muir, D.C.; Koöck, G.; Iqaluk, D.; Wang, X. Climatic influence on temporal trends of polychlorinated biphenyls and organochlorine pesticides in landlocked char from lakes in the Canadian High Arctic. Environ. Sci. Technol. 2018, 52, 10380–10390. [Google Scholar] [CrossRef]
- Adams, E.M.; von Hippel, F.A.; Hungate, B.A.; Buck, C.L. Polychlorinated biphenyl (PCB) contamination of subsistence species on Unalaska Island in the Aleutian Archipelago. Heliyon 2019, 5, e02989. [Google Scholar] [CrossRef]
- Masset, T.; Frossard, V.; Perga, M.E.; Cottin, N.; Piot, C.; Cachera, S.; Naffrechoux, E. Trophic position and individual feeding habits as drivers of differential PCB bioaccumulation in fish populations. Sci. Total Environ. 2019, 674, 472–481. [Google Scholar] [CrossRef]
- Halldorsson, T.I.; Thorsdottir, I.; Meltzer, H.M.; Nielsen, F.; Olsen, S.F. Linking exposure to polychlorinated biphenyls with fatty fish consumption and reduced fetal growth among Danish pregnant women: A cause for concern? Am. J. Epidemiol. 2008, 168, 958–965. [Google Scholar] [CrossRef]
- Bellehumeur, K.; Lapointe, D.; Cooke, S.J.; Moon, T.W. Exposure to sublethal levels of PCB−126 impacts fuel metabolism and swimming performance in rainbow trout. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 199, 97–104. [Google Scholar] [CrossRef]
- U.S. FDA (Food and Drug Administration). Electronic Code of Federal Regulations, Title 21, Chapter I, Subchapter B, Part 109, Subpart B, §109.30. Title 21: Food and Drugs. PART 109—Unavoidable Contaminants in Food for Human Consumption and Food-Packaging Material. Subpart B—Tolerances for Unavoidable Poisonous or Deleterious Substances. §109.30 Tolerances for Polychlorinated Biphenyls (PCB’s). 2019. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=109.30 (accessed on 8 March 2020).
- Bright, D.A.; Dushenko, W.T.; Grundy, S.L.; Reimer, K.J. Evidence for short-range transport of polychlorinated biphenyls in the Canadian Arctic using congener signatures of PCBs in soils. Sci. Total Environ. 1995, 160, 251–263. [Google Scholar] [CrossRef]
- Weist, P.; Schade, F.M.; Damerau, M.; Barth, J.M.I.; Dierking, J.; André, C.; Petereit, C.; Reusch, T.; Jentoft, S.; Hanel, R.; et al. Assessing SNP-markers to study population mixing and ecological adaptation in Baltic cod. PLoS ONE 2019, 14, e0218127. [Google Scholar] [CrossRef] [PubMed]
- McKinney, G.J.; Seeb, J.E.; Seeb, L.W. Managing mixed-stock fisheries: Genotyping multi-SNP haplotypes increases power for genetic stock identification. Can. J. Fish. Aquat. Sci. 2017, 74, 429–434. [Google Scholar] [CrossRef]
- Lougheed, S.; Driver, H.; (Queen’s University, Kingston, ON, Canada). Personal Communication, 2020.





| Site Name & Number A | GPS Location | Geographic Group | Water Type, Fishing Gear, Contaminants &/Or Genetics |
|---|---|---|---|
| 1. Port Parry | 69.55799, −97.43719 | 1 | fresh/nets/C and G |
| 26. GKWI lake 4 | 69.48518, −97.5351 | 1 | fresh/nets/G |
| 21. GKWI lake 1 | 68.85708, −96.46545 | 2 | fresh/nets/G |
| 22. GKWI lake 2 | 68.88999, −96.27868 | 2 | fresh/nets/G |
| 23. GKWI lake 3 | 68.88233, −96.25877 | 2 | fresh/nets/G |
| 17. KWI Weir | 68.93280, −96.2195 | 2 | fresh/spears/nets/C and G |
| 2. Swan Lake | 68.67045, −95.94928 | 2 | fresh/nets/G |
| 3. Koka Lake | 68.53475, −96.21275 | 2 | fresh/nets/C and G |
| 10. Merilik Lake | 68.57388, −97.32687 | 2 | fresh/nets/C |
| 24. Gjoa Haven | 68.63374, −95.80885 | 2 | saline/nets/G |
| 8/14. Murchison R. | 68.56700, −93.37707 | 3 | fresh/nets/C and G |
| 6. Backhouse Pt. | 67.45756, −95.36072 | 4 | saline/nets/C and G |
| 7. Legendary R. | 67.52161, −96.43939 | 4 | saline/nets/C and G |
| 13/18/19. Back R. | 66.95853, −95.30144 | 4 | fresh/nets/C |
| 20. Hayes R. | 67.13852, −95.2964 | 4 | fresh/nets/G |
| 28. W. of Chantry 3 | 67.81159, −97.04374 | 4 | fresh/nets/C |
| 29. W. of Chantry 4 | 67.86958, −96.7187 | 4 | fresh/nets/C |
| 31. Cambridge Bay B | 69.40600, −06.31393 | - | fresh/nets/G |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walker, V.K.; Das, P.; Li, P.; Lougheed, S.C.; Moniz, K.; Schott, S.; Qitsualik, J.; Koch, I. Identification of Arctic Food Fish Species for Anthropogenic Contaminant Testing Using Geography and Genetics. Foods 2020, 9, 1824. https://doi.org/10.3390/foods9121824
Walker VK, Das P, Li P, Lougheed SC, Moniz K, Schott S, Qitsualik J, Koch I. Identification of Arctic Food Fish Species for Anthropogenic Contaminant Testing Using Geography and Genetics. Foods. 2020; 9(12):1824. https://doi.org/10.3390/foods9121824
Chicago/Turabian StyleWalker, Virginia K., Pranab Das, Peiwen Li, Stephen C. Lougheed, Kristy Moniz, Stephan Schott, James Qitsualik, and Iris Koch. 2020. "Identification of Arctic Food Fish Species for Anthropogenic Contaminant Testing Using Geography and Genetics" Foods 9, no. 12: 1824. https://doi.org/10.3390/foods9121824
APA StyleWalker, V. K., Das, P., Li, P., Lougheed, S. C., Moniz, K., Schott, S., Qitsualik, J., & Koch, I. (2020). Identification of Arctic Food Fish Species for Anthropogenic Contaminant Testing Using Geography and Genetics. Foods, 9(12), 1824. https://doi.org/10.3390/foods9121824






